Abstract
Plant roots have been an intriguing subject to plant biologists for a long time. Roots serve a wide range of functions in plants, including anchorage and absorption of water and nutrients. Roots are highly plastic in their development and directional growth to facilitate better absorption and assimilation of water and nutrients under different environmental conditions. Plasticity of root system architecture (RSA) confers different architecture in response to immediate soil-based and endogenous signals. Nutrient heterogeneity in the soil is a major factor determining RSA. Plants sense local nutrient availability and facilitates directional root development to enhance nutrient acquisition. Among the mineral nutrients, Nitrogen (N) and Phosphate (Pi) are major macronutrients important for plant development. N dissolves in water and accumulates in deep soil, thus compelling plants for producing deeper roots. Pi retains in topsoil and during pi deficiency, root system grows more laterally to enhance pi uptake. Thus, RSA is highly dynamic in nature which varies according to soil condition and nutrient availability and enhancing the plasticity of RSA will improve the nutrient uptake and use efficiency of crops. In this chapter, we are discussing how N and pi availability remodel different aspects of RSA in model plants and crops.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Ammonium
- Lateral root differentiation
- Lateral root primordia
- Nitrate
- Nutrient acquisition
- Nutrient crosstalk
- Nutrient use efficiency
- Phosphate
- Primary root growth
- Root system architecture
- Root plasticity
- Root hairs
- Root meristem
- Soil heterogeneity
- Topsoil foraging
1 Introduction
Roots played a crucial role in the evolutionary success of land plants. They search for minerals and water in the soil and supply it to the above-ground parts of the plant. Plants need mineral nutrients for optimal growth. Roots forage the soil to maximize nutrient acquisition. Plants require a range of essential nutrients, among them, nitrogen (N), phosphate (Pi), potassium (K) are required in large quantities. The inorganic forms of phosphate (HPO4−, Pi) and nitrate (NO3−) or ammonium (NH4+) are taken up from the soil (Bouain et al. 2019). The heterogeneous distribution of these nutrients in the soil limits their acquisition. Moreover the assimilation of pi in plant is also poor as inorganic phosphate reacts with iron (Fe3+), aluminum (Al3+), and calcium (Ca2+) ions present in soil to form an insoluble complex. Limitation of phosphate and nitrogen in the plant severely affects plant growth and productivity and nitrogen and phosphate are externally supplied to the crop in form of fertilizers to enhance productivity. Excessive input of fertilizers causes water and air pollution. Ongoing soil degradation, water pollution, and climatic changes negatively affect sustainable agriculture (Li et al. 2016). An increasing population and limited agricultural space aggravate this problem. One effort in mitigating the situation would be to improve nutrient use efficiency (NUE) of crop plants. Global agriculture demands nutrient efficient crops with improved nutrient uptake even under nutrient-deficient conditions. Plants employ multi-level pathways for nutrient uptake, such as modification of root system, enhancing the activity of nutrient transporters, releasing organic acids in soil, and developing symbiosis with fungi or bacteria (Chen and Liao 2017).
The intricate three-dimensional web of roots in the soil is known as root system architecture (RSA) (Satbhai et al. 2015). Enhancing the resilience and plasticity of RSA is one of the ways to improve the nutrient uptake of crops. Root length, branching, angle, and surface area, are major tenets of RSA. The RSA of a plant is the cumulative product of genetic and environmental interaction (Lynch 2019). Plants sharing the same genotype might develop different root systems depending on the immediate requirement. This interaction of genome and environment provides plasticity to the plant roots system. Root plasticity is more advanced in higher plants to support much complex physiology (Satbhai et al. 2015). Phenotypic plasticity of roots shapes root system in response to local root environment which includes, soil texture and compactness, water and nutrient availability, soil fauna, and gravity (Satbhai et al. 2015). As nitrogen and phosphate both are involved in RSA remodeling in response to their availability in soil, but the phenotypical changes in roots are contrasting to each other in terms of primary root growth and lateral root formation. Plants faces deficiencies of multiple nutrients in soil at a time and in such combinatorial stress, a crosstalk between the nutrients signaling is required (Bouain et al. 2019). In this chapter, we will briefly discuss the idea of RSA in model and crop plant then we will move forward to study the role of nitrogen in mediating RSA. We will zoom into the molecular machinery of plant RSA in response to phosphate, and progress in utilizing the underlying machinery to improve the nutrient efficiency of crops.
2 RSA and Nitrogen Mediated Root Remodeling
Nutrient efficiency has been an important trait in the evolution of roots in land plants (Lynch 2019). Plants utilize the root system for the acquisition of mineral nutrients such as nitrogen and phosphate from the soil. Plants need to forage the soil in search of nutrients which varies from place, time, and depth of soil. Exploration of soil is mediated by roots in terms of root growth, branching, root hair formation, and microbial symbiosis. Dicots plants such as Arabidopsis have tap root system, which consists of the main primary root and several branched lateral roots along with root hairs (Satbhai et al. 2015). Monocots such as maize and rice develop fibrous roots primarily consisting of adventitious roots. Adventitious roots rising from any non-root tissue, such as junction roots, crown roots, etc. (Del Bianco and Kepinski 2018). Root system architecture (RSA) has been considered essential for soil exploration and NUE. Modifying the RSA for improving the nutrient acquisition of the plants is the most basic yet complicated trait for crop improvement.
In general, the distribution of nutrients in soil depends on the amount of nutrients, soil composition, nutrient solubility, etc. Nitrogen (N) is an essential structural and functional component of primary and secondary organic compounds. Limitation of nitrogen in plants constraints plant growth and development with subsequent reductions of plant productivity (Luo et al. 2020). Owing to Haber’s process of artificial nitrogen fixation, the production of ammonium has increased (Jenkinson 2001). However, the high solubility of nitrate and ammonium in water causes nitrogen to leach out with the groundwater, causing poor nitrogen acquisition and environmental pollution. The most logical and accessible option to reduce wastage and pollution is to improve nitrogen utilization efficiency (Xu and Takahashi 2020). In dicots, such as model plant Arabidopsis, four methods of root remodeling have been studied in the presence of nitrogen, (i) presence of nitrate promotes localized elongation of lateral root growth, (ii) high tissue nitrate systemically inhibits the lateral root meristems, (iii) exogenously supplied L-glutamate inhibits primary root growth and promotes root branching, (iv) high carbon to nitrogen ratio inhibits the lateral root initiation (Zhang et al. 2007).
The effect of normal nitrate on primary root growth was either insensitive or modestly promoting the primary root growth (Forde 2014). Although moderately higher concentration of KNO3 inhibited the primary root growth and this was mediated via the miR393/AFB3 regulatory module (Vidal et al. 2010). microRNA393 is induced by nitrate and it specifically cleaves transcripts of AFB3, an auxin receptor which has a role in auxin dependent root growth in response to nitrate (Vidal et al. 2010). L-glutamate, an amino acid when exogenously provided specifically inhibited the primary root growth (Forde 2014). Nitrate was seen to monitor the stem cell dynamics by regulating the cell division genes and differentiation of distal stem cells (Guan et al. 2017; Wang et al. 2017). TCP20, a component of nitrate signaling interacts with NIN-like proteins, NLP6 and NLP7, and upregulates the expression of nitrate-dependent genes and downregulates the G2/M cell cycle gene CYCB1;1 in the root meristem (Guan et al. 2017).
Local patches of nitrate promote preferential lateral root growth (Mounier et al. 2014). Nitrate being the major source of nitrogen has an intrinsic role as a nutrient and signaling molecule perceived by plant nitrate transporter 1.1 (NRT1.1) (Maghiaoui et al. 2020b). This nitrate-dependent lateral root growth is the outcome of nitrate transporter/sensor NRT1.1 (also known as NPF6.3 or CHL1) and auxin interaction (Krouk et al. 2010). NRT1.1 regulates lateral root growth by orchestrating the basipetal transport of auxin out of lateral root primordia (LRPs) (Krouk et al. 2010). In a recent study, it was found that NRT1.1 also regulates the auxin biosynthesis (Maghiaoui et al. 2020a), NRT1.1 negatively regulated the expression of auxin biosynthetic gene, TAR2 and auxin influx carrier, LAX3 to reduce acropetal transport of auxin in LRPs (Maghiaoui et al. 2020a). Interestingly, the supply of NH4+a a preferential source of nitrogen for crops, stimulate lateral root branching (Lima et al. 2010; Jia and von Wirén 2020). In Arabidopsis, it was found that local NH4+ promoted branching with the help of NH4+ transporter AMT1;3. This branching was found absent in quadruple mutant of AMMONIUM TRANSPORTER (amt1;1, amt1;2, amt1;3, amt2;1), reconstituting the expression of AMT1;3 in quadruple mutant restored the branching (Lima et al. 2010). Ammonium uptake releases protons in the apoplast, this acidification of apoplast modifies the auxin mobility which in turn increased the lateral root density (Meier et al. 2020; Pélissier et al. 2021). A detailed overview of nitrogen-dependent lateral root formation is published recently (Pélissier et al. 2021).
Along with local signaling, systemic signaling also regulates root architecture in foraging soil nitrogen. Systemic signals of nitrogen deficiency shape the root architecture by modulating the phytohormones such as auxin, cytokinin, and Brassinosteroids (Kiba et al. 2011). The steep, cheap, and deep roots in crops attain root architecture exploring deeper sections of soil in search of mobile nitrate (Lynch 2019). This architecture includes steep lateral growth angle, fewer branching of root or cheaper in carbon units, and deeper roots in foraging nitrogen. Several reports in maize and rice support steep, cheap, and deep root ideotype of crops in low nitrogen condition (Lynch 2013; Trachsel et al. 2013; Ju et al. 2015; Chen and Liao 2017; Lynch 2019). Recent studies have suggested an integration of phosphate and nitrate signaling in plant root development (Medici et al. 2015, 2019a; Bouain et al. 2019). GARP type transcription factors, HRS1 and NIGT1.2 both have been shown to involved in nitrate and phosphate signaling (Medici et al. 2015; Wang et al. 2020). A crosstalk between Nitrate receptor NRT1.1 and PHOSPHATE STARVATION RESPONSE1 (PHR1) was observed (Medici et al. 2019a). PHOSPHATE2 (PHO2) integrated the N availability into the phosphate starvation responses (PSR). Nitrogen mediated PSR were significantly affected in pho2 mutants (Medici et al. 2019a).
3 Root System Architecture in Response to Phosphate (Pi)
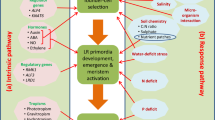
Phosphate (HPO4−, Pi) is another essential nutrient required for plant growth and development. Being a component of DNA, protein, and cell membrane, it is requisite along with carbon and nitrogen for optimal growth. The acquisition of phosphate in plants is also inefficient, which is supported by the fact that only 10–25% of applied phosphate is taken up by the plant (Crombez et al. 2019). Limitation of pi in soil affects plant growth and reduces plant productivity. Unlike nitrogen, the sources of phosphorus are non-renewable and has been predicted to deplete in the next few decades (Lynch 2013, 2019). The predominant form of phosphorus in soil is orthophosphate (HPO4−), which reacts with cations such as Mg2+ and Ca2+ (Chen and Liao 2017). Low mobility of orthophosphate or pi in the soil makes the topsoil rich in phosphate. To increase the uptake of pi, plants forage the topsoil (Lynch 2019). Topsoil foraging in crops generally reorganizes the root system architecture with more production of axial roots, shallower axial root angle, greater lateral root density, and longer and denser root hairs (Lynch 2019). In Arabidopsis shallow root system is optimized with reduced primary root length, increasing lateral root length and density, and producing denser root hairs (Fig. 1) (Huang and Zhang 2020).
RSA of Arabidopsis wildtype in phosphate deficient (10 µM) and sufficient medium (625 µM). 7 DAG old Col-0 subjected to pi deficiency for five days showed inhibition of primary root growth, an increase in lateral root density (panel a), and root hair (panel b). Dots represent the primary root growth kinetics for a period of five days
3.1 Primary Root Growth Under Pi Deficiency
Inhibition of primary root growth in low phosphate is orchestrated in a coordinated fashion, by arresting cell division, inhibiting the cell elongation, and stimulating the cell differentiation (Péret et al. 2014). Numerous genetic screenings identified the role of several mutants in regulating the root meristem under pi deficiency. In Arabidopsis, LOW PHOSPHATE ROOT 1 (LPR1) and PHOSPHATE DEFICIENCY RESPONSE 2 (PDR2) appeared in regulating the meristem activity (Ticconi et al. 2009; Müller et al. 2015). PDR2 encodes a single P5-type ATPase which supports the expression of Scarecrow (SCR) in maintaining root patterning (Ticconi et al. 2009). LPR1 (Ferroxidase) and PDR2 both works together under pi deficiency to arrest root apical meristem (RAM). LPR1-PDR2 module promotes iron deposition specifically in the cell wall of RAM. Accumulation of Fe stimulates callose deposition which interfere the cell to cell communication and inhibiting transport of SHORTROOT (SHR) from stele into the quiescent center (Müller et al. 2015). Inhibition of primary root growth in pi deficiency was completely abolished in lpr1lpr2 double mutant. In contrast, the primary root of pdr2 mutant was hypersensitive to pi deficiency (Müller et al. 2015). Phosphate-dependent callose deposition in RAM was due to enhanced reactive oxygen species (ROS) production in stem cells, which increased in pdr2 mutant (Müller et al. 2015). Reduced cell elongation and stimulation of cell differentiation are two other very important characteristics of pi-mediated inhibition of PR growth. STOP (SENSITIVE TO PROTON TOXICITY1) and ALMT1 (ALUMINUM-ACTIVATED MALATE TRANSPORTER1) module coordinate the cell expansion in Arabidopsis (Balzergue et al. 2017). STOP is a transcription factor that controls the cell elongation by modulating the expression of its direct target gene, ALMT which codes for malate channel. Together they mediate cell wall stiffening with the help of iron and peroxidase (Balzergue et al. 2017). SIZ1 a SUMO E3 ligase regulates PSR, siz1 mutant exhibited reduced primary root and extensive lateral root and root hair development in pi deficient conditions (Miura et al. 2005).
In crops, the effect of primary root growth inhibition in pi deficiency is less pronounced (Péret et al. 2014). Different crops and cultivars showed a different response to primary root growth under pi deficiency (Péret et al. 2014). Crop plants favor proliferation of adventitious roots in pi deficiency. Maize genotypes with more crown roots and lateral root branching have higher topsoil foraging, growth, and a better yield under low pi conditions (Jia et al. 2018; Lynch 2019). In addition to that, different plant families such as Proteaceae and Leguminosae develop denser lateral roots in cluster are known as proteoid roots in response to low pi (Li et al. 2016). Pi deficiency in plants stepwise coordinates the reduction in primary root growth to mobilize the carbon allocation from the primary root to lateral root density and root hair formation.
3.2 Lateral Root Growth in Pi Deficiency
Lateral roots in plants have an important role in the acquisition of pi from the topsoil by increasing the total surface area (Waidmann et al. 2020). In pi deficiency, plants have to minimize the resources allocation as well as it has to explore the soil. As a result, the primary root growth needs to be reduced along with the elongation of lateral roots and the formation of higher-order LRs (Waidmann et al. 2020). A repertoire of genes gets differentially activated in response to lateral root formation. These genes are majorly involved in auxin biosynthesis, transport, and signaling (Banda et al. 2019; Crombez et al. 2019). Systemic responses of phosphate sensing and signaling are largely managed by the transcription factor PHR1 (Rubio et al. 2001; Gutiérrez-Alanís et al. 2018). PHR1 binds on PHR1-binding sites (P1BS) and regulates the expression of phosphate starvation responses (PSR) genes (Fig. 2) (Rubio et al. 2001; Castrillo et al. 2017). Auxin signaling interacts with phosphate signaling in response to lateral root formation and elongation.
Phosphate signaling in plant cell. Phosphate (pi) is taken up by phosphate transporters present on the cell membrane (PHTs). Vacuolar efflux transporters situated on tonoplast controls cytosolic pi concentration by transporting the pi in and out of the vacuole. PHOSPHATE STARVATION RESPONSE (PHR) is sequestered by SPX domain harboring proteins in the presence of Inositol Phosphate (InsPs). In the deficiency of phosphate SPX allows PHR to bind to cis-regulatory elements P1BS to regulate the expression of phosphate starvation-response (PSR) genes. Image created in BioRender.com
Lateral root initiation begins from the cells present in the pericycle known as lateral root founder cells (LRFCs). Auxin is known to mark the pre-branch site for future lateral roots (Banda et al. 2019). Pi starvation increases auxin level and sensitivity in root tip and lateral root primordia of Arabidopsis. Auxin receptor TRANSPORT INHIBITOR RESPONSE1 (TIR1) expression increases under pi deficiency, causing the degradation of AUX/IAA proteins, which overall increases the auxin sensitivity in pi deficiency (Pérez-Torres et al. 2008). TIR1 expression in low phosphate was found to be directly regulated by PHR1 (Castrillo et al. 2017). Auxin-response factors (ARFs) are downstream to the auxin signaling controlling various aspects of auxin signaling. ARF7 and ARF19 play important role in lateral root priming, lateral root initiation, patterning, and initiation (Banda et al. 2019; Crombez et al. 2019). Interestingly, expression of PHR1 was also positively regulated by ARF7/19, and the impairment of lateral root growth in arf7arf19 double mutant was also rescued by overexpressing the PHR1 (Huang et al. 2018). PIN-FORMED (PIN) transporters of auxin and several ARFs are also possible targets of PHR1 (Castrillo et al. 2017). Along with auxin several other mutants such as lpr1, pnp, plt1;1plt1;4, alf3, siz1, and wrky75 showed differential regulation of lateral roots in low pi (Niu et al. 2013). (Huang and Zhang 2020) recently reviewed the role of different phytohormone in pi deficiency.
The response to low pi conforms to differential root growth in crops. Crop plants such as maize and rice have a fibrous root system that is generally deeper and shallower than the taproot system of Arabidopsis, and bean. Several genes have been identified in maize, rice, tomato, and bean that showed robust remodeling of root in varying pi conditions (Yang et al. 2007; Zhou et al. 2008; Dai et al. 2012; Postma et al. 2014; Zhou et al. 2014; Jia et al. 2018; Gonçalves et al. 2020). Simulation modeling of Zea mays roots using SimRoot suggested densely spaced but shorter lateral roots were more optimal to phosphorus acquisition (Postma et al. 2014). Maize with greater lateral root branching outperformed others in terms of phosphate acquisition and crop productivity. Higher uptake of phosphate led to 14% greater grain yield than control maize plants (Jia et al. 2018). Considering the above-mentioned studies of lateral root growth in pi deficiency, lateral roots show a promising role in battling pi deficiency. Branching of lateral roots overall improves the phosphate uptake and thus the growth of the plant.
3.3 Role of Root Hairs in Pi Deficiency
Root hairs are specialized tubular epidermal cells of plant roots that play a significant role in nutrient and water absorption (Salazar-Henao et al. 2016). The presence of root hairs near the root tip is considered a hotspot for pi assimilation as root hairs can be responsible for up to 90% of phosphate uptake by the plants (Brown et al. 2013). Restricted cell elongation in the primary root is the prerequisite for increasing root hair frequency (Salazar-Henao et al. 2016). Pi deficiency-induced callose deposition inhibits the cell to cell signaling in epidermal cells leading to shorter epidermal cells and increased expression of ENHANCER OF TRY AND CPC 1 (ETC), which results in a higher frequency of hairs per unit root length (Savage et al. 2013; Salazar-Henao et al. 2016b). Auxin has a significant role in root hair initiation and elongation (Knox et al. 2003; Salazar-Henao et al. 2016a; Bhosale et al. 2018; Giri et al. 2018). Pi deficiency-induced root hair growth is determined by bHLH transcription factor ROOT HAIR DEFECTIVE 6-LIKE 4 (RSL4). RSL4 protein synthesis increases in low pi and initiates the hair elongation and it gets gradually degraded by 26S proteasomal pathway. The amount of RSL4 synthesized directly determines the final size of differentiated root hair cells (Datta et al. 2015). Rise in auxin level ultimately promotes downstream ARFs. ARF19 induction in root apex shown to induce RSL4 in the root differentiation zone (Bhosale et al. 2018). Notably, AXR3/IAA17 and SHY2/IAA3 module are also involved in root hair initiation and elongation (Knox et al. 2003).
Improving RH density is an important agronomic trait (Brown et al. 2013). In rice and Brachypodium, overexpression of ROOT HAIR DEFECTIVE SIX‐LIKE (RSL) class I bHLH transcription factor improved the root hair length (Kim et al. 2017; Zhang et al. 2018). Genome-wide association study (GWAS) of desi chickpea, maize, common bean identified several genetic loci associated with RH length (Yan et al. 2004; Zhu et al. 2005; Kohli et al. 2020). These QTLs analysis will help in modifying the surface area of root system architecture to improve crop productivity.
4 Conclusion and Future Perspective
Root system architecture is the underground three-dimensional arrangement of roots. RSA of a plant is the cumulative output of genomic and the immediate environmental conditions. Root plasticity reorganizes the RSA in response to water, mineral, microorganisms present in the soil. Nutrients are present in a heterogeneous manner in the soil. The limitation of nutrients in plants drastically affects the yield of the plant. Plants adapt to local patches of nutrients in the soil by proliferating the root growth for greater nutrient acquisition. Nitrogen and phosphorus are two essential nutrients that employ differential root systems for respective nutrient absorption. Deeper RSA with longer primary root and sparsely spaced lateral roots favor nitrogen uptake, while shorter primary root with higher lateral root branching with denser root hairs promotes phosphate acquisition. Molecular integration of nitrate and phosphate signaling also affected the RSA. Expression of nitrate transporter NRT1.5 was strongly induced by Pi starvation, while its mutants observed an significant increase in primary root and reduced lateral roots in pi deficiency (Cui et al. 2019). Inhibition of cell division in phosphate deficiency is largely determined by the Fe-stimulated callose deposition in the root meristem. Promotion of lateral root increases the root surface that assists in topsoil foraging. Also, while increasing the foraged soil volume, the higher number of lateral roots also leads to a greater number of root tips. Root tips along with the root hairs are hotspots for pi uptake (Brown et al. 2013; Crombez et al. 2019). Integration of various nutrient signaling the in response to combinatorial stress project to develop the smart and sustainable agriculture.
Most studies done in Arabidopsis have been carried out under the artificial system by adding and reducing exogenous nutrient to “mimic” natural environmental conditions. These conditions are much more informative than the studies done in natural conditions. However, such studies propose the question of translation efficiency of nature mimicked studies to the natural conditions (Shahzad and Amtmann 2017). Buffered delivery of phosphate to Arabidopsis roots showed a non-canonical phenotype of pi deficiency. Phosphate buffered with Al2O3 particles supplied realistic low phosphate to plants. This buffered pi delivery resulted in smaller plants with reduced lateral root branching density, longer root hair, and differential expression of canonical phosphate starvation genes (Hanlon et al. 2018). This inefficient translation of the lab generated information to the field calls for much robust and updated experimental design.
An interconnected hub underlies RSA remodeling, wherein different phytohormone, signal peptides, and nutrient signals integrate to regulate primary root, lateral root, and root hair formation and elongation. Several reports have identified genes and QTLs to develop a smart root system. Different nutrients interact with each other in the remodeling of the root system (Bouain et al. 2019; Medici et al. 2019a, b). In future scientists need to consider a much practical soil conditions with multiple nutrient deficiency. Our challenge would be to develop a smarter network root system to coordinate mineral nutrient homeostasis and root growth.
Abbreviations
- RSA:
-
Root system architecture
- N:
-
Nitrogen
- Pi:
-
Phosphate
- NUE:
-
Nutrient use efficiency
- LRP:
-
Lateral root primordia
- PHR1:
-
Phosphate starvation response
- PSR:
-
Phosphate starvation responses
- LR:
-
Lateral roots
- PR:
-
Primary root
- P1BS:
-
PHR1-binding sites
- ARFs:
-
Auxin-response factors
- GWAS:
-
Genome-wide association study
- QTLs:
-
Quantitative trait locus
References
Balzergue C, Dartevelle T, Godon C, Laugier E, Meisrimler C, Teulon JM, Creff A, Bissler M, Brouchoud C, Hagège A et al (2017) Low phosphate activates STOP1-ALMT1 to rapidly inhibit root cell elongation. Nat Commun. https://doi.org/10.1038/ncomms15300
Banda J, Bellande K, Wangenheim D, Goh T, Guyomarch S, Laplaze L, Bennett MJ (2019) Lateral root formation in arabidopsis: a well-ordered LRexit. Trends Plant Sci 24:826–839
Bhosale R, Giri J, Pandey BK, Giehl RFH, Hartmann A, Traini R, Truskina J, Leftley N, Hanlon M, Swarup K et al (2018) A mechanistic framework for auxin dependent Arabidopsis root hair elongation to low external phosphate. Nat Commun 9:1409
Del Bianco M, Kepinski S (2018) Building a future with root architecture. J Exp Bot 69:5319–5323
Bouain N, Krouk G, Lacombe B, Rouached H (2019) Getting to the root of plant mineral nutrition: combinatorial nutrient stresses reveal emergent properties. Trends Plant Sci 24:542–552
Brown LK, George TS, Dupuy LX, White PJ (2013) A conceptual model of root hair ideotypes for future agricultural environments: what combination of traits should be targeted to cope with limited P availability? Ann Bot 112:317–330
Castrillo G, Teixeira PJPL, Paredes SH, Law TF, de Lorenzo L, Feltcher ME, Finkel OM, Breakfield NW, Mieczkowski P, Jones CD et al (2017) Root microbiota drive direct integration of phosphate stress and immunity. Nature 543:513–518
Chen L, Liao H (2017) Engineering crop nutrient efficiency for sustainable agriculture. J Integr Plant Biol 59:710–735
Crombez H, Motte H, Beeckman T (2019) Tackling plant phosphate starvation by the roots. Dev Cell 48:599–615
Cui YN, Li XT, Yuan JZ, Wang FZ, Wang SM, Ma Q (2019) Nitrate transporter NPF7.3/NRT1.5 plays an essential role in regulating phosphate deficiency responses in Arabidopsis. Biochem Biophys Res Commun 508:314–319
Dai X, Wang Y, Yang A, Zhang WH (2012) OsMYB2P-1, an R2R3 MYB transcription factor, is involved in the regulation of phosphate-starvation responses and root architecture in rice. Plant Physiol 159:169–183
Datta S, Prescott H, Dolan L (2015) Intensity of a pulse of RSL4 transcription factor synthesis determines Arabidopsis root hair cell size. Nat Plants 1:1–6
Forde BG (2014) Nitrogen signalling pathways shaping root system architecture: an update. Curr Opin Plant Biol 21:30–36
Giri J, Bhosale R, Huang G, Pandey BK, Parker H, Zappala S, Yang J, Dievart A, Bureau C, Ljung K et al (2018) Rice auxin influx carrier OsAUX1 facilitates root hair elongation in response to low external phosphate. Nat Commun 9:1–7
Goncalves BX, Lima-Melo Y, Maraschin F dos S, Margis-Pinheiro M (2020) Phosphate starvation responses in crop roots: from well-known players to novel candidates. Environ Exp Bot 178:104162
Guan P, Ripoll JJ, Wang R, Vuong L, Bailey-Steinitz LJ, Ye D, Crawford NM (2017) Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc Natl Acad Sci U S A 114:2419–2424
Gutiérrez-Alanís D, Ojeda-Rivera JO, Yong-Villalobos L, Cárdenas-Torres L, Herrera-Estrella L (2018) Adaptation to phosphate scarcity: tips from arabidopsis roots. Trends Plant Sci 23:721–730
Hanlon MT, Ray S, Saengwilai P, Luthe D, Lynch JP, Brown KM (2018) Buffered delivery of phosphate to Arabidopsis alters responses to low phosphate. J Exp Bot 69:1207–1219
Huang G, Zhang D (2020) The plasticity of root systems in response to external phosphate. Int J Mol Sci 21:1–12
Huang K-L, Ma G-J, Zhang M-L, Xiong H, Wu H, Zhao C-Z, Liu C-S, Jia H-X, Chen L, Kjorven JO et al (2018) The ARF7 and ARF19 transcription factors positively regulate PHOSPHATE STARVATION RESPONSE1 in Arabidopsis roots. Plant Physiol 178:413–427
Jenkinson DS (2001) The impact of humans on the nitrogen cycle, with focus on temperate arable agriculture. Plant Soil 228:3–15
Jia X, Liu P, Lynch JP (2018) Greater lateral root branching density in maize improves phosphorus acquisition from low phosphorus soil. J Exp Bot 69:4961–4970
Jia Z, von Wirén N (2020) Signaling pathways underlying nitrogen-dependent changes in root system architecture: from model to crop species. J Exp Bot 71:4393–4404
Ju C, Buresh RJ, Wang Z, Zhang H, Liu L, Yang J, Zhang J (2015) Root and shoot traits for rice varieties with higher grain yield and higher nitrogen use efficiency at lower nitrogen rates application. F Crop Res 175:47–55
Kiba T, Kudo T, Kojima M, Sakakibara H (2011) Hormonal control of nitrogen acquisition: roles of auxin, abscisic acid, and cytokinin. J Exp Bot 62:1399–1409
Kim CM, Han C, Dolan L (2017) RSL class I genes positively regulate root hair development in Oryza sativa. New Phytol 213:314–323
Knox K, Grierson CS, Leyser O (2003) AXR3 and SHY2 interact to regulate root hair development. Development 130:5769–5777
Kohli PS, Kumar Verma P, Verma R, Parida SK, Thakur JK, Giri J (2020) Genome-wide association study for phosphate deficiency responsive root hair elongation in chickpea. Funct Integr Genomics 20:775–786
Krouk G, Lacombe B, Bielach A, Perrine-Walker F, Malinska K, Mounier E, Hoyerova K, Tillard P, Leon S, Ljung K et al (2010) Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell 18:927–937
Li X, Zeng R, Liao H (2016) Improving crop nutrient efficiency through root architecture modifications. J Integr Plant Biol 58:193–202
Lima JE, Kojima S, Takahashi H, von Wirén N (2010) Ammonium triggers lateral root branching in Arabidopsis in an AMMONIUM TRANSPORTER1;3-dependent manner. Plant Cell 22:3621–3633
Luo L, Zhang Y, Xu G (2020) How does nitrogen shape plant architecture? J Exp Bot 71:4415–4427
Lynch JP (2019) Root phenotypes for improved nutrient capture: an underexploited opportunity for global agriculture. New Phytol 223:548–564
Lynch JP (2013) Steep, cheap and deep: an ideotype to optimize water and N acquisition by maize root systems. Ann Bot 112:347–357
Maghiaoui A, Bouguyon E, Cuesta C, Perrine-Walker F, Alcon C, Krouk G, Benková E, Nacry P, Gojon A, Bach L (2020a) The arabidopsis NRT1.1 transceptor coordinately controls auxin biosynthesis and transport to regulate root branching in response to nitrate. J Exp Bot 71:4480–4494
Maghiaoui A, Gojon A, Bach L (2020b) NRT1.1-centered nitrate signaling in plants. J Exp Bot 71:6226–6237
Medici A, Marshall-Colon A, Ronzier E, Szponarski W, Wang R, Gojon A, Crawford NM, Ruffel S, Coruzzi GM, Krouk G (2015) AtNIGT1/HRS1 integrates nitrate and phosphate signals at the Arabidopsis root tip. Nat Commun 6:1–11
Medici A, Szponarski W, Dangeville P, Safi A, Dissanayake IM, Saenchai C, Emanuel A, Rubio V, Lacombe B, Ruffel S et al (2019a) Identification of molecular integrators shows that nitrogen actively controls the phosphate starvation response in plants. Plant Cell 31:1171–1184
Medici A, Szponarski W, Dangeville P, Safi A, Dissanayake IM, Saenchai C, Emanuel A, Rubio V, Lacombe B, Ruffel S et al (2019b) Nitrogen actively controls the phosphate starvation response in plants. Plant Cell Adv Publ Publ. https://doi.org/10.1105/tpc.18.00656
Meier M, Liu Y, Lay-Pruitt KS, Takahashi H, von Wirén N (2020) Auxin-mediated root branching is determined by the form of available nitrogen. Nat Plants 6:1136–1145
Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA et al (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci 102:7760–7765
Mounier E, Pervent M, Ljung K, Gojon A, Nacry P (2014) Auxin-mediated nitrate signalling by NRT1.1 participates in the adaptive response of Arabidopsis root architecture to the spatial heterogeneity of nitrate availability. Plant Cell Environ 37:162–174
Müller J, Toev T, Heisters M, Teller J, Moore KL, Hause G, Dinesh DC, Bürstenbinder K, Abel S (2015) Iron-dependent callose deposition adjusts root meristem maintenance to phosphate availability. Dev Cell 33:216–230
Niu YF, Chai RS, Jin GL, Wang H, Tang CX, Zhang YS (2013) Responses of root architecture development to low phosphorus availability: a review. Ann Bot 112:391–408
Pélissier P-M, Motte H, Beeckman T (2021) Lateral root formation and nutrients: nitrogen in the spotlight. Plant Physiol. https://doi.org/10.1093/plphys/kiab145
Peret B, Desnos T, Jost R, Kanno S, Berkowitz O, Nussaume L (2014) Root architecture responses. In: Search of phosphate. Plant Physiol 166:1713–1723
Pérez-Torres CA, López-Bucio J, Cruz-Ramírez A, Ibarra-Laclette E, Dharmasiri S, Estelle M, Herrera-Estrella L (2008) Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell 20:3258–3272
Postma JA, Dathe A, Lynch JP (2014) The optimal lateral root branching density for maize depends on nitrogen and phosphorus availability. Plant Physiol 166:590–602
Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15:2122–2133
Salazar-Henao JE, Vélez-Bermúdez IC, Schmidt W (2016) The regulation and plasticity of root hair patterning and morphogenesis. Development 143:1848–1858
Satbhai SB, Ristova D, Busch W (2015) Underground tuning: quantitative regulation of root growth. J Exp Bot 66:1099–1112
Savage N, Yang TJW, Chen CY, Lin KL, Monk NAM, Schmidt W (2013) Positional signaling and expression of enhancer of try and CPC1 are tuned to increase root hair density in response to phosphate deficiency in Arabidopsis thaliana. PLoS ONE. https://doi.org/10.1371/journal.pone.0075452
Shahzad Z, Amtmann A (2017) Food for thought: how nutrients regulate root system architecture. Curr Opin Plant Biol 39:80–87
Ticconi CA, Lucero RD, Sakhonwasee S, Adamson AW, Creff A, Nussaume L, Desnos T, Abel S (2009) ER-resident proteins PDR2 and LPR1 mediate the developmental response of root meristems to phosphate availability. Proc Natl Acad Sci 106:14174–14179
Trachsel S, Kaeppler SM, Brown KM, Lynch JP (2013) Maize root growth angles become steeper under low N conditions. F Crop Res 140:18–31
Vidal EA, Araus V, Lu C, Parry G, Green PJ, Coruzzi GM, Gutiérrez RA (2010) Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc Natl Acad Sci U S A 107:4477–4482
Waidmann S, Sarkel E, Kleine-Vehn J (2020) Same same, but different: growth responses of primary and lateral roots. J Exp Bot 71:2397–2411
Wang J, Pei L, Jin Z, Zhang K, Zhang J (2017) Overexpression of the protein phosphatase 2A regulatory subunit a gene ZmPP2AA1 improves low phosphate tolerance by remodeling the root system architecture of maize. PLoS One 12:e0176538
Wang X, Wang HF, Chen Y, Sun MM, Wang Y, Chen YF (2020) The transcription factor NIGT1.2 modulates both phosphate uptake and nitrate influx during phosphate starvation in arabidopsis and maize. Plant Cell 32:3519–3534
Xu G, Takahashi H (2020) Improving nitrogen use efficiency: from cells to plant systems. J Exp Bot 71:4359–4364
Yan X, Liao H, Beebe SE, Blair MW, Lynch JP (2004) QTL mapping of root hair and acid exudation traits and their relationship to phosphorus uptake in common bean. Plant Soil 265:17–29
Yang H, Knapp J, Koirala P, Rajagopal D, Peer WA, Silbart LK, Murphy A, Gaxiola RA (2007) Enhanced phosphorus nutrition in monocots and dicots over-expressing a phosphorus-responsive type I H+-pyrophosphatase. Plant Biotechnol J 5:735–745
Zhang C, Simpson RJ, Kim CM, Warthmann N, Delhaize E, Dolan L, Byrne ME, Wu Y, Ryan PR (2018) Do longer root hairs improve phosphorus uptake? Testing the hypothesis with transgenic Brachypodium distachyon lines overexpressing endogenous RSL genes. New Phytol 217:1654–1666
Zhang H, Rong H, Pilbeam D (2007) Signalling mechanisms underlying the morphological responses of the root system to nitrogen in Arabidopsis thaliana. J Exp Bot 2329–2338
Zhou J, Jiao FC, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P (2008) OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 146:1673–1686
Zhou J, Xie J, Liao H, Wang X (2014) Overexpression of β-expansin gene GmEXPB2 improves phosphorus efficiency in soybean. Physiol Plant 150:194–204
Zhu J, Kaeppler SM, Lynch JP (2005) Mapping of QTL controlling root hair length in maize (Zea mays L.) under phosphorus deficiency. Plant Soil 270:299–310
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Awasthi, P., Laxmi, A. (2021). Root Architectural Plasticity in Changing Nutrient Availability. In: Mukherjee, S., Baluška, F. (eds) Rhizobiology: Molecular Physiology of Plant Roots. Signaling and Communication in Plants. Springer, Cham. https://doi.org/10.1007/978-3-030-84985-6_2
Download citation
DOI: https://doi.org/10.1007/978-3-030-84985-6_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-84984-9
Online ISBN: 978-3-030-84985-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)