Abstract
Aberrant control of acid–base homeostasis is an emerging cancer hallmark that results from adaptation to oncogenic metabolism and tumor microenvironment. It has dramatic consequences, contributing to acquisition of aggressive tumor phenotype and disease progression. Regulation of pH is executed by transporters, exchangers, and pumps mediating ion fluxes in rates dictated by actual physiological needs of tumor cells. However, full performance of the pH-regulation machinery requires carbonic anhydrases (CAs) catalyzing a reaction key to acid-base balance: reversible conversion of carbon dioxide to bicarbonate ion and proton. Here, we provide an overview of known and predicted links of the human CA isoforms to various types of neoplasms and summarize the mechanisms, through which they functionally contribute to cancer. We use the approach, in which data from published papers are complemented by publicly available metadata processed by the GEPIA2 instrument. This allows not only for aligning bioinformatics to experimental evidence (supporting a prominent position of CA IX), but also for prediction of novel correlations that remain to be experimentally proven (such as for CA XIV). With a continuous accumulation of the knowledge, it is now becoming clear that CAs are broadly expressed in tumors and actively participate in cancer development and/or progression.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
7.1 Introduction
Vital functions of cells, tissues, and organs in the human body crucially depend, among other factors, on proper pH regulation, especially in situations of high metabolic performance, gas exchange, and/or fluid production. Such processes occur under physiological conditions in virtually all organs and are particularly intense in stomach, intestine, liver, pancreas, kidney, lung, and brain. However, many pathological conditions including cancer are characterized by aberrant control of acid–base homeostasis as a result of perturbed ion transport and abnormal metabolism. Especially in cancer cells, this has very dramatic consequences, supporting acquisition of an aggressive tumor phenotype and contributing to disease progression (Corbet and Feron 2017).
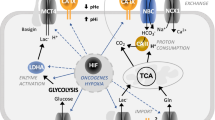
Regulation of pH is executed by a number of ion transporters, exchangers, and pumps that carry out ion extrusion and/or ion import in a rate dictated by actual physiological needs of the cell and by status of its intracellular and extracellular microenvironment. Among these, lactate and proton exporters, and bicarbonate importers play a major role as pH-modulating mediators of ion fluxes across the plasma membrane, while proton pumps also control pH of certain intracellular organelles (endosomes, lysosomes). However, in addition to these “ion transport managers,” full performance of the pH regulation machinery requires enzymes of the carbonic anhydrase family catalyzing the reaction that is fundamental for acid-base balance, namely, the reversible conversion of carbon dioxide to carbonic acid that spontaneously dissociates to bicarbonate ion and proton (Supuran 2016). The human body contains 15 carbonic anhydrase (CA) isoforms of the α-CA family, namely CA I-IV, CA VA and CA VB, CA VI-XIV. Twelve isoforms are active enzymes, with catalytic activity ranging from low to very high (CA II and CA IX belonging to the most active enzymes in the nature), while three isoforms are inactive (CA VIII, X, and XI). CAs also differ by subcellular localization: CA I-III, VII, VIII, X, XI, XIII are cytoplasmic, CA VA and VB are mitochondrial, CA IV, IX, XII, XIV are membrane-associated, and CA VI is secreted (Pastorekova et al. 2004; Mboge et al. 2018). The diversity of the human CAs is further widened by their variable expression levels and heterogeneous expression pattern in organs and tissues ranging from almost ubiquitous (e.g., CA II and CA XIII), to limited to certain tissue (e.g., CA VA and CA VI). This makes their complex understanding very difficult and development of general statements very challenging.
It has been long believed that the nature “invented” these enzymes primarily to drive pH-related physiological processes in normal tissues, since the initial efforts to link CAs to cancer did not bring any clear-cut data. However, everything changed in the early 90 's of the twentieth century, with the discovery of two, at that time novel, CA isoforms. First it was CA IX, which showed strong association with a broad range of tumors contrasting to limited expression in normal tissues (Pastorek et al. 1994) followed by CA XII, which was detected in many normal tissues, but showed overexpression in certain tumors (Tureci et al. 1998). Later on, thorough examination of some of the “old” isoforms (CA I, II, IV) revealed that their expression patterns exhibit either positive or negative correlations with cancer within specific contexts. Moreover, inactive isoforms CA X and CA XI, have also been implicated in tumor phenotype. In fact, with continuous accumulation of data, it is now becoming clearly apparent that CAs can actively participate in cancer development and/or progression.
Here we present an overview of known and predicted links of CA isoforms to various types of neoplasms and summarize the mechanisms through which they functionally contribute to cancer. In this overview, data from published papers are complemented by publicly available RNA-seq metadata from TCGA and GTEx databases processed by the GEPIA2 instrument, which uses Transcripts Per Million (TPM) values of transcription levels normalized to transcript's length and divided by the scaling factor to allow for comparison of gene expression levels among different types (Tang et al. 2019). This combined approach allows not only for aligning bioinformatics to experimental evidence, but also for prediction of some correlations that still remain to be experimentally proven. In this way, it is also possible to demonstrate that the cancer-related expression pattern, in some cases, does not necessarily translate to clinically relevant information, suggesting that the experimental evidence including functional studies is an imperative prerequisite for understanding the roles of genes/proteins including carbonic anhydrases in cancer biology and of their potential usefulness for clinical applications. Note that CA genes are written in italic style with Arabic numbers, while CA proteins are written in normal style with Roman numbers, in line with the official nomenclature, and that abbreviations used to designate different cancer types were used according to GEPIA2 (Table 7.1).
7.2 Expression of Cytoplasmic CA Isoforms in Neoplasms
Metadata analysis revealed that all of the five genes encoding the cytoplasmic CAs are primarily expressed in normal tissues, with CA2 exhibiting the highest transcription levels followed by CA1, CA3, CA7, and CA13 (Fig. 7.1). Notably, CA II protein displays also the highest catalytic activity, while the activity of CA III is the lowest among all active CA isoforms (Mboge et al. 2018). In line with these facts, most of the research related to cytoplasmic CAs in cancer has been focused on CA II as described in more detail further below.
Expression of genes coding for the cytoplasmic carbonic anhydrases in tumors and normal samples. Data extracted from TCGA/GTEx databases were analyzed and converted to graphics using GEPIA2 instrument (Tang et al. 2019). Data in the graphs relate to TPM units and intensities of colors in the bodymaps correspond to Log/TPM+1 Scores in particular tissues. Numbers above the columns highlight the highest TPMs for each CA isoform. Kcat/KM values in the upper left corners inform about the catalytic efficiency of CA isoforms {adopted from (Mboge et al. 2018)}
As shown in Figs. 7.1 and 7.5, CA1 gene transcription is relatively low, reaching TPM values of around 30 in normal colon, B-cells, and T-cells, and is not expressed in their neoplastic counterparts. However, CA1 expression is increased in acute myeloid leukemia (LAML, TPM 40.57) compared to normal myeloid cells (TPM 7). In spite of this differential expression, GEPIA2-performed Mantel-Cox Log-rank test of metadata from LAML failed to show a significantly better overall survival in the high CA1 expression cohort, irrespective of whether cutoff was set to median (50%) or whether cutoff-high was set to 75% and cutoff-low to 25% (Fig. 7.6). Nevertheless, the literature contains dozens of papers dealing with the relationship of CA1 to cancer. CA I protein expression was demonstrated by immunohistochemistry (IHC) in both normal and malignant endocrine tissue of pancreas, with intense positive staining observed particularly in the cells expressing glucagon (Parkkila et al. 1995b). IHC comparison of CA I distribution in normal large intestine versus colorectal tumors showed high CA I expression in normal colon, decreased intensity in benign lesions, and very weak staining in malignant tumors (Kivela et al. 2001). This finding was confirmed by an independent proteomic study suggesting that downregulation of CA I is an early event in colorectal carcinogenesis (Wang et al. 2012). In contrary, no CA1 expression-based difference in overall survival was predicted by log-rank test of COAD cohorts from TCGA database. CA I protein was also detected by proteomic methods in sera of patients with non-small cell lung carcinoma (Wang et al. 2016). Noteworthy, sera of patients suffering from both acute myeloid leukemia and chronic lymphocytic leukemia were found to contain CA I (and CA II) autoantibodies (Mentese et al. 2017, 2018). Lakota and colleagues observed elevated CA I autoantibodies in sera of patients with spontaneous regression of tumors developing an aplastic anemia-type syndrome after a high-dose therapy with autologous stem cell transplantation (Skultety et al. 2010; Jankovicova et al. 2013). Silencing of CA I in prostatic (PC3) tumor cells was shown to change the composition of exosomes secreted by these cells indicating their enhanced malignant potential (Banova Vulic et al. 2019). However, due to many open questions and generally inconsistent data, it is currently impossible to make any conclusion on the relationship of CA I to cancer.
The CA3 gene is mostly transcribed in several normal tissues (mainly breast, lung, and uterus), with a TPM value up-to 15 (Fig. 7.1). It is generally not expressed in tumors, except relatively low expression in glioblastoma (GBM, TPM 8.8), which is about 8-times higher than in normal brain (Fig. 7.5). However, this does not translate into the correlation to overall survival of GBM patients, but such correlation exists in LAML (Fig. 7.6). Interestingly, there are only two published studies dealing with CA III expression or function in cancer. Despite metadata showing only very low expression of CA III in liver and liver-derived neoplasms (1 TPM vs 0.1 TPM), an immunohistochemical analysis suggests that CA III is expressed in normal hepatocytes and that it is reduced in hepatocellular carcinoma (Kuo et al. 2003). The second study explored the effects of manipulated CA III expression on invasiveness of HCC (in TCGA abbreviation LIHC) cell line SK-Hep1 (Dai et al. 2008). The authors found that CA III suppression decreases, while CA III overexpression increases cell invasiveness possibly via activation of FAK signaling and extracellular acidification. Taking into account that there is no extension/confirmation of this observation and that CA III activity is very low, biological relevance of that observation remains elusive.
CA7 transcription is detectable only in normal tissues (namely intestine, brain, and testes, TPM up to 8.5) but not in neoplasms (Figs. 7.1 and 7.5). Albeit absent or low, CA7 levels in tumors derived from these tissues (COAD, READ, GBM, LGG) do not correlate with overall survival in any group of patients. In the published study of Yang and colleagues (Yang et al. 2015), reduced CA7 mRNA and protein expression in colorectal carcinoma was significantly correlated with poor differentiation, positive lymph node metastasis, advanced TNM stage, and unfavorable clinical outcome. However, no such correlation was confirmed by immunostaining of colorectal carcinoma specimens from an independent cohort of patients, when both extent and intensity of staining were taken into account (Viikila et al. 2016). Thus, further studies are needed to resolve these conflicting results.
CA13 gene transcription is detected both in normal tissues and in tumors, although it appears to be increased in some normal tissues and decreased in other ones when compared to tumors (Figs. 7.1 and 7.5). The significance of this differential expression remains unknown, but taking into account the fact that the levels of CA13 transcription are rather low (up to 9 TPM), differences are too minor to have a biological relevance. There is only a single paper using immunohistochemistry to detect CA XIII protein expression in colorectal carcinoma (Kummola et al. 2005). The study results suggest that the expression of CA XIII is downregulated in tumor cells compared to the normal tissue, which is actually opposite to RNA-seq data acquired from TCGA database where significant correlation to overall survival was found in KIRC, LAML, LGG, and SKCM.
Lastly, the apparently most important cytoplasmic isoform is encoded by the CA2 gene. The catalytic activity of CA II is among the highest not only within the CA family but also generally among all known enzymes. Its expression assessed by immunodetection was found to correlate with intracellular CA activity measured in a range of intact human cancer- and fibroblast-derived cells and in their membrane-free lysates. Moreover, genetic knockdown of CA II in HCT116 colon carcinoma cells demonstrated that majority of intracellular CA activity was attributable to CA II. These data support the view that CA II is a crucial intracellular CA isoenzyme (Hulikova et al. 2014). Studies based on manipulated CA II expression and on use of small molecule inhibitors suggested that CA II activity participates in intracellular pH regulation (irrespective of normal or tumor cell phenotype) through cooperation with ion transporters, namely, by facilitating proton or bicarbonate transport across membranes, or mediating proton diffusion in cytoplasm. Initially, it was proposed that CA II forms a transport metabolon with chloride/bicarbonate exchangers that exploit the CA catalytic activity (Sterling et al. 2001). Later on, this concept was extended to the sodium/proton exchanger NHE1 (Li et al. 2002) and to the sodium/bicarbonate co-transporter (Villafuerte et al. 2014). CA II was also shown to cooperate in a non-catalytic manner with the monocarboxylate transporter MCT1, acting as an antenna collecting and transferring protons to MCT1 in order to facilitate its transport activity (Becker et al. 2005; Noor et al. 2018).
However, these scenarios are partly questioned by the finding of Swietach and colleagues who demonstrated that intracellular CA activity in cancer cell lines does not correlate with resting intracellular pH (pHi), NHE1 flux, or bicarbonate transporter flux measured in these cells, and that proton and bicarbonate fluxes produced by cells over the physiological pHi range are not of sufficient magnitude to require intracellular CA catalysis, with the exception of acid loading by bicarbonate export at high pHi (over 7.4) that is not typical for cancer cells (Hulikova et al. 2014). This study also suggests that high intracellular CA activity (mostly attributable to CA II) is associated with faster and larger pHi oscillations, larger pH-dependent intracellular calcium ion oscillations, and stronger inhibition of mTORC1 pathway in response to extracellular pCO2 fluctuations (Hulikova et al. 2014). In contrast, pHi of cells exhibiting low intracellular CA activity is less responsive to pCO2 fluctuations, which might be of key importance for cells’ survival especially in tumor tissues with dynamic temporal and/or regional changes of blood flow, delivery of oxygen and nutrients as well as removal of metabolic waste due to aberrant tumor vasculature (Gillies et al. 2018).
Actually, the above-described view can explain why expression of the CA2 gene (as well as the levels of other genes encoding the cytoplasmic CAs with lesser contribution to overall intracellular CA activity) is primarily detected in non-cancerous tissues (with TPM values up to 245) and downregulated in related tumors. In line with this, TCGA/GTEx databases processed by GEPIA2 show that CA II levels are decreased in KIRC, KIRP, SARC, STAD, and TGCT when compared to their normal counterparts. In contrary, CA2 expression is increased in GBM and KICH (with TPM of 126 and 495, see Figs. 7.1 and 7.5). While this increased tumor expression does not predict overall survival of cancer patients in case of GBM and KICH, reduced CA2 transcription is significantly associated with reduced overall survival in patients with KIRC and SARC (Figs. 7.6 and 7.7), but not KIRP, STAD, and TGCT. Interestingly, significantly worse overall survival predicted by GEPIA2 is also linked with decreased CA II levels in COAD (with cutoff-high 75% and cutoff-low 25%) that is principally in line with IHC and proteomic studies of independent patients’ cohorts showing that loss of expression of CA II (and CA I) accompanies progression to malignant phenotype (Kivela et al. 2001; Wang et al. 2012). Moreover, CA II is the most widely distributed isoform in the central nervous system, and its expression was also detected by IHC in different brain tumors (Parkkila et al. 1995a), medulloblastomas and neuroectodermal tumors (Nordfors et al. 2010), hematological malignancies (Leppilampi et al. 2002), esophageal carcinoma (Nortunen et al. 2018), pancreatic tumors (Parkkila et al. 1995b), gastrointestinal stromal tumors (Parkkila et al. 2010; Liu et al. 2013), hepatocellular carcinomas (Kuo et al. 2003), uterine tumors (Hynninen et al. 2012), and urinary bladder cancers (Tachibana et al. 2017). In meningiomas, CA II was found in endothelial cells in association with increasing malignancy grade and tumor proliferation rates (Korhonen et al. 2009). CA II expression also was observed in tumor vessel endothelia of melanoma and esophageal, renal, and lung cancers (Yoshiura et al. 2005).
There are also several papers describing the functional aspects of CA II. It was shown that forced CA II expression in colorectal cancer cells remarkably suppresses tumor cell growth both in vitro and in vivo (Zhou et al. 2013). Similarly, CA II overexpression inhibits cell migration and invasion by reversing EMT in hepatocellular carcinoma cells, while its downregulation promoted invasiveness and metastasis (Zhang et al. 2018). Thus, the link of CA II to neoplastic phenotype is tumor tissue- and cell type-dependent and may also reflect the physiology of tumor microenvironment.
7.3 Expression of Mitochondrial CA Isoforms in Cancer
Mitochondrial CA isoforms are encoded by two phylogenetically-related genes CA5A and CA5B. Since mitochondria are impermeable to bicarbonate ions, activity of CA VA and CA VB enzymes is required for production of bicarbonate that is utilized by mitochondrial enzymes for metabolic processes, namely, by pyruvate carboxylase for gluconeogenesis and carbamoyl phosphate synthase I for ureagenesis. Targeted disruption of the murine Car5A and Car5B genes and their double knock-out implicated both enzymes in ammonia detoxication and glucose metabolism (Shah et al. 2013). Despite glucose metabolism being of high importance for tumor biology, so far there are no thorough studies linking CA5A and/or CA5B to cancers, except a single paper identifying the CA5A gene within a signature of genes predicting decreased overall survival in patients with head and neck carcinoma (Bornstein et al. 2016).
Thus, publicly available RNA-seq metadata can provide at least basic insight into possible relationships of CA5A and CA5B expression to human tumors. GEPIA2 testing showed that CA5A gene is transcribed in normal liver (in accord with expectation) at the TPM level of 23.2 and was slightly decreased in liver neoplasms (TPM 16.3). In addition, CA5A expression was detected in bile ducts, but not in cholangiocarcinoma (Figs. 7.2 and 7.5). In contrary, CA5B is transcribed in many normal tissues (namely in the breast, ovaria, uterus, cervix, testes, TPM up to 9.9) and at lower levels also in the corresponding cancers. The highest CA5B expression is present in acute myeloid leukemia (Fig. 7.2), but it has no impact on overall survival (Fig. 7.6). However, these data do not take into consideration the potential role of CA VA/CA VB catalytic activities, which might be involved in adaptation to metabolic demands of tumor cells. Clearly, additional studies are warranted to resolve whether mitochondrial CAs are involved in tumor biology.
Expression of genes coding for the mitochondrial carbonic anhydrases in tumors and normal samples. Data extracted from TCGA/GTEx databases were analyzed and converted to graphics using GEPIA2 instrument (Tang et al. 2019). Data in the graphs relate to TPM units and intensities of colors in the bodymaps correspond to Log/TPM+1 Scores in particular tissues. Numbers above the columns highlight the highest TPMs for each CA isoform. Kcat/KM values in the upper left corners inform about the catalytic efficiency of CA isoforms {adopted from (Mboge et al. 2018)}
7.4 Expression of Membrane-Bound and Secreted CA Isoforms in Cancer
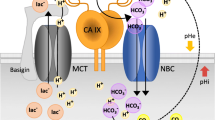
Four membrane-bound isoforms (CA IV, IX, XII, and XIV) and one secreted isoform (CA VI) are active enzymes with the catalytic sites facing the extracellular space. Their catalytic performance is relatively high, with CA IX exhibiting the highest activity (similar to CA II), then followed by CA VI, CA IV, CA XIV, and CA XII, activity of which is approximately 4-times lower than that of CA IX. All these enzymes appear to support extracellular CO2 diffusion, and the membrane-bound isoforms also participate in bicarbonate import and proton extrusion.
While CA IV is bound to the plasma membrane by the GPI anchor, three other exofacial CAs are type I membrane proteins, with the N-terminal side at the cell surface and C-terminal side in the cytoplasm (Pastorekova et al. 2004). In addition, CA IX has an extra N-terminal proteoglycan-like domain (PG) linked by the hinge region to the CA domain (Opavsky et al. 1996). This PG domain contains a repetitive stretch of proline-perturbed basic amino acids, and has an intrinsically disordered structure with conformational flexibility that is implicated in non-catalytic functions of CA IX relevant for tumor biology (Zavada et al. 2000; Langella et al. 2018). CA IX isoform was shown to cooperate with extracellular structures of bicarbonate transporters and sodium/proton exchangers facilitating ion transport via its catalytic activity (Morgan et al. 2007; Orlowski et al. 2012; Svastova et al. 2012), and with monocarboxylate transporters MCT1 and MCT4 via a non-catalytic mechanism, in which the PG domain serves as a proton-conducting antenna interacting with MCT4 chaperone CD147 (Jamali et al. 2015; Ames et al. 2018, 2019). Similar model was demonstrated for CA IV, which facilitates lactate export via interaction with chaperons of MCTs (Klier et al. 2014; Forero-Quintero et al. 2019).
TCGA/GTEx metadata-derived expression patterns of the membrane-bound and secreted CAs provided by GEPIA2 instrument are very distinct and remarkable (Figs. 7.3 and 7.5), and generally have a lot of support in the published papers, albeit some previously unknown and potentially interesting connections have also been disclosed as described below.
Expression of genes coding for the membrane-bound and secreted carbonic anhydrases in tumors and normal samples. Data extracted from TCGA/GTEx databases were analyzed and converted to graphics using GEPIA2 instrument (Tang et al. 2019). Data in the graphs relate to TPM units and intensities of colors in the bodymaps correspond to Log/TPM+1 Scores in particular tissues. Numbers above the columns highlight the highest TPMs for each CA isoform. Kcat/KM values in the upper left corners inform about the catalytic efficiency of CA isoforms {adopted from (Mboge et al. 2018)}
Noteworthy, the CA4 gene is mostly expressed in normal tissues, and only occasionally and at a low level in some tumors (Figs. 7.3 and 7.5). The highest TPM values of CA4 transcription (up to 81.7) are exhibited by normal counterparts of LUAD, LUSC, THCA, BRCA, SARC, DLBC, KIRP, KIRC, KICH, and THYM. However, only KIRC and LUAD show a highly significant correlation between high expression of CA4 and better overall survival, with hazard risk reduced up to 3-times compared to tumors with low CA4 (Figs. 7.6 and 7.7). Interestingly, these significant correlations exist independently of whether the analysis includes all CA4 transcript variants together, or only individual protein-coding transcript variants identified by GEPIA2, namely, CA4-001, CA4-005 (both in KIRC and LUAD), and CA4-006 (only in KIRC). In addition, both median and quartile cutoffs produced similar probability values.
Experimental data are in agreement with predictions based on bioinformatics. In patients with renal clear cell carcinoma, decreased expression of CA4 mRNA in tumors was associated with poor survival (Takenawa et al. 1998). Loss of CA4 was also proposed as a biomarker distinguishing follicular thyroid carcinomas from follicular adenomas (Davidov et al. 2014). CA4 expression determined by Q PCR was also found to be significantly downregulated in non-small cell lung carcinomas (NSCLC) as well as in six NSCLC-derived cell lines, and lower CA4 levels were correlated with lymph node metastasis and shorter overall survival (Chen et al. 2017). Importantly, CA4 was proposed to be a novel tumor suppressor in colorectal cancer (CRC). This proposal was based on the observation that CA4 gene is silenced in more than 90% of CRC tumors primarily by the promoter hypermethylation (Zhang et al. 2016). In support of the tumor suppressor concept, re-expression of CA4 in CRC cells inhibits cell proliferation and induces apoptosis via downregulation of the WNT pathway and degradation of β-catenin (Zhang et al. 2016).
Based on GEPIA2 analysis, CA6 expression is confined to normal skin (TPM 17, Figs. 7.3 and 7.5), and its reduced level in SKCM predicts worse overall survival (Figs. 7.6 and 7.7). In the published literature, CA VI isoform is primarily known as a component of saliva expressed in serous acinar cells of human salivary glands. Immunostaining revealed its potential diagnostic utility for discrimination of acinic cell carcinoma from mammary analogue secretory carcinoma of the salivary gland (Hsieh et al. 2016). No further information on the relationship of CA VI to cancer is available to date.
In contrast to the other CA genes, CA9 is predominantly expressed in tumor tissues (CESC, CHOL, COAD, ESCA, GBM, HNSC, KIRC, LUSC, OV, PAAD, READ, UCEC, UCS, TPM up to 305.3) and virtually not in their normal counterparts (Figs. 7.3 and 7.5). According to RNA-seq data from TCGA/GTEx databases, the only normal tissues that exhibit CA9 transcription levels higher than tumors are stomach (TPM 216) and testes (TPM 35.7), albeit published IHC studies detected CA IX protein also in the epithelia of pancreas, gallbladder, intestinal crypts, and basal skin cells (Pastorekova et al. 1997; Kivela et al. 2001).
In tumors, expression of CA9 is directed primarily by a HIF-1 transcription factor composed of an oxygen-regulated α subunit and a constitutive β subunit. The α subunit is degraded in proteasome following hydroxylation of its critical prolyl residues and recognition by pVHL tumor suppressor protein acting as E3 ubiquitin ligase, in conditions of normal oxygen delivery (Kaelin and Ratcliffe 2008). When metabolic and proliferative demands of tumor cells exceed the availability of oxygen, which is reduced due to aberrant tumor vasculature, resulting hypoxia inhibits prolyl hydroxylases (including the asparaginyl hydroxylase that hampers HIF-α transcriptional activity) and leads to escape of an α subunit from pVHL recognition and degradation. As a consequence, HIF-α is stabilized and activated, accumulates in the cytoplasm, enters the nucleus, dimerizes with HIF-β, and forms a competent transcription factor that induces expression of hundreds of genes coding for proteins involved in adaptation of tumor cells to hypoxia (Ratcliffe 2013; Semenza 2012). CA9 is one of the key targets of HIF-1 that drives its transcription in hypoxic tumor cells to a very high magnitude through binding closely to the transcription initiation site of the CA9 gene (Wykoff et al. 2000). Thus, expression of CA9 in the majority of tumor types is heterogeneous and regionally distributed in hypoxic or post-hypoxic areas. However, in some cancer entities, CA9 expression is induced due to activation of pathways driven by oncogenes (such as SRC or RET, Takacova et al. 2010, 2014).
The situation is rather different in KIRC, which express the highest levels of CA9 transcript as well as CA IX protein (this can be observed both in metadata and IHC studies). These renal clear cell carcinomas are characterized by inactivating mutations of VHL tumor suppressor gene, resulting in constitutive stabilization of HIF-α subunit and sustained activation of the HIF-governed hypoxia-related pathways even in the absence of physiological hypoxia (Wiesener et al. 2001). Since CA9 is one of the most highly HIF-1-induced genes, it is expressed in a very high percentage of VHL-defective renal cancer clear cells (Ivanov et al. 1998; Bui et al. 2003; Stillebroer et al. 2010).
When including all four CA9 alternative transcripts (two protein-coding and two non-translated) into GEPIA2 survival analysis, CA9 expression shows significant associations with overall survival of patients with several types of cancers, namely, GBM, LUAD, PAAD, and SKCM (Fig. 7.6). However, when solely the full-length transcript CA9-001 is taken into account, the spectrum of tumor types is extended to KIRC and SARC (Fig. 7.7). CA9-001 is the only full-length transcript, which is induced by hypoxia and translated to a functional and active CA IX protein (Barathova et al. 2008).
In all mentioned tumor types except KIRC, higher CA9 expression is related to significantly poorer prognosis (Figs. 7.6 and 7.7). In the case of KIRC, higher CA9-001 levels (but not total CA9 levels) show correlation with significantly better overall survival (Figs. 7.6 and 7.7). This appears to be a consequence of a shift from HIF-1 that principally governs the CA9 transcription in early stages of this disease, towards HIF-2, which drives a kidney cancer progression and determines malignant tumor phenotype in renal clear cell carcinoma, but induces CA9 to much lower degree (Raval et al. 2005).
Numerous immunohistochemical studies of various tumor tissue specimens suggest significant relationships to clinical variables in a broad spectrum of tumors other than those revealed by GEPIA2 analysis, including BRCA, ESCA, COAD, MESO, OV, READ, STAD, UCEC, etc. {reviewed in (Pastorek and Pastorekova 2015; Pastorekova and Gillies 2019)}. Thorough meta-analysis of the results of 147 publicly available clinical studies (excluding KIRC) reveals a strong significant association between CA IX expression and all survival endpoints: overall, disease-free, loco-regional, disease-specific, metastasis-free, and progression-free survival (van Kuijk et al. 2016). Absence of correlations of TGCT/GTEx CA9 data to survival in these additional cancer types can be explained by a regionally and temporally heterogeneous, hypoxia-induced CA9 gene expression pattern, as well as by relatively short half-time of the CA9 transcripts contrasting with very long half-time of the CA IX protein (Rafajova et al. 2004). Moreover, the CA IX ectodomain can be cleaved by metalloproteinases and released to extracellular space and body fluid, which can affect the CA IX protein expression in tumors (Zatovicova et al. 2005).
In addition to the value of CA9 gene and/or CA IX protein expression as a surrogate marker of tumor hypoxia {which is the topic of extensive translational research, e.g., for bioimaging (Tafreshi et al. 2012)}, a lot of attention has been paid to the role of CA IX protein in tumor biology. Numerous studies employing genetic manipulation or pharmacologic inhibition of CA IX show that it is a key component of the pH-regulating machinery that enables tumor cells to survive hostile hypoxic and acidotic conditions in the tumor microenvironment and to gain invasiveness and metastatic propensity (Svastova et al. 2004, 2012; Swietach et al. 2009; Chiche et al. 2009; Radvak et al. 2013; Csaderova et al. 2013; Chafe et al. 2015; Swayampakula et al. 2017; Lee et al. 2018; Debreova et al. 2019). In addition, CA IX was found to contribute to stemness and chemoresistance (Ledaki et al. 2015; Vidlickova et al. 2016; Gibadulinova et al. 2020). Interestingly, CA IX has been associated with invasive phenotype pioneering in a novel and sometimes hostile environment (Lloyd et al. 2016). Moreover, CA IX interacts or communicates with a number of regulatory proteins (besides ion transporters mentioned above), which are involved in important cellular processes, such as β-catenin, α2β1 integrin, CD98hc, MMP14, and other actin-regulatory proteins, hERG1 potassium channel, NCX1 sodium/calcium exchanger, PKA, PI3 kinase, etc. (Svastova et al. 2003; Swayampakula et al. 2017; Debreova et al. 2019; Lastraioli et al. 2019; Liskova et al. 2019; Ditte et al. 2011; Dorai et al. 2005). These interactions can have profound effects on enzymatic and signaling functions of CA IX and also on its accessibility to antibodies and small molecules.
The expression pattern in a broad range of tumors as well as functional involvement of CA IX in tumor biology have raised enormous interest in the development of CA IX-targeting drugs, including inhibitors and specific monoclonal antibodies for immunotherapeutic applications {reviewed in (Neri and Supuran 2011; Oosterwijk-Wakka et al. 2013; Pastorek and Pastorekova 2015; Singh et al. 2018; Pastorekova and Gillies 2019)}. These efforts are continuously ongoing and novel combination strategies are being investigated (McIntyre et al. 2012; Dubois et al. 2013; Boyd et al. 2017; Chafe et al. 2019).
CA12 expression evaluated by GEPIA2 is present in several tumor types including KICH, KIRC, BRCA, and HNSC, but also in the normal counterparts of KIRC, KIRP, KICH, SARC, and SKCM (Figs. 7.3 and 7.5). The highest level is detected in KICH (TPM 572), but the relationship of a high CA12 level to better overall survival is significant only in SKCM (both for sum of all three protein-coding transcripts and solely for CA12-001 transcript) (Fig. 7.6). In KIRC, only the CA12-002 transcript relates to better overall survival while the other transcript variants do not show any significant correlations. Based on the available experimental data, CA12 expression is regulated by hypoxia, albeit to a lower magnitude than CA9, and a direct role of the HIF transcription factor in its regulation has not been clearly defined (Wykoff et al. 2000). However, CA12 expression appears to be driven by differentiation factors at least in some tumor types (Barnett et al. 2008; Waheed and Sly 2017; Franke et al. 2019). In invasive BRCA, CA XII protein expression assessed by IHC is associated with a lower grade, a lower relapse rate, and a better overall survival (Wykoff et al. 2001; Watson et al. 2003; Li et al. 2019). The link between CA XII expression and better prognosis was demonstrated also in cervical cancer {in association with superior disease-free survival, (Yoo et al. 2010) and resectable NSCLC (Ilie et al. 2011)}. In contrast, IHC staining for CA XII increases with increasing grade of colorectal tumors (Kivela et al. 2000), high CA XII expression is linked to lower survival rate of patients with esophageal squamous cell carcinoma (Ochi et al. 2015). Interestingly, a shorter, alternatively spliced CA12 transcript has been associated with poor prognosis of patients with diffusely infiltrating astrocytic gliomas (Haapasalo et al. 2008). In cell culture models with genetically manipulated expression, CA XII was shown to participate in pH regulation, tumor growth, and chemoresistance (Chiche et al. 2009; Kopecka et al. 2015). Based on these relationships, also CA XII is investigated as therapy target for CA inhibitors and monoclonal antibodies (Gondi et al. 2013; Kopecka et al. 2015; Boyd et al. 2017).
Expression of CA14 has not been experimentally linked to cancer so far, but GEPIA2 analysis disclosed its increased levels in GBM, LGG, and SKCM (TPM up to 34.3), Figs. 7.3 and 7.5. Even more interestingly, higher CA14 expression (for all transcript variants together and also separately for CA14-001 and CA14-005) shows a significant correlation to worse overall survival of patients with LGG and SKCM, but to better prognosis in case of GBM (Figs. 7.6 and 7.7). This is an unexpected finding that does not have any supporting clinical data in the published literature. Additional links of CA XIV to cancer coming from other publicly available databases are described by Mboge and colleagues (Mboge et al. 2018), but those data also require experimental/clinical validation.
7.5 Expression of Inactive CA Isoforms in Cancer
Three genes coding for inactive CA isoforms, namely, CA8, CA10, and CA11 contain mutations affecting the critical residues involved in the coordination of a zinc ion, which is crucial for the CA catalytic activity. This precludes participation of these isoforms in the canonic pH regulation-related metabolons involving bicarbonate transporters and sodium/proton exchangers. However, the non-catalytic cooperation with MCTs can still be feasible and was recently demonstrated for all three proteins CA VIII, CA X, and CA XI (Aspatwar et al. 2019). The authors suggest that the inactive CAs may function as a proton antenna for MCT1, to drive proton-coupled lactate transport across cell membranes.
GEPIA2 shows that the CA8 gene is differentially transcribed particularly in LAML and TGCT (TPM up to 30.3) where it is strongly decreased in comparison to normal counterparts, but does not show any relationship to overall survival (Figs. 7.4, 7.5, 7.6). On the other hand, CA8 transcription is elevated in tumor versus normal tissues in UCEC, THYM, OV, PAAD, and PCPG (TPM up to 9.4). Out of these tumor types, only in SKCM higher CA8 relates to poor prognosis, whereas in UCEC and PAAD (with quartile cutoffs), higher CA8 expression is linked to better overall survival (Fig. 7.6). This is corresponding to the observation that CA VIII immunopositivity in 13% of astrocytomas and 9% of oligodendrogliomas is associated with more benign behavior (Karjalainen et al. 2018). However, the opposite relationship was found in colorectal and lung carcinomas (Miyaji et al. 2003; Akisawa et al. 2003), and experiments using cell models suggest that CA VIII promotes progression and invasiveness of lung cancer cells (Lu et al. 2004; Ishihara et al. 2006) and growth of colon cancer cells (Nishikata et al. 2007).
Expression of genes coding for the inactive carbonic anhydrases in tumors and normal samples. Data extracted from TCGA/GTEx databases were analyzed and converted to graphics using GEPIA2 instrument (Tang et al. 2019). Data in the graphs relate to TPM units and intensities of colors in the bodymaps correspond to Log/TPM + 1 Scores in particular tissues. Numbers above the columns highlight the highest TPMs for each CA isoform
Heatmap visualizing differential expression of the genes coding for CA isoforms in all GEPIA2-analyzed tumors compared to normal samples. Tumor/normal ratio is expressed in color scale shown on the right side, with tumor-related expression illustrated in red shades and normal sample-related expression in grey shades. Lack of expression in both tumor and normal samples is represented by white color and similar expression level in tumor versus normal samples is represented by dotted pattern. The differential expression does not automatically translate to prognostic value as specified in the body text for each CA isoform
Selection of carbonic anhydrase-related neoplasms with depicted expression levels of CA isoforms (expressed as TPM) in the particular cancer type. Graphs do not show expression of the genes in the corresponding normal samples. Data extracted from TCGA/GTEx databases were analyzed using GEPIA2 instrument (Tang et al. 2019). Stars indicate significant relationship to overall survival (* p < 0.05, **p < 0.01, ***p < 0.001) without discriminating positive and negative correlations
Selected examples of Kaplan-Meier survival plots generated by the analysis of overall survival performed by GEPIA2 instrument (Tang et al. 2019) based on expression of CA genes. GEPIA2 uses Long-rank test (Mantel-Cox test) for hypothesis test. The Cox proportional hazard ratio (HR) and the 95% confidence interval are included in the survival plot
CA10 gene expression is increased in PCPG (TPM 19.7) and decreased in GBM, LGG, TGCT, KIRC, KIRP, SARC, and KICH, with no association to overall survival, except a significant correlation of high CA10 transcription with a better overall survival in LGG patients (Figs. 7.4–7.7). The CA11 gene expression pattern (tumor vs normal) is similar to CA10 and includes increased levels in PCPG (TPM 83) and decreased levels in GBM, LGG, OV, UCEC, and UCS (Figs. 7.4,7.5). A significant relationship between high CA11 level and better overall survival was observed only in LGG (Fig. 7.6). In line with this, levels of CA X and CA XI isoforms (that are secreted synaptic proteins inhibiting the growth of glioma cell lines) were reduced in clinical glioma samples and negatively associated with high histological grade (Tao et al. 2019). In preclinical experiments of the same study, CA11 knockdown promoted cell growth, clone formation and migration, and increased tumor size in xenografted mice. In contrast, CA XI immunostaining was observed in the small fraction of the astrocytic and oligodendroglial tumor specimens, but not in the most benign pilocytic astrocytomas (Karjalainen et al. 2018). These scarce data indicate that our knowledge related to expression and functions of inactive CA isoforms is still limited.
7.6 Carbonic Anhydrase-Related Neoplasms
Taking together all the information extracted from the TCGA/GTEx datasets by the GEPIA2 tool, we can identify few tumor entities that are characterized by expression of carbonic anhydrases, which also show relationship to overall survival (Figs. 7.6 and 7.7).
GBM (Glioblastoma multiforme) is a malignant brain tumor that accounts for about 15% of all brain tumors in adults. GBM patients have a poor prognosis and survive less than 15 months following diagnosis (information extracted from The Cancer Genome Atlas (TCGA), National Cancer Institute at the National Institutes of Health, USA). Recent integrated analysis of genetic alterations in main signaling pathways in tumors suggests that the most frequent alterations in GBM affect the pathways of RTK/Ras, cell cycle, PI3K, and p53 which control cell proliferation and survival and affect diverse aspect of tumor phenotype including metabolism and adaptation to stresses (Sanchez-Vega et al. 2018; Vander Heiden et al. 2009). Moreover, GBM tumors are characterized by hypoxia and acidosis, and thus, it is not surprising that these tumors express several carbonic anhydrases (Figs. 7.5 and 7.6), of which increased expression of CA14 is linked to better prognosis, and increased expression of CA9 is linked to poor prognosis, while the other isoforms’ genes (including the highly expressed CA2) do not show any significant relationship to overall survival (Fig. 7.6).
LGG (Low grade glioma). Glioma develops in the brain’s glial cells, which support the brain’s nerve cells and keep them healthy. Tumors are classified into grades I, II, III, or IV based on standards set by the World Health Organization. Lower grade glioma consists of grades II and III. Regardless of grade, growing glioma tumor compresses the normal brain tissue, frequently leading to disabling or fatal effects (The Cancer Genome Atlas, NIH, USA). There are three molecular subtypes of LGG with distinct clinical outcomes depending on presence of mutations in IDH1/IDH2 genes and a co-deletion of a short arm of chromosome 1 and longer arm of chromosome 19. The subtype with the poorest outcomes contains wild-type IDH and appears to be a precursor of the more aggressive GBM. IDH (isocitrate dehydrogenase) is an NADP+-dependent enzyme that catalyzes a conversion of isocitrate to α-ketoglutarate and CO2. It is a tumor suppressor protein that functions at a crossroad of cellular metabolism in lipid synthesis, cellular defense against oxidative stress, oxidative respiration, and oxygen-sensing signal transduction (Reitman and Yan 2010). IDH mutations cause stabilization of HIF transcription factor and hypoxic reprogramming. Glioma development in IDH-mutant and IDH wild-type tumors is driven by different oncogenic pathways that include mainly RTK/RAS pathway (IDHwt), p53 and Hippo pathways (IDHmut), and Wnt, TGFβ, and Hippo (IDHMut-codel), as illustrated in integrative analysis performed by Sanchez-Vega and colleagues (Sanchez-Vega et al. 2018). Interestingly, compared to IDHwt gliomas, IDHmut gliomas have distinct metabolic and microenvironmental characteristics correlated with tumor acidity and hypoxia (Yao et al. 2019). This might be one of the reasons, why LGG display variable expression of CA genes, including CA2, CA4, CA5B, CA8, CA10, CA11, CA12, CA13, and CA14 (Fig. 7.5) and all of them, except CA8, are significantly associated with prognosis (Fig. 7.6). Tumor-related increases of CA4, CA10, and CA11 correlate with better overall survival, increases of CA2, CA5B, CA12, CA13, and CA14 correlate with worse prognosis. Because LGGs are strongly related with carbonic anhydrases, these neoplasms might be a suitable target for anticancer strategies exploiting CA inhibitors.
KICH (Chromophobe renal cell carcinoma) is a rare type cancer that originates in the distal regions of kidney and forms in the cells lining the small tubules, which help filter waste from the blood, making urine. It accounts for 5% of all kidney cancer cases and can have either hereditary or sporadic basis (The Cancer Genome Atlas, NIH, USA). Pathways altered in KICH relate to cell cycle, PI3K, and p53 and often translate to oncogenic metabolism and adaptation to hypoxia and acidosis (Sanchez-Vega et al. 2018; Linehan and Ricketts 2013). Only two CA isoforms show high expression in KICH and decreased expression in the corresponding normal tissues: CA2 and CA12, but none of them is significantly associated with overall survival.
KIRC (Renal cell carcinoma) is the most common type of kidney cancer that forms in the cells lining the small proximal tubules in the kidney, which filter waste from the blood and make urine. When detected early, most cases of KIRC can be treated effectively, but survival rates are low when the cancer has spread from the kidney to other parts of the body. About 92% of KIRC are clear cell carcinoma (The Cancer Genome Atlas, NIH, USA). According to analysis of Sanchez-Vega and colleagues (Sanchez-Vega et al. 2018), altered pathways include RTK/RAS, cell cycle, and PI3K, but the primary genetic event is an inactivating mutation in VHL gene, which affects cellular oxygen sensing and leads to constitutive activation of the HIF pathway even in the absence of physiologic hypoxia (Kaelin and Ratcliffe 2008; Wiesener et al. 2001). This also includes metabolic changes associated with poor outcomes. Since CA9 and CA12 are upregulated by hypoxia, it is not surprising that these are the main isoforms present in this type of tumor, accompanied by CA2. As explained above, due to a HIF-1 to HIF-2 shift in the course of cancer progression, a higher level of HIF-1-regulated CA9 is associated with a better prognosis, albeit its expression in later stages still remains relatively high and suitable for immunotherapeutic targeting (Bui et al. 2003). CA12 exhibits a similar relationship, but only when CA12-002 splicing isoform is tested, otherwise there is no clear relationship to overall survival. Similarly, increased CA2 expression is associated with a better prognosis, while CA4 shows a highly significant association with longer overall survival when absent or low (Figs. 7.6 and 7.7).
LAML (Acute myeloid leukemia) is a cancer of the blood and bone marrow, which can quickly worsen and result in death within months, when untreated. Survival decreases with older age because standard treatments are less tolerated (The Cancer Genome Atlas, NIH, USA). LAML cancers carry relatively few mutations compared to other frequently occurring solid tumors. About a half of AMLs include activation of the RTK/RAS pathway, and about one-sixth is affected in the cell cycle and Notch pathways (Sanchez-Vega et al. 2018). CA1, CA2, and CA5B are the most highly expressed CA genes in LAML, but they do not display any link to prognosis in contrast to less expressed CA3 as well as CA13, both being positively associated with poor prognosis (Figs. 7.6 and 7.7).
SKCM (Skin cutaneous melanoma) is a cancer of melanocytes, skin cells that produce melanin. Melanoma is most often discovered because it has metastasized, or spread, to another organ, such as the lymph nodes. There are four major subtypes of cutaneous melanoma: BRAF mutant (most common), RAS mutant, NF1 mutant, and Triple Wild-Type. Mutations in each of the identified driver genes, BRAF, RAS, and NF1 lead to uncontrolled cell growth (The Cancer Genome Atlas, NIH, USA). According to integrated analysis of genetic alterations in cancer, RTK/RAS and cell cycle oncogenic pathways predominate in SKCM (Sanchez-Vega et al. 2018). Based on experimental data, hypoxia and acidosis appear to be important components of melanoma progression (Moellering et al. 2008; Marino et al. 2012). Out of CA isoforms, only CA14 exhibits a tumor-related expression linked to worse prognosis in SKCM (Figs. 7.6 and 7.7). The other CAs (CA2, CA3, CA4, CA6, CA9, CA11, CA12, and CA13) are expressed at higher levels in normal tissues compared to tumors. In a survival analysis, higher levels of CA8 and CA9 predict worse overall survival, while CA12 and CA13 show opposite relationships (Fig. 7.6).
Additional CA-related tumor types can be recognized on the basis of immunohistochemical staining of human tumor tissues that were published in hundreds of studies. These include, for example, breast carcinomas that express CA IX as a poor prognostic factor and CA XII as a good prognostic factor; colorectal carcinomas that express CA I, CA II, CA VII, CA XIII (all decreasing as cancer progresses), CA VIII, CA IX, CA XII (increasing with cancer progression); lung carcinomas (mainly NSCLC) expressing CA I and CA IV as good prognostic factors, CA VIII and CA IX as poor prognostic factors, and CA XII as a good prognostic factor (see the references related to the particular CA isoforms).
7.7 Conclusions
Both genomic metadata and immunohistochemical studies discussed here and described elsewhere in the literature indicate that there is virtually no cancer devoid of carbonic anhydrase, apparently because CA-facilitated pH regulation, ion transport, and/or CO2 diffusion are really fundamental for cell survival, proliferation, and metabolic processes in tumor tissues {see also (Mboge et al. 2018)}. Naturally, histological and mutational diversity combined with all abnormalities characteristic for malignant cancer, including oncogenic metabolism, aberrant proliferation, deregulated signaling, and adaptations to stresses in tumor microenvironment cause that the expression patterns of diverse CA isoforms in tumor tissues differ from those in normal tissues and are generally very complicated and context-dependent. This complexity of CA isoforms is further elaborated by the differences in the expression dynamics and stability of their transcripts compared to proteins, by the existence of alternative splicing variants of CA transcripts (with only some variants coding for functional proteins), by the posttranslational modifications of CA proteins affecting their functions, subcellular localizations, and regional tissue distribution, etc. Another level of complexity is added by a diversity of CA activities and interactomes. These numerous factors are behind the lack of full agreement between the data from RNA-seq, IHC/proteomic analyses, and clinical parameters. Despite this intricate picture, there are few clearly evident and undisputable relationships between certain CA isoforms and particular tumor types. Out of them, the most prominent position belongs to CA IX, which indicates an aggressive phenotype, poor prognosis, and poor response to therapy in a broad range of tumors (except renal clear cell carcinomas, as explained above), followed by CA XII (in a majority of cases indicating good prognosis), and CA IV that was proposed to be a tumor suppressor. Since these relationships are not new, the bioinformatic data included here can serve to support the existing studies.
In contrast, data mining and their integrated analysis using GEPIA2 instrument brought up very interesting and novel relationships, such as for CA XIV, which has not been previously linked to cancer through other than bioinformatic approaches (Mboge et al. 2018). In this case, but also regarding other in silico identified relationships of CAs to cancer, it is clearly evident that the RNA-seq data have to be supplemented and supported by the experimental evidence in order to obtain reliable and clinically useful information. To fulfill such requirement, the field of carbonic anhydrases requires further exploration followed by translation of knowledge to meaningful anticancer strategies.
Abbreviations
- BRAF:
-
B-Raf oncoprotein
- CA:
-
carbonic anhydrase
- HIF:
-
hypoxia-inducible factor
- IDH:
-
isocitrate dehydrogenase
- IHC:
-
immunohistochemistry
- MCT:
-
monocarboxylate transporter
- MET:
-
hepatocyte growth factor receptor
- NF1:
-
neurofibromatosis 1
- NHE:
-
sodium-proton exchanger
- NSCLC:
-
non-small cell lung carcinoma
- PG:
-
proteoglycan
- RTK:
-
receptor
tyrosine kinase
- RAS:
-
c-Ras oncoprotein
- TPM:
-
transcripts per million
- VHL:
-
von Hippel Lindau
References
Akisawa Y, Nishimori I, Taniuchi K, Okamoto N, Takeuchi T, Sonobe H, Ohtsuki Y, Onishi S (2003) Expression of carbonic anhydrase-related protein CA-RP VIII in non-small cell lung cancer. Virchows Arch: Int J Pathol 442(1):66–70. https://doi.org/10.1007/s00428-002-0721-y
Ames S, Pastorekova S, Becker HM (2018) The proteoglycan-like domain of carbonic anhydrase IX mediates non-catalytic facilitation of lactate transport in cancer cells. Oncotarget 9(46):27940–27957. https://doi.org/10.18632/oncotarget.25371
Ames S, Andring JT, McKenna R, Becker HM (2019) CAIX forms a transport metabolon with monocarboxylate transporters in human breast cancer cells. Oncogene. https://doi.org/10.1038/s41388-019-1098-6
Aspatwar A, Tolvanen MEE, Schneider HP, Becker HM, Narkilahti S, Parkkila S, Deitmer JW (2019) Catalytically inactive carbonic anhydrase-related proteins enhance transport of lactate by MCT1. FEBS Open Bio 9(7):1204–1211. https://doi.org/10.1002/2211-5463.12647
Banova Vulic R, Zduriencikova M, Tyciakova S, Benada O, Dubrovcakova M, Lakota J, Skultety L (2019) Silencing of carbonic anhydrase I enhances the malignant potential of exosomes secreted by prostatic tumour cells. J Cell Mol Med 23(5):3641–3655. https://doi.org/10.1111/jcmm.14265
Barathova M, Takacova M, Holotnakova T, Gibadulinova A, Ohradanova A, Zatovicova M, Hulikova A, Kopacek J, Parkkila S, Supuran CT, Pastorekova S, Pastorek J (2008) Alternative splicing variant of the hypoxia marker carbonic anhydrase IX expressed independently of hypoxia and tumour phenotype. Br J Cancer 98(1):129–136. https://doi.org/10.1038/sj.bjc.6604111
Barnett DH, Sheng S, Charn TH, Waheed A, Sly WS, Lin CY, Liu ET, Katzenellenbogen BS (2008) Estrogen receptor regulation of carbonic anhydrase XII through a distal enhancer in breast cancer. Can Res 68(9):3505–3515. https://doi.org/10.1158/0008-5472.CAN-07-6151
Becker HM, Hirnet D, Fecher-Trost C, Sultemeyer D, Deitmer JW (2005) Transport activity of MCT1 expressed in Xenopus oocytes is increased by interaction with carbonic anhydrase. J Biol Chem 280(48):39882–39889. https://doi.org/10.1074/jbc.M503081200
Bornstein S, Schmidt M, Choonoo G, Levin T, Gray J, Thomas CR Jr, Wong M, McWeeney S (2016) IL-10 and integrin signaling pathways are associated with head and neck cancer progression. BMC Genomics 17:38. https://doi.org/10.1186/s12864-015-2359-6
Boyd NH, Walker K, Fried J, Hackney JR, McDonald PC, Benavides GA, Spina R, Audia A, Scott SE, Libby CJ, Tran AN, Bevensee MO, Griguer C, Nozell S, Gillespie GY, Nabors B, Bhat KP, Bar EE, Darley-Usmar V, Xu B, Gordon E, Cooper SJ, Dedhar S, Hjelmeland AB (2017) Addition of carbonic anhydrase 9 inhibitor SLC-0111 to temozolomide treatment delays glioblastoma growth in vivo. JCI insight 2(24). https://doi.org/10.1172/jci.insight.92928
Bui MH, Seligson D, Han KR, Pantuck AJ, Dorey FJ, Huang Y, Horvath S, Leibovich BC, Chopra S, Liao SY, Stanbridge E, Lerman MI, Palotie A, Figlin RA, Belldegrun AS (2003) Carbonic anhydrase IX is an independent predictor of survival in advanced renal clear cell carcinoma: implications for prognosis and therapy. Clin Cancer Res: off J Am Assoc Cancer Res 9(2):802–811
Chafe SC, Lou Y, Sceneay J, Vallejo M, Hamilton MJ, McDonald PC, Bennewith KL, Moller A, Dedhar S (2015) Carbonic anhydrase IX promotes myeloid-derived suppressor cell mobilization and establishment of a metastatic niche by stimulating G-CSF production. Can Res 75(6):996–1008. https://doi.org/10.1158/0008-5472.CAN-14-3000
Chafe SC, McDonald PC, Saberi S, Nemirovsky O, Venkateswaran G, Burugu S, Gao D, Delaidelli A, Kyle AH, Baker JHE, Gillespie JA, Bashashati A, Minchinton AI, Zhou Y, Shah SP, Dedhar S (2019) Targeting hypoxia-induced carbonic anhydrase IX enhances immune-checkpoint blockade locally and systemically. Cancer Immunol Res 7(7):1064–1078. https://doi.org/10.1158/2326-6066.CIR-18-0657
Chen J, Hu L, Zhang F, Wang J, Chen J, Wang Y (2017) Downregulation of carbonic anhydrase IV contributes to promotion of cell proliferation and is associated with poor prognosis in non-small cell lung cancer. Oncol Lett 14(4):5046–5050. https://doi.org/10.3892/ol.2017.6740
Chiche J, Ilc K, Laferriere J, Trottier E, Dayan F, Mazure NM, Brahimi-Horn MC, Pouyssegur J (2009) Hypoxia-inducible carbonic anhydrase IX and XII promote tumor cell growth by counteracting acidosis through the regulation of the intracellular pH. Can Res 69(1):358–368. https://doi.org/10.1158/0008-5472.CAN-08-2470
Corbet C, Feron O (2017) Tumour acidosis: from the passenger to the driver’s seat. Nat Rev Cancer 17(10):577–593. https://doi.org/10.1038/nrc.2017.77
Csaderova L, Debreova M, Radvak P, Stano M, Vrestiakova M, Kopacek J, Pastorekova S, Svastova E (2013) The effect of carbonic anhydrase IX on focal contacts during cell spreading and migration. Front Physiol 4:271. https://doi.org/10.3389/fphys.2013.00271
Dai HY, Hong CC, Liang SC, Yan MD, Lai GM, Cheng AL, Chuang SE (2008) Carbonic anhydrase III promotes transformation and invasion capability in hepatoma cells through FAK signaling pathway. Mol Carcinog 47(12):956–963. https://doi.org/10.1002/mc.20448
Davidov T, Nagar M, Kierson M, Chekmareva M, Chen C, Lu SE, Lin Y, Chernyavsky V, Potdevin L, Arumugam D, Barnard N, Trooskin S (2014) Carbonic anhydrase 4 and crystallin alpha-B immunoreactivity may distinguish benign from malignant thyroid nodules in patients with indeterminate thyroid cytology. J Surg Res 190(2):565–574. https://doi.org/10.1016/j.jss.2014.03.042
Debreova M, Csaderova L, Burikova M, Lukacikova L, Kajanova I, Sedlakova O, Kery M, Kopacek J, Zatovicova M, Bizik J, Pastorekova S, Svastova E (2019) CAIX regulates invadopodia formation through both a pH-dependent mechanism and interplay with actin regulatory proteins. Int J Mol Sci 20(11). https://doi.org/10.3390/ijms20112745
Ditte P, Dequiedt F, Svastova E, Hulikova A, Ohradanova-Repic A, Zatovicova M, Csaderova L, Kopacek J, Supuran CT, Pastorekova S, Pastorek J (2011) Phosphorylation of carbonic anhydrase IX controls its ability to mediate extracellular acidification in hypoxic tumors. Can Res 71(24):7558–7567. https://doi.org/10.1158/0008-5472.CAN-11-2520
Dorai T, Sawczuk IS, Pastorek J, Wiernik PH, Dutcher JP (2005) The role of carbonic anhydrase IX overexpression in kidney cancer. Eur J Cancer 41(18):2935–2947. https://doi.org/10.1016/j.ejca.2005.09.011
Dubois L, Peeters SG, van Kuijk SJ, Yaromina A, Lieuwes NG, Saraya R, Biemans R, Rami M, Parvathaneni NK, Vullo D, Vooijs M, Supuran CT, Winum JY, Lambin P (2013) Targeting carbonic anhydrase IX by nitroimidazole based sulfamides enhances the therapeutic effect of tumor irradiation: a new concept of dual targeting drugs. Radiother Oncol: J Eur Soc Ther Radiol Oncol 108(3):523–528. https://doi.org/10.1016/j.radonc.2013.06.018
Forero-Quintero LS, Ames S, Schneider HP, Thyssen A, Boone CD, Andring JT, McKenna R, Casey JR, Deitmer JW, Becker HM (2019) Membrane-anchored carbonic anhydrase IV interacts with monocarboxylate transporters via their chaperones CD147 and GP70. J Biol Chem 294(2):593–607. https://doi.org/10.1074/jbc.RA118.005536
Franke CM, Gu VW, Grimm BG, Cassady VC, White JR, Weigel RJ, Kulak MV (2019) TFAP2C regulates carbonic anhydrase XII in human breast cancer. Oncogene. https://doi.org/10.1038/s41388-019-1062-5
Gibadulinova A, Bullova P, Strnad H, Pohlodek K, Jurkovicova D, Takacova M, Pastorekova S, Svastova E (2020) CAIX-mediated control of LIN28/let-7 axis contributes to metabolic adaptation of breast cancer cells to hypoxia. Int J Mol Sci 21(12):4299. https://doi.org/10.3390/ijms21124299
Gillies RJ, Brown JS, Anderson ARA, Gatenby RA (2018) Eco-evolutionary causes and consequences of temporal changes in intratumoural blood flow. Nat Rev Cancer 18(9):576–585. https://doi.org/10.1038/s41568-018-0030-7
Gondi G, Mysliwietz J, Hulikova A, Jen JP, Swietach P, Kremmer E, Zeidler R (2013) Antitumor efficacy of a monoclonal antibody that inhibits the activity of cancer-associated carbonic anhydrase XII. Can Res 73(21):6494–6503. https://doi.org/10.1158/0008-5472.CAN-13-1110
Haapasalo J, Hilvo M, Nordfors K, Haapasalo H, Parkkila S, Hyrskyluoto A, Rantala I, Waheed A, Sly WS, Pastorekova S, Pastorek J, Parkkila AK (2008) Identification of an alternatively spliced isoform of carbonic anhydrase XII in diffusely infiltrating astrocytic gliomas. Neuro Oncol 10(2):131–138. https://doi.org/10.1215/15228517-2007-065
Hsieh MS, Jeng YM, Jhuang YL, Chou YH, Lin CY (2016) Carbonic anhydrase VI: a novel marker for salivary serous acinar differentiation and its application to discriminate acinic cell carcinoma from mammary analogue secretory carcinoma of the salivary gland. Histopathology 68(5):641–647. https://doi.org/10.1111/his.12792
Hulikova A, Aveyard N, Harris AL, Vaughan-Jones RD, Swietach P (2014) Intracellular carbonic anhydrase activity sensitizes cancer cell pH signaling to dynamic changes in CO2 partial pressure. J Biol Chem 289(37):25418–25430. https://doi.org/10.1074/jbc.M114.547844
Hynninen P, Parkkila S, Huhtala H, Pastorekova S, Pastorek J, Waheed A, Sly WS, Tomas E (2012) Carbonic anhydrase isozymes II, IX, and XII in uterine tumors. APMIS: Acta Pathol, Microbiol, Et Immunol Scand 120(2):117–129. https://doi.org/10.1111/j.1600-0463.2011.02820.x
Ilie MI, Hofman V, Ortholan C, Ammadi RE, Bonnetaud C, Havet K, Venissac N, Mouroux J, Mazure NM, Pouyssegur J, Hofman P (2011) Overexpression of carbonic anhydrase XII in tissues from resectable non-small cell lung cancers is a biomarker of good prognosis. Int J Cancer 128(7):1614–1623. https://doi.org/10.1002/ijc.25491
Ishihara T, Takeuchi T, Nishimori I, Adachi Y, Minakuchi T, Fujita J, Sonobe H, Ohtsuki Y, Onishi S (2006) Carbonic anhydrase-related protein VIII increases invasiveness of non-small cell lung adenocarcinoma. Virchows Arch: Int J Pathol 448(6):830–837. https://doi.org/10.1007/s00428-006-0199-0
Ivanov SV, Kuzmin I, Wei MH, Pack S, Geil L, Johnson BE, Stanbridge EJ, Lerman MI (1998) Down-regulation of transmembrane carbonic anhydrases in renal cell carcinoma cell lines by wild-type von Hippel-Lindau transgenes. Proc Natl Acad Sci USA 95(21):12596–12601. https://doi.org/10.1073/pnas.95.21.12596
Jamali S, Klier M, Ames S, Barros LF, McKenna R, Deitmer JW, Becker HM (2015) Hypoxia-induced carbonic anhydrase IX facilitates lactate flux in human breast cancer cells by non-catalytic function. Sci Rep 5:13605. https://doi.org/10.1038/srep13605
Jankovicova B, Skultety L, Dubrovcakova M, Stern M, Bilkova Z, Lakota J (2013) Overlap of epitopes recognized by anti-carbonic anhydrase I IgG in patients with malignancy-related aplastic anemia-like syndrome and in patients with aplastic anemia. Immunol Lett 153(1–2):47–49. https://doi.org/10.1016/j.imlet.2013.07.006
Kaelin WG Jr, Ratcliffe PJ (2008) Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell 30(4):393–402. https://doi.org/10.1016/j.molcel.2008.04.009
Karjalainen SL, Haapasalo HK, Aspatwar A, Barker H, Parkkila S, Haapasalo JA (2018) Carbonic anhydrase related protein expression in astrocytomas and oligodendroglial tumors. BMC Cancer 18(1):584. https://doi.org/10.1186/s12885-018-4493-4
Kivela A, Parkkila S, Saarnio J, Karttunen TJ, Kivela J, Parkkila AK, Waheed A, Sly WS, Grubb JH, Shah G, Tureci O, Rajaniemi H (2000) Expression of a novel transmembrane carbonic anhydrase isozyme XII in normal human gut and colorectal tumors. Am J Pathol 156(2):577–584. https://doi.org/10.1016/S0002-9440(10)64762-1
Kivela AJ, Saarnio J, Karttunen TJ, Kivela J, Parkkila AK, Pastorekova S, Pastorek J, Waheed A, Sly WS, Parkkila TS, Rajaniemi H (2001) Differential expression of cytoplasmic carbonic anhydrases, CA I and II, and membrane-associated isozymes, CA IX and XII, in normal mucosa of large intestine and in colorectal tumors. Dig Dis Sci 46(10):2179–2186. https://doi.org/10.1023/a:1011910931210
Klier M, Andes FT, Deitmer JW, Becker HM (2014) Intracellular and extracellular carbonic anhydrases cooperate non-enzymatically to enhance activity of monocarboxylate transporters. J Biol Chem 289(5):2765–2775. https://doi.org/10.1074/jbc.M113.537043
Kopecka J, Campia I, Jacobs A, Frei AP, Ghigo D, Wollscheid B, Riganti C (2015) Carbonic anhydrase XII is a new therapeutic target to overcome chemoresistance in cancer cells. Oncotarget 6(9):6776–6793. https://doi.org/10.18632/oncotarget.2882
Korhonen K, Parkkila AK, Helen P, Valimaki R, Pastorekova S, Pastorek J, Parkkila S, Haapasalo H (2009) Carbonic anhydrases in meningiomas: association of endothelial carbonic anhydrase II with aggressive tumor features. J Neurosurg 111(3):472–477. https://doi.org/10.3171/2008.10.17672
Kummola L, Hamalainen JM, Kivela J, Kivela AJ, Saarnio J, Karttunen T, Parkkila S (2005) Expression of a novel carbonic anhydrase, CA XIII, in normal and neoplastic colorectal mucosa. BMC Cancer 5:41. https://doi.org/10.1186/1471-2407-5-41
Kuo WH, Chiang WL, Yang SF, Yeh KT, Yeh CM, Hsieh YS, Chu SC (2003) The differential expression of cytosolic carbonic anhydrase in human hepatocellular carcinoma. Life Sci 73(17):2211–2223. https://doi.org/10.1016/s0024-3205(03)00597-6
Langella E, Buonanno M, Vullo D, Dathan N, Leone M, Supuran CT, De Simone G, Monti SM (2018) Biochemical, biophysical and molecular dynamics studies on the proteoglycan-like domain of carbonic anhydrase IX. Cell Mol Life Sci: CMLS 75(17):3283–3296. https://doi.org/10.1007/s00018-018-2798-8
Lastraioli E, Pillozzi S, Mari A, Tellini R, Duranti C, Baldazzi V, Venturini S, Minervini A, Lapini A, Nesi G, Carini M, Arcangeli A (2019) hERG1 and CA IX expression are associated with disease recurrence in surgically resected clear cell renal carcinoma. Eur J Surg Oncol. https://doi.org/10.1016/j.ejso.2019.10.031
Ledaki I, McIntyre A, Wigfield S, Buffa F, McGowan S, Baban D, Li JL, Harris AL (2015) Carbonic anhydrase IX induction defines a heterogeneous cancer cell response to hypoxia and mediates stem cell-like properties and sensitivity to HDAC inhibition. Oncotarget 6(23):19413–19427. https://doi.org/10.18632/oncotarget.4989
Lee SH, McIntyre D, Honess D, Hulikova A, Pacheco-Torres J, Cerdan S, Swietach P, Harris AL, Griffiths JR (2018) Carbonic anhydrase IX is a pH-stat that sets an acidic tumour extracellular pH in vivo. Br J Cancer 119(5):622–630. https://doi.org/10.1038/s41416-018-0216-5
Leppilampi M, Koistinen P, Savolainen ER, Hannuksela J, Parkkila AK, Niemela O, Pastorekova S, Pastorek J, Waheed A, Sly WS, Parkkila S, Rajaniemi H (2002) The expression of carbonic anhydrase II in hematological malignancies. Clin Cancer Res: off J Am Assoc Cancer Res 8(7):2240–2245
Li X, Alvarez B, Casey JR, Reithmeier RA, Fliegel L (2002) Carbonic anhydrase II binds to and enhances activity of the Na+/H+ exchanger. J Biol Chem 277(39):36085–36091. https://doi.org/10.1074/jbc.M111952200
Linehan WM, Ricketts CJ (2013) The metabolic basis of kidney cancer. Semin Cancer Biol 23(1):46–55. https://doi.org/10.1016/j.semcancer.2012.06.002
Liskova V, Hudecova S, Lencesova L, Iuliano F, Sirova M, Ondrias K, Pastorekova S, Krizanova O (2019) Type 1 sodium calcium exchanger forms a complex with carbonic anhydrase IX and via reverse mode activity contributes to pH control in hypoxic tumors. Cancers 11(8). https://doi.org/10.3390/cancers11081139
Liu LC, Xu WT, Wu X, Zhao P, Lv YL, Chen L (2013) Overexpression of carbonic anhydrase II and Ki-67 proteins in prognosis of gastrointestinal stromal tumors. World J Gastroenterol 19(16):2473–2480. https://doi.org/10.3748/wjg.v19.i16.2473
Li Y, Lei B, Zou J, Wang W, Chen A, Zhang J, Fu Y, Li Z (2019) High expression of carbonic anhydrase 12 (CA12) is associated with good prognosis in breast cancer. Neoplasma 66(3):420–426. https://doi.org/10.4149/neo_2018_180819N624
Lloyd MC, Cunningham JJ, Bui MM, Gillies RJ, Brown JS, Gatenby RA (2016) Darwinian dynamics of intratumoral heterogeneity: not solely random mutations but also variable environmental selection forces. Can Res 76(11):3136–3144. https://doi.org/10.1158/0008-5472.CAN-15-2962
Lu SH, Takeuchi T, Fujita J, Ishida T, Akisawa Y, Nishimori I, Kohsaki T, Onishi S, Sonobe H, Ohtsuki Y (2004) Effect of carbonic anhydrase-related protein VIII expression on lung adenocarcinoma cell growth. Lung Cancer 44(3):273–280. https://doi.org/10.1016/j.lungcan.2003.12.011
Marino ML, Pellegrini P, Di Lernia G, Djavaheri-Mergny M, Brnjic S, Zhang X, Hagg M, Linder S, Fais S, Codogno P, De Milito A (2012) Autophagy is a protective mechanism for human melanoma cells under acidic stress. J Biol Chem 287(36):30664–30676. https://doi.org/10.1074/jbc.M112.339127
Mboge MY, Mahon BP, McKenna R, Frost SC (2018) Carbonic anhydrases: role in pH control and cancer. Metabolites 8(1). https://doi.org/10.3390/metabo8010019
McIntyre A, Patiar S, Wigfield S, Li JL, Ledaki I, Turley H, Leek R, Snell C, Gatter K, Sly WS, Vaughan-Jones RD, Swietach P, Harris AL (2012) Carbonic anhydrase IX promotes tumor growth and necrosis in vivo and inhibition enhances anti-VEGF therapy. Clin Cancer Res: off J Am Assoc Cancer Res 18(11):3100–3111. https://doi.org/10.1158/1078-0432.CCR-11-1877
Mentese A, Fidan E, Alver A, Demir S, Yaman SO, Sumer A, Fidan S, Kavgaci H, Turan I (2017) Detection of autoantibodies against carbonic anhydrase I and II in the plasma of patients with gastric cancer. Cent-Eur J Immunol 42(1):73–77. https://doi.org/10.5114/ceji.2017.67320
Mentese A, Erkut N, Demir S, Yaman SO, Sumer A, Erdem M, Alver A, Sonmez MG (2018) Serum carbonic anhydrase I and II autoantibodies in patients with chronic lymphocytic leukaemia. Cent-Eur J Immunol 43(3):276–280. https://doi.org/10.5114/ceji.2018.80046
Miyaji E, Nishimori I, Taniuchi K, Takeuchi T, Ohtsuki Y, Onishi S (2003) Overexpression of carbonic anhydrase-related protein VIII in human colorectal cancer. J Pathol 201(1):37–45. https://doi.org/10.1002/path.1404
Moellering RE, Black KC, Krishnamurty C, Baggett BK, Stafford P, Rain M, Gatenby RA, Gillies RJ (2008) Acid treatment of melanoma cells selects for invasive phenotypes. Clin Exp Metas 25(4):411–425. https://doi.org/10.1007/s10585-008-9145-7
Morgan PE, Pastorekova S, Stuart-Tilley AK, Alper SL, Casey JR (2007) Interactions of transmembrane carbonic anhydrase, CAIX, with bicarbonate transporters. Am J Physiol Cell Physiol 293(2):C738-748. https://doi.org/10.1152/ajpcell.00157.2007
Neri D, Supuran CT (2011) Interfering with pH regulation in tumours as a therapeutic strategy. Nat Rev Drug Discov 10(10):767–777. https://doi.org/10.1038/nrd3554
Nishikata M, Nishimori I, Taniuchi K, Takeuchi T, Minakuchi T, Kohsaki T, Adachi Y, Ohtsuki Y, Onishi S (2007) Carbonic anhydrase-related protein VIII promotes colon cancer cell growth. Mol Carcinog 46(3):208–214. https://doi.org/10.1002/mc.20264
Noor SI, Jamali S, Ames S, Langer S, Deitmer JW, Becker HM (2018) A surface proton antenna in carbonic anhydrase II supports lactate transport in cancer cells. eLife 7. https://doi.org/10.7554/eLife.35176
Nordfors K, Haapasalo J, Korja M, Niemela A, Laine J, Parkkila AK, Pastorekova S, Pastorek J, Waheed A, Sly WS, Parkkila S, Haapasalo H (2010) The tumour-associated carbonic anhydrases CA II, CA IX and CA XII in a group of medulloblastomas and supratentorial primitive neuroectodermal tumours: an association of CA IX with poor prognosis. BMC Cancer 10:148. https://doi.org/10.1186/1471-2407-10-148
Nortunen M, Huhta H, Helminen O, Parkkila S, Kauppila JH, Karttunen TJ, Saarnio J (2018) Carbonic anhydrases II, IX, and XII in Barrett’s esophagus and adenocarcinoma. Virchows Arch: Int J Pathol 473(5):567–575. https://doi.org/10.1007/s00428-018-2424-z
Ochi F, Shiozaki A, Ichikawa D, Fujiwara H, Nakashima S, Takemoto K, Kosuga T, Konishi H, Komatsu S, Okamoto K, Kishimoto M, Marunaka Y, Otsuji E (2015) Carbonic anhydrase XII as an independent prognostic factor in advanced esophageal squamous cell carcinoma. J Cancer 6(10):922–929. https://doi.org/10.7150/jca.11269
Oosterwijk-Wakka JC, Boerman OC, Mulders PF, Oosterwijk E (2013) Application of monoclonal antibody G250 recognizing carbonic anhydrase IX in renal cell carcinoma. Int J Mol Sci 14(6):11402–11423. https://doi.org/10.3390/ijms140611402
Opavsky R, Pastorekova S, Zelnik V, Gibadulinova A, Stanbridge EJ, Zavada J, Kettmann R, Pastorek J (1996) Human MN/CA9 gene, a novel member of the carbonic anhydrase family: structure and exon to protein domain relationships. Genomics 33(3):480–487. https://doi.org/10.1006/geno.1996.0223
Orlowski A, De Giusti VC, Morgan PE, Aiello EA, Alvarez BV (2012) Binding of carbonic anhydrase IX to extracellular loop 4 of the NBCe1 Na+/HCO3- cotransporter enhances NBCe1-mediated HCO3- influx in the rat heart. Am J Physiol Cell Physiol 303(1):C69-80. https://doi.org/10.1152/ajpcell.00431.2011
Parkkila AK, Herva R, Parkkila S, Rajaniemi H (1995a) Immunohistochemical demonstration of human carbonic anhydrase isoenzyme II in brain tumours. Histochem J 27(12):974–982
Parkkila S, Parkkila AK, Juvonen T, Lehto VP, Rajaniemi H (1995b) Immunohistochemical demonstration of the carbonic anhydrase isoenzymes I and II in pancreatic tumours. Histochem J 27(2):133–138. https://doi.org/10.1007/bf00243908
Parkkila S, Lasota J, Fletcher JA, Ou WB, Kivela AJ, Nuorva K, Parkkila AK, Ollikainen J, Sly WS, Waheed A, Pastorekova S, Pastorek J, Isola J, Miettinen M (2010) Carbonic anhydrase II: a novel biomarker for gastrointestinal stromal tumors. Mod Pathol: Off J U S Can Acad Pathol, Inc 23(5):743–750. https://doi.org/10.1038/modpathol.2009.189
Pastorekova S, Gillies RJ (2019) The role of carbonic anhydrase IX in cancer development: links to hypoxia, acidosis, and beyond. Cancer Metastasis Rev 38(1–2):65–77. https://doi.org/10.1007/s10555-019-09799-0
Pastorekova S, Parkkila S, Parkkila AK, Opavsky R, Zelnik V, Saarnio J, Pastorek J (1997) Carbonic anhydrase IX, MN/CA IX: analysis of stomach complementary DNA sequence and expression in human and rat alimentary tracts. Gastroenterology 112(2):398–408. https://doi.org/10.1053/gast.1997.v112.pm9024293
Pastorekova S, Parkkila S, Pastorek J, Supuran CT (2004) Carbonic anhydrases: current state of the art, therapeutic applications and future prospects. J Enzyme Inhib Med Chem 19(3):199–229. https://doi.org/10.1080/14756360410001689540
Pastorek J, Pastorekova S (2015) Hypoxia-induced carbonic anhydrase IX as a target for cancer therapy: from biology to clinical use. Semin Cancer Biol 31:52–64. https://doi.org/10.1016/j.semcancer.2014.08.002
Pastorek J, Pastorekova S, Callebaut I, Mornon JP, Zelnik V, Opavsky R, Zat’ovicova M, Liao S, Portetelle D, Stanbridge EJ et al (1994) Cloning and characterization of MN, a human tumor-associated protein with a domain homologous to carbonic anhydrase and a putative helix-loop-helix DNA binding segment. Oncogene 9(10):2877–2888
Radvak P, Repic M, Svastova E, Takacova M, Csaderova L, Strnad H, Pastorek J, Pastorekova S, Kopacek J (2013) Suppression of carbonic anhydrase IX leads to aberrant focal adhesion and decreased invasion of tumor cells. Oncol Rep 29(3):1147–1153. https://doi.org/10.3892/or.2013.2226
Rafajova M, Zatovicova M, Kettmann R, Pastorek J, Pastorekova S (2004) Induction by hypoxia combined with low glucose or low bicarbonate and high posttranslational stability upon reoxygenation contribute to carbonic anhydrase IX expression in cancer cells. Int J Oncol 24(4):995–1004
Ratcliffe PJ (2013) Oxygen sensing and hypoxia signalling pathways in animals: the implications of physiology for cancer. J Physiol 591(8):2027–2042. https://doi.org/10.1113/jphysiol.2013.251470
Raval RR, Lau KW, Tran MG, Sowter HM, Mandriota SJ, Li JL, Pugh CW, Maxwell PH, Harris AL, Ratcliffe PJ (2005) Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol Cell Biol 25(13):5675–5686. https://doi.org/10.1128/MCB.25.13.5675-5686.2005
Reitman ZJ, Yan H (2010) Isocitrate dehydrogenase 1 and 2 mutations in cancer: alterations at a crossroads of cellular metabolism. J Natl Cancer Inst 102(13):932–941. https://doi.org/10.1093/jnci/djq187
Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, Dimitriadoy S, Liu DL, Kantheti HS, Saghafinia S, Chakravarty D, Daian F, Gao Q, Bailey MH, Liang WW, Foltz SM, Shmulevich I, Ding L, Heins Z, Ochoa A, Gross B, Gao J, Zhang H, Kundra R, Kandoth C, Bahceci I, Dervishi L, Dogrusoz U, Zhou W, Shen H, Laird PW, Way GP, Greene CS, Liang H, Xiao Y, Wang C, Iavarone A, Berger AH, Bivona TG, Lazar AJ, Hammer GD, Giordano T, Kwong LN, McArthur G, Huang C, Tward AD, Frederick MJ, McCormick F, Meyerson M, Cancer Genome Atlas Research N, Van Allen EM, Cherniack AD, Ciriello G, Sander C, Schultz N (2018) Oncogenic signaling pathways in the cancer genome atlas. Cell 173(2):321–337 e310. https://doi.org/10.1016/j.cell.2018.03.035
Semenza GL (2012) Hypoxia-inducible factors: mediators of cancer progression and targets for cancer therapy. Trends Pharmacol Sci 33(4):207–214. https://doi.org/10.1016/j.tips.2012.01.005
Shah GN, Rubbelke TS, Hendin J, Nguyen H, Waheed A, Shoemaker JD, Sly WS (2013) Targeted mutagenesis of mitochondrial carbonic anhydrases VA and VB implicates both enzymes in ammonia detoxification and glucose metabolism. Proc Natl Acad Sci USA 110(18):7423–7428. https://doi.org/10.1073/pnas.1305805110
Singh S, Lomelino CL, Mboge MY, Frost SC, McKenna R (2018) Cancer drug development of carbonic anhydrase inhibitors beyond the active site. Molecules 23(5). https://doi.org/10.3390/molecules23051045
Skultety L, Jankovicova B, Svobodova Z, Mader P, Rezacova P, Dubrovcakova M, Lakota J, Bilkova Z (2010) Identification of carbonic anhydrase I immunodominant epitopes recognized by specific autoantibodies which indicate an improved prognosis in patients with malignancy after autologous stem cell transplantation. J Proteome Res 9(10):5171–5179. https://doi.org/10.1021/pr1004778
Sterling D, Reithmeier RA, Casey JR (2001) A transport metabolon: functional interaction of carbonic anhydrase II and chloride/bicarbonate exchangers. J Biol Chem 276 (51):47886–47894. https://doi.org/10.1074/jbc.M105959200
Stillebroer AB, Mulders PF, Boerman OC, Oyen WJ, Oosterwijk E (2010) Carbonic anhydrase IX in renal cell carcinoma: implications for prognosis, diagnosis, and therapy. Eur Urol 58(1):75–83. https://doi.org/10.1016/j.eururo.2010.03.015
Supuran CT (2016) Structure and function of carbonic anhydrases. Biochem J 473(14):2023–2032. https://doi.org/10.1042/BCJ20160115
Svastova E, Zilka N, Zat’ovicova M, Gibadulinova A, Ciampor F, Pastorek J, Pastorekova S (2003) Carbonic anhydrase IX reduces E-cadherin-mediated adhesion of MDCK cells via interaction with beta-catenin. Exp Cell Res 290(2):332–345. https://doi.org/10.1016/s0014-4827(03)00351-3
Svastova E, Hulikova A, Rafajova M, Zat’ovicova M, Gibadulinova A, Casini A, Cecchi A, Scozzafava A, Supuran CT, Pastorek J, Pastorekova S (2004) Hypoxia activates the capacity of tumor-associated carbonic anhydrase IX to acidify extracellular pH. FEBS Lett 577(3):439–445. https://doi.org/10.1016/j.febslet.2004.10.043
Svastova E, Witarski W, Csaderova L, Kosik I, Skvarkova L, Hulikova A, Zatovicova M, Barathova M, Kopacek J, Pastorek J, Pastorekova S (2012) Carbonic anhydrase IX interacts with bicarbonate transporters in lamellipodia and increases cell migration via its catalytic domain. J Biol Chem 287(5):3392–3402. https://doi.org/10.1074/jbc.M111.286062
Swayampakula M, McDonald PC, Vallejo M, Coyaud E, Chafe SC, Westerback A, Venkateswaran G, Shankar J, Gao G, Laurent EMN, Lou Y, Bennewith KL, Supuran CT, Nabi IR, Raught B, Dedhar S (2017) The interactome of metabolic enzyme carbonic anhydrase IX reveals novel roles in tumor cell migration and invadopodia/MMP14-mediated invasion. Oncogene 36(45):6244–6261. https://doi.org/10.1038/onc.2017.219
Swietach P, Patiar S, Supuran CT, Harris AL, Vaughan-Jones RD (2009) The role of carbonic anhydrase 9 in regulating extracellular and intracellular ph in three-dimensional tumor cell growths. J Biol Chem 284(30):20299–20310. https://doi.org/10.1074/jbc.M109.006478
Tachibana H, Gi M, Kato M, Yamano S, Fujioka M, Kakehashi A, Hirayama Y, Koyama Y, Tamada S, Nakatani T, Wanibuchi H (2017) Carbonic anhydrase 2 is a novel invasion-associated factor in urinary bladder cancers. Cancer Sci 108(3):331–337. https://doi.org/10.1111/cas.13143
Tafreshi NK, Bui MM, Bishop K, Lloyd MC, Enkemann SA, Lopez AS, Abrahams D, Carter BW, Vagner J, Grobmyer SR, Gillies RJ, Morse DL (2012) Noninvasive detection of breast cancer lymph node metastasis using carbonic anhydrases IX and XII targeted imaging probes. Clin Cancer Res: off J Am Assoc Cancer Res 18(1):207–219. https://doi.org/10.1158/1078-0432.CCR-11-0238
Takacova M, Holotnakova T, Barathova M, Pastorekova S, Kopacek J, Pastorek J (2010) Src induces expression of carbonic anhydrase IX via hypoxia-inducible factor 1. Oncol Rep 23(3):869–874
Takacova M, Bullova P, Simko V, Skvarkova L, Poturnajova M, Feketeova L, Babal P, Kivela AJ, Kuopio T, Kopacek J, Pastorek J, Parkkila S, Pastorekova S (2014) Expression pattern of carbonic anhydrase IX in Medullary thyroid carcinoma supports a role for RET-mediated activation of the HIF pathway. Am J Pathol 184(4):953–965. https://doi.org/10.1016/j.ajpath.2014.01.002
Takenawa J, Kaneko Y, Kishishita M, Higashitsuji H, Nishiyama H, Terachi T, Arai Y, Yoshida O, Fukumoto M, Fujita J (1998) Transcript levels of aquaporin 1 and carbonic anhydrase IV as predictive indicators for prognosis of renal cell carcinoma patients after nephrectomy. Int J Cancer 79(1):1–7. https://doi.org/10.1002/(sici)1097-0215(19980220)79:1%3c1::aid-ijc1%3e3.0.co;2-5
Tang Z, Kang B, Li C, Chen T, Zhang Z (2019) GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 47(W1):W556–W560. https://doi.org/10.1093/nar/gkz430
Tao B, Ling Y, Zhang Y, Li S, Zhou P, Wang X, Li B, Jun Z, Zhang W, Xu C, Shi J, Wang L, Zhang W, Li S (2019) CA10 and CA11 negatively regulate neuronal activity-dependent growth of gliomas. Mol Oncol 13(5):1018–1032. https://doi.org/10.1002/1878-0261.12445
Tureci O, Sahin U, Vollmar E, Siemer S, Gottert E, Seitz G, Parkkila AK, Shah GN, Grubb JH, Pfreundschuh M, Sly WS (1998) Human carbonic anhydrase XII: cDNA cloning, expression, and chromosomal localization of a carbonic anhydrase gene that is overexpressed in some renal cell cancers. Proc Natl Acad Sci USA 95(13):7608–7613. https://doi.org/10.1073/pnas.95.13.7608
Vander Heiden MG, Cantley LC, Thompson CB (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324(5930):1029–1033. https://doi.org/10.1126/science.1160809
van Kuijk SJ, Yaromina A, Houben R, Niemans R, Lambin P, Dubois LJ (2016) Prognostic Significance of Carbonic Anhydrase IX Expression in Cancer Patients: A Meta-Analysis. Front Oncol 6:69. https://doi.org/10.3389/fonc.2016.00069
Vidlickova I, Dequiedt F, Jelenska L, Sedlakova O, Pastorek M, Stuchlik S, Pastorek J, Zatovicova M, Pastorekova S (2016) Apoptosis-induced ectodomain shedding of hypoxia-regulated carbonic anhydrase IX from tumor cells: a double-edged response to chemotherapy. BMC Cancer 16:239. https://doi.org/10.1186/s12885-016-2267-4
Viikila P, Kivela AJ, Mustonen H, Koskensalo S, Waheed A, Sly WS, Pastorek J, Pastorekova S, Parkkila S, Haglund C (2016) Carbonic anhydrase enzymes II, VII, IX and XII in colorectal carcinomas. World J Gastroenterol 22(36):8168–8177. https://doi.org/10.3748/wjg.v22.i36.8168
Villafuerte FC, Swietach P, Youm JB, Ford K, Cardenas R, Supuran CT, Cobden PM, Rohling M, Vaughan-Jones RD (2014) Facilitation by intracellular carbonic anhydrase of Na+ -HCO3- co-transport but not Na+/H+ exchange activity in the mammalian ventricular myocyte. J Physiol 592(5):991–1007. https://doi.org/10.1113/jphysiol.2013.265439
Waheed A, Sly WS (2017) Carbonic anhydrase XII functions in health and disease. Gene 623:33–40. https://doi.org/10.1016/j.gene.2017.04.027
Wang N, Chen Y, Han Y, Zhao Y, Liu Y, Guo K, Jiang Y (2012) Proteomic analysis shows down-regulations of cytoplasmic carbonic anhydrases, CAI and CAII, are early events of colorectal carcinogenesis but are not correlated with lymph node metastasis. Tumori 98(6):783–791. https://doi.org/10.1700/1217.13504
Wang DB, Lu XK, Zhang X, Li ZG, Li CX (2016) Carbonic anhydrase 1 is a promising biomarker for early detection of non-small cell lung cancer. Tumour Biol: J Int Soc Oncodevelopmental Biol Med 37(1):553–559. https://doi.org/10.1007/s13277-015-3834-z
Watson PH, Chia SK, Wykoff CC, Han C, Leek RD, Sly WS, Gatter KC, Ratcliffe P, Harris AL (2003) Carbonic anhydrase XII is a marker of good prognosis in invasive breast carcinoma. Br J Cancer 88(7):1065–1070. https://doi.org/10.1038/sj.bjc.6600796
Wiesener MS, Munchenhagen PM, Berger I, Morgan NV, Roigas J, Schwiertz A, Jurgensen JS, Gruber G, Maxwell PH, Loning SA, Frei U, Maher ER, Grone HJ, Eckardt KU (2001) Constitutive activation of hypoxia-inducible genes related to overexpression of hypoxia-inducible factor-1alpha in clear cell renal carcinomas. Can Res 61(13):5215–5222
Wykoff CC, Beasley N, Watson PH, Campo L, Chia SK, English R, Pastorek J, Sly WS, Ratcliffe P, Harris AL (2001) Expression of the hypoxia-inducible and tumor-associated carbonic anhydrases in ductal carcinoma in situ of the breast. Am J Pathol 158(3):1011–1019. https://doi.org/10.1016/S0002-9440(10)64048-5
Wykoff CC, Beasley NJ, Watson PH, Turner KJ, Pastorek J, Sibtain A, Wilson GD, Turley H, Talks KL, Maxwell PH, Pugh CW, Ratcliffe PJ, Harris AL (2000) Hypoxia-inducible expression of tumor-associated carbonic anhydrases. Can Res 60(24):7075–7083
Yang GZ, Hu L, Cai J, Chen HY, Zhang Y, Feng D, Qi CY, Zhai YX, Gong H, Fu H, Cai QP, Gao CF (2015) Prognostic value of carbonic anhydrase VII expression in colorectal carcinoma. BMC Cancer 15:209. https://doi.org/10.1186/s12885-015-1216-y
Yao J, Chakhoyan A, Nathanson DA, Yong WH, Salamon N, Raymond C, Mareninov S, Lai A, Nghiemphu PL, Prins RM, Pope WB, Everson RG, Liau LM, Cloughesy TF, Ellingson BM (2019) Metabolic characterization of human IDH mutant and wild type gliomas using simultaneous pH- and oxygen-sensitive molecular MRI. Neuro Oncol. https://doi.org/10.1093/neuonc/noz078
Yoo CW, Nam BH, Kim JY, Shin HJ, Lim H, Lee S, Lee SK, Lim MC, Song YJ (2010) Carbonic anhydrase XII expression is associated with histologic grade of cervical cancer and superior radiotherapy outcome. Radiat Oncol 5:101. https://doi.org/10.1186/1748-717X-5-101
Yoshiura K, Nakaoka T, Nishishita T, Sato K, Yamamoto A, Shimada S, Saida T, Kawakami Y, Takahashi TA, Fukuda H, Imajoh-Ohmi S, Oyaizu N, Yamashita N (2005) Carbonic anhydrase II is a tumor vessel endothelium-associated antigen targeted by dendritic cell therapy. Clin Cancer Res: off J Am Assoc Cancer Res 11(22):8201–8207. https://doi.org/10.1158/1078-0432.CCR-05-0816
Zatovicova M, Sedlakova O, Svastova E, Ohradanova A, Ciampor F, Arribas J, Pastorek J, Pastorekova S (2005) Ectodomain shedding of the hypoxia-induced carbonic anhydrase IX is a metalloprotease-dependent process regulated by TACE/ADAM17. Br J Cancer 93(11):1267–1276. https://doi.org/10.1038/sj.bjc.6602861
Zavada J, Zavadova Z, Pastorek J, Biesova Z, Jezek J, Velek J (2000) Human tumour-associated protein MN/CA IX: identification of M75 epitope and of the region mediating cell adhesion. Br J Cancer 82(11):1808–1813. https://doi.org/10.1054/bjoc.2000.1111
Zhang J, Tsoi H, Li X, Wang H, Gao J, Wang K, Go MY, Ng SC, Chan FK, Sung JJ, Yu J (2016) Carbonic anhydrase IV inhibits colon cancer development by inhibiting the Wnt signalling pathway through targeting the WTAP-WT1-TBL1 axis. Gut 65(9):1482–1493. https://doi.org/10.1136/gutjnl-2014-308614
Zhang C, Wang H, Chen Z, Zhuang L, Xu L, Ning Z, Zhu Z, Wang P, Meng Z (2018) Carbonic anhydrase 2 inhibits epithelial-mesenchymal transition and metastasis in hepatocellular carcinoma. Carcinogenesis 39(4):562–570. https://doi.org/10.1093/carcin/bgx148
Zhou R, Huang W, Yao Y, Wang Y, Li Z, Shao B, Zhong J, Tang M, Liang S, Zhao X, Tong A, Yang J (2013) CA II, a potential biomarker by proteomic analysis, exerts significant inhibitory effect on the growth of colorectal cancer cells. Int J Oncol 43(2):611–621. https://doi.org/10.3892/ijo.2013.1972
Acknowledgements
The authors would like to thank Prof. Jaromir Pastorek (Biomedical Research Center of the Slovak Academy of Sciences and MABRO, a.s. Bratislava, Slovakia) for the critical reading of the manuscript. The authors would like to thank the TCGA Research Network (https://www.cancer.gov/tcga) for providing useful information. The authors’ research is supported by grants from: Slovak Research and Development Agency (APVV-19-0098), Slovak Scientific Grant Agency (VEGA 2/0076/20), European Regional Development Fund (University Science Park for Biomedicine in Bratislava, ITMS26240220087), and The George Schwab and Leona Lauder Foundation.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Takacova, M., Pastorekova, S. (2021). An Overview of Carbonic Anhydrase-Related Neoplasms. In: Chegwidden, W.R., Carter, N.D. (eds) The Carbonic Anhydrases: Current and Emerging Therapeutic Targets. Progress in Drug Research, vol 75. Springer, Cham. https://doi.org/10.1007/978-3-030-79511-5_7
Download citation
DOI: https://doi.org/10.1007/978-3-030-79511-5_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-79510-8
Online ISBN: 978-3-030-79511-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)