Abstract
In recent years, 3D segmentation of the pancreas has received a lot of attention from researchers because of its importance for clinical diagnosis and treatment. However, there are many problems with 3D pancreatic segmentation: 1) because the shape of the pancreas is not regular enough compared to other organs in the abdomen, it has a relatively small shape and there is also an excessive background of interference, resulting in inaccurate segmentation results for the pancreas; 2) one of the main drawbacks of 3D convolutional neural networks for segmentation is the excessive memory occupation, which requires censoring of the network structure to fit a given memory budget. To address the above issues, this paper proposes a new coarse-to-fine method based on convolutional neural networks (CNNs). In the first stage, the segmentation is trained to obtain candidate regions. In the second stage, the approximate location of the pancreas is obtained after the first stage, and then the pancreas is finely segmented in this approximate location. The convolutional neural network used in this paper is a modified 3DUnet network, which is improved to require less memory and higher segmentation accuracy compared to the traditional 3DUnet network. This segmentation method requires a less demanding experimental environment than other algorithms, and can also improve accuracy by eliminating a large amount of irrelevant background interference. The combination of our proposed network structure and the two-stage segmentation method achieves advanced performance.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Pancreatic cancer is a very deadly malignancy. Automatic and accurate segmentation of the pancreas is an important prerequisite for computer-assisted clinicians to diagnose pancreatic cancer. In anatomy, the pancreas is an elongated gland with a length of about 14 cm –8 cm, a width of 3 cm –9 cm, and a thickness of 1.5 cm –5 cm. In addition, the pancreas is located in the upper abdomen area and the left quarter rib area, the back of the stomach and the retrospections space is approximately equal to the first lumbar vertebral body, lying on the posterior abdominal wall. The pancreas are divided into four parts: head, neck, body, and tail, and there is no obvious boundary between them. This paper focuses on the segmentation of the pancreas in a 3D computed tomography (CT) volume, which is more difficult than the segmentation of other abdominal organs such as the liver and kidney [1] because of its small size and excessive interference background. Pancreatic dataset used in this paper is the MSD (Medical Segmentation Decathlon) pancreatic dataset. The first place winner in the MSD competition, nn-Unet [2], had an accuracy of 80% for pancreatic segmentation dice.
With the rapid development of deep learning, methods based on convolutional neural networks (CNN) have excellent performance in the field of medical image segmentation, and 3D medical image segmentation has slowly entered the public view and shown good segmentation results [3]. Compared with 2D medical image segmentation [7], 3D medical image segmentation has the advantage of better access to image contextual information, which enables tighter correlation of segmentation results and thus optimises the segmentation results. The best approach is to use 3D neural networks such as V-Net [4] and 3D U-Net [5] directly. However, one of the main difficulties encountered with 3D convolution is that it requires a large amount of memory, which prevents 3D segmentation from being studied by the general public.
To solve the problem of excessive memory required for 3D convolution, the mainstream methods are to do 3D patch [5] or pseudo-3D convolution [6]. Although 3D patch does not require high computer performance, it ignores the global information, so the final recombination segmentation result will not be too satisfactory. In the traditional pseudo-3D convolutional operation [14], it still essentially uses 2D convolutional blocks to integrate contextual information, so it is still not as powerful as 3D convolution. Although majority voting [15] can enhance the pseudo-3D contextual information from different angles through 2D segmentation, it is still not as powerful as 3D convolution.
In addition, two-stage segmentation is increasingly used in the field of 3D segmentation [14]. For example, we were inspired by the automatic pancreatic segmentation proposed by Zhao et al. [7], but we still found that Zhao did not normalise the size of the training set in his experiments, and it did not work on computers with small memory. There is also the one proposed by Yu Q et al. [14], which is more efficient but actually uses 2D convolution to solve the 3D segmentation problem, while 3D convolution is more powerful, so this method is not desirable.
To address the above problems, this paper proposes a two-stage segmentation method and an improved 3DUnet, which not only solve the problem of excessive memory consumption, but also reduces excessive background interference, and also improves the dice accuracy. In this experiment, in the first stage of segmentation in this paper, the resolution size of the image is reduced to 1/4 of the original size, and then roughly determined to the location of the pancreas, and then in the second stage, the target region is finely segmented to obtain the final segmentation result.
In this paper, we evaluate the capabilities of U-Net, one of the most popular and successful deep learning models for biomedical image segmentation, while also identifying some potential areas for improvement after a closer look at the network architecture. We propose and hypothesise that the U-Net architecture may lack certain criteria and suggest some modifications based on current developments in deep computer vision. As a result, we have developed a new model called ResConv-3DUnet, which is an enhanced version of U-Net. In this paper, experiments are conducted using a public pancreas segmentation dataset, and the experimental results show that the method proposed in this paper can effectively solve the pancreas segmentation problem.
The contributions of this paper can be summarized as follows:
-
The two-stage segmentation method proposed in this paper can greatly solve the problem of excessive memory requirement in 3D segmentation.
-
ResConv-3DUnet, which we used in the experiment, not only consumes less computational memory resources, but also has a positive effect on the segmentation of small target pancreas.
2 Method
2.1 Segmentation Method Overview
Since the pancreas is relatively small in the whole CT image, there is a large amount of irrelevant background information that affects the segmentation results, so a better approach is to roughly locate the pancreas first, then remove the irrelevant background interference, and finally perform the segmentation.
The entire pancreas segmentation process can be roughly divided into two stages: the first stage is the coarse segmentation, which consists of down-sampling the 3D volumes and then initial localisation of the pancreas; the second stage is the fine segmentation stage, in which the approximate location of the pancreas is obtained in the first stage of segmentation, then the irrelevant background of the pancreas is removed, and finally the remaining regions of interest of the pancreas is finely segmented. The segmentation result is then obtained. The neural network used in the two-stage segmentation is the ResConv-3DUnet proposed in this paper.
2.2 Pre-processing
Because of the different number of slices of the pancreatic dataset used in this study, the z-axis spacing of the datasets varied, and they ranged from 0.7 mm to 7.5 mm. As showed in Fig. 1, this is a lateral view of the z-axis of the 3D CT scans of four patients who were not pre-treated. It can be seen that without processing, they are too different for segmentation, so our treatment is to interpolate in the z-axis so that their z-axis spacing is all specified as 1 mm. Because the number of z-axis slices increases after interpolation, the memory consumption requirement for training individual patients also increases, so training cannot be done directly. This paper addresses this problem by randomly selecting 48 consecutive slices for training in each epoch training.
Before training, the CT value is intercepted to [−100, 240] through HU constraints and normalized to [−1, 1]; Ground Truth only performs [0, 1] normalized. For 3D CT scans, the number of slices is not uniform which have more or fewer slices, which means that the size is not unified, so we cannot train together.
In view of the above problems, the solutions given in this paper are as follows: 1) The z-axis interval between the CT volume and the label mask was interpolated to 1 mm, specifically by using cubic interpolations for the CT volume and the nearest interpolation for the label mask. 2) After the above interpolation process, the number of slices for a single patient was above 100 slices. In order to eliminate more slices that did not contain the pancreas, we used the label map to find slices that contained the pancreas. In order not to lose relevant information, we expanded 10 slices up and down in the pancreatic region, respectively, and then kept these slices, discarding all other CT slices, and labelled this preserved slice region as V0. 3) In the first stage of segmentation training, we interpolated the length and width of V0 to 128 × 128 and recorded it as V1. 4) Since the pancreas in the MSD pancreas dataset is less than 256 in length and 192 in width, we used labels to find the pancreas region in V0 and then expanded its length and width region outward to 256 × 192, recording this region as V2.
2.3 Training
The First Stage: Coarse Segmentation and Positioning.
The length and width of each 3D CT scans decreased from 512 × 512 to 128 × 128, and sample down to a quarter of the original size. Since the number of slices varies and the size is not uniform, it is still necessary to randomly select 48 consecutive slices for training in each period. After many times of training, we obtained the final segmentation result, then obtained the approximate location of the pancreas, and obtained the minimum enclosing rectangle of the predicted area of the pancreas. Then we multiplied the obtained enclosing rectangle by 4 to obtain the minimum enclosing rectangle of the pancreas in the original 3D CT scans size.
The Second Stage: Fine Segmentation.
In this pancreas datasets, the length of the pancreas is less than 256 and the width is less than 192. The label of the CT image was used to find the smallest external rectangular box of the pancreas, and then margins were added to extend the length and width to 256 and 192. And then each epoch training is randomly selected consecutively 48 slices, so when training after processing, the length, width and height of the data are unified to 256 × 192 × 48. Compared with the entire CT volume, the organs considered in this paper tend to occupy a relatively small area, so the final segmentation performance relies heavily on the coarse localization results. When we have this approximate region of pancreatic coordinates, we select this region and segment the pancreas only in this region. This has the advantage of not only reducing memory consumption but also removing a lot of background information that is not relevant to the pancreas.
1) The pre-processed V0 is downsampled to V1; 2) We find the rectangular box with the smallest pancreas in V0 by the label of the pancreas, then expand the edges of the rectangular box, and finally mark this rectangular box region as V2 and perform segmentation. M1 and M2 are the segmentation models obtained after two training segmentation, respectively.
Training Details.
In the first stage of training, we use the ResConv-3DUnet proposed in this paper to train V1. During training, 48 consecutive slices of the CT volume are randomly taken in each epoch, so its length, width and height are unified to 128 × 128 × 48. In this paper, the model obtained after training is denoted as M1. In the second stage of training, ResConv-3DUnet is also used to train V2, and 48 consecutive slices are randomly taken for training in each epoch, and its length, width and height are unified to 256 × 192 × 48. We denote the model obtained after training as M2. The overall flow of the segmentation is shown in Fig. 2. It can be seen that the volume of the CT image in the two experiments is not large, and it does not require much memory at all.
(a) shows the entire testing process. We took 48 consecutive slices of the CT scans and tested them in 12 steps, then fused these test results to obtain the final segmentation results. M1 and M2 are the models obtained after two stages of training, respectively. (b) shows the detailed steps of the single test: 1) CT slices were reduced to 1/4 of the original image and then roughly segmented to simply locate the pancreas; 2) the segmented feature map was enlarged by 4 times to locate the approximate position of the pancreas in the original image; 3) the boundary of the target region was expanded to 256 × 192 and then it was cut; 4) the cut region was finely segmented to obtain the final segmentation result.
2.4 Testing
The test in this paper also requires two stages. In the first stage, we first reduced the 3D CT scans to a quarter of its original size, then we use M1 to simply locate the pancreas, and then the minimum boundary rectangle of the region is obtained and its maximum and minimum length are recorded and record the position of the enclosing rectangular box, which is \({x}_{max}, {x}_{min},{y}_{max}\) and \({y}_{min}\) respectively, and then multiply them by 4 to predict the approximate pancreas position in the original 3D CT scans, which were called\({X}_{max}=4\times {x}_{max},{X}_{min}=4\times {x}_{min} ,{Y}_{max}=4\times {y}_{max} , {Y}_{min}=4\times {y}_{min}\). In the second stage, because the approximate position of the pancreas was obtained, so we can figure out its length and width:\({X}_{len}={X}_{max}-{X}_{min} ,{Y}_{len}={Y}_{max}-{Y}_{min}\), then we extended its length and width to 256 × 192, which is a unified standard, and then cut out this area, and finally used M2 to accurately cut out the pancreas.
In two stages of testing a single patient CT image, we took 48 consecutive slices sequentially, with a step length of 12, and finally fused the test results together. The test flow is shown in Fig. 3. In addition, we still need to interpolate the z-axis interval of the data to 1mm before this test process, and then interpolate the z-axis of the prediction label finally obtained by the test to the original interval, and finally compare it with the ground truth to get the accuracy.
2.5 Network Architecture
Our network is based on the 3DUNet [5] architecture. Because the pancreas is small in the CT image, and some have tumors, so its shape and size are different, which is not conducive to segmentation. In response to the above problems, we have added the ResConv block to the encoder and decoder. In order to enhance the semantic expression ability of features at different scales. It also has four layers of encoders and four layers of decoders. For each layer of encoders, max pooling is used to decrease the resolution while increasing the feature map. At the same time, the function of the decoder is to increase the resolution while decreasing the feature map. In order to get more adequate training in the shallow layer and avoid the disappearance of the gradient, ResConv-3DUnet introduces a deep supervision mechanism [11]. The network architecture is shown in Fig. 4. Generally speaking, increasing the depth of a neural network can improve the representation ability of the network to a certain extent, but as the depth deepens, the neural network gradually appears difficult to train, including the gradient disappearance, gradient explosion and other phenomena. The deep supervision mechanism [11] can reduce gradient disappearance and promote network fusion when training deep neural networks, as well as to capture more meaningful features, and also to improve the directness and transparency of the hidden layer learning process. Deep supervision is the technique of adding a secondary classifier to some intermediate hidden layer of a deep neural network as a kind of network branch to supervise the backbone network. The deep supervision mechanism in this paper uses ground truth to supervise feature maps of different scales, which can conclude that the training speed of multi-scale convolution neural network model is faster and the convergence is more stable [19]. Finally, we use the concatenate operation to fuse the features of four scales and let them undergo 1 × 1 × 1 convolution to get the final output.
The ResConv block is composed of three consecutive 3 × 3 × 3 convolution blocks, show in Fig. 5. Two consecutive 3 × 3 × 3 convolution operations can be similar to a 5 × 5 × 5 convolution operation, and three consecutive 3 × 3 × 3 convolution operations can be roughly equivalent the convolution operation with a convolution kernel of 7 [9]. The advantage of this is that if the convolution kernel is 5 or 7 convolution operations will greatly consume huge memory, which is very unfavorable for 3D segmentation, so we can replace these operations with three consecutive convolution blocks, which can also save memory consumption. Connect them together to extract spatial features of different scales. In this way, the convolution kernel not only expands the receptive field, but also processes the input and increases the kernel size in parallel to capture different levels of details [13].
We also add an additional residual connection, which adds a 1 × 1 × 1 convolutional layer, which allows us to get additional spatial information. After each convolution operation in this res block, there is a Prelu activation function and a Group Normalization.
Group Normalization [10] is proposed by He Kaiming’s team. Group Normalization (GN) is an improved algorithm for the higher error rate of Batch Normalization (BN) when the batch size is small. Although the general GPU video memory in the classification algorithm can cover larger batch settings, the input image in 3D segmentation is large, and the memory requirement is huge, so the batch size is generally set relatively small, and the proposed GN has a significant improvement on this algorithm.
3 Experiments
3.1 Datasets
MSD (short for Medical Segmentation Decathlon challenge) provides 281 volumes of CT with labelled pancreas mask which have a spatial resolution of 512 x 512 and a slice count ranging from 37 to 751. We randomly split them into 201 volumes for training and 80 for testing.
3.2 Implementation Details
We adopted PyTorch 12 to implement all models. Our workstation was equipped with NVIDIA RTX 2080 Ti 11GB GPUs. And the viewable software for looking at 3D modeling is ITK-SNAP [18].
In the training network, the two-stage network is based on ResConv-3DUnet proposed in this paper. Moreover, the loss during training is dice loss [4]. During training, the batch size of the first stage is 3 and the batch size of the second stage is 1. Although the two network models are the same, the size of the input network image is different. In addition, the initial learning rate of the two experiments was lr = 1e4. Meanwhile, Adam was used as the optimizer, and the attenuation coefficient of deep supervision during the experiment was 0.33, which was attenuated once every 40 epoch. The training duration of the experiment was about 52 h.
3.3 Results
Our method has been successfully verified on MSD pancreas datasets, achieving state-of-the-art performance, and the dice accuracy can reach 84.6%, which is more accurate than the first place in the MSD competition. As the MSD competition is now closed and its submission entry has been closed, we are unable to submit the results to the official website for verification.
In order to demonstrate the effectiveness of our method, we compared it with state-of-the-art volumetric segmentation algorithms, in addition to comparing it with the accuracy of the pancreas segmentation that came first in the competition. The final comparison results are shown in Table 1. According to Table 1, it can be seen that our comparison experiments are roughly divided into the following categories: predicting segmentation results directly using 3D networks [3, 16]; segmenting 2D slices using a 2D model and then fusing the segmentation results into 3D results [17, 14]; and segmenting using a pseudo-3D (P3D) segmentation model [15, 14]. It can be easily seen from the table that our method outperforms them. Both the 2D segmentation and the P3D segmentation have a lower average DSC than ours. In addition, the standard deviation of the dice score in our method is also relatively small, suggesting that the method proposed in this paper would be more comprehensive in the segmentation of the experimental pancreas. More importantly, it can be seen from this comparison experiment that both 2D segmentation method and P3D segmentation method have reached a terrible number of parameters. However, as our method uses the two-stage segmentation method proposed in this paper, the number of parameters is greatly reduced and the memory pressure is alleviated.
In order to further illustrate the effectiveness of our proposed network, we conducted ablation experiments under the two-stage method proposed in this paper. The initial baseline network was 3DUNet [5]. As the different modules are added, we record the accuracy of their dice separately so that they can be compared. Table 2 shows the final experimental comparison results. The evaluation metrics are the dice score and the number of parameters, respectively. The results show that the accuracy of our network is indeed higher than that of the baseline, and it can be seen from the number of parameters that our network is more efficient. It can be seen that the accuracy of 3D UNet improves with the addition of the ResConv block module. In addition, deep supervision also plays a role in improving the accuracy, although it is not as obvious as the former, but it can also be improved in practice.
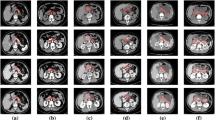
Figure 6 shows the test results of pancreas_088 and pancreas_299. The first row of the image shows a diseased pancreas, which is already severely diseased in the caudal part. From the figure, it can be seen that 3DUnet and baseline + DS can only roughly segment the diseased part of the pancreas, and the dice accuracy is not satisfactory. In contrast, the ResConv-3DUnet segmentation proposed in this paper was the best, and was able to segment the pancreas more completely, with a dice score of 77.47%. The shape of the pancreas in the second row was relatively normal, but the original baseline could only segment one point, and the accuracy was as low as 2.85%, while the ResConv-3DUnet segmentation had a more complete shape, which reflected the robustness of our network more. As can be seen from the figure, the segmentation result of the baseline + Rb (ResConv block) is relatively complete, but most importantly compared to the other three, the dice score obtained from our network architecture segmentation is the highest, indicating that the results of ResConv-3DUnet segmentation proposed in this paper are more consistent with the ground truth. Combined with Table 1, it can be seen that ResConv block can indeed increase the accuracy.
4 Conclusions
In this paper, we propose an end-to-end 3D segmentation method for the pancreas to reduce memory consumption, which includes both our proposed two-stage segmentation method and the ResConv-3DUnet presented in this paper. This experiment successfully completed the segmentation of MSD pancreas 3D datasets under the environment of 11G computer memory, and the accuracy of the proposed method outperformed those state-of-the-art volumetric segmentation algorithms, not only that, but also much higher than the score of winning the first place in the MSD competition. The experimental results show that this method can accomplish the pancreas segmentation task well and is expected to be applied in clinical practice.
References
Roth, H.R., et al.: DeepOrgan: multi-level deep convolutional networks for automated pancreas segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9349, pp. 556–564. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24553-9_68
Isensee, F., Petersen, J., Kohl, S.A.A, et al.: Nnu-net: breaking the spell on successful medical image segmentation (2019)
Roth, H.R., Oda, H., Zhou, X., et al.: An application of cascaded 3D fully convolutional networks for medical image segmentation. Comput. Med. Imaging Graph. Off. J. Comput. Med. Imaging Soc. 66, 90 (2018)
Milletari, F., Navab, N., Ahmadi, S.A.: V-net: Fully convolutional neural networks for volumetric medical image segmentation. In: 2016 Fourth International Conference on 3D Vision (3DV), pp. 565–571 (2016)
Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T., Ronneberger, O.: 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 424–432. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_49
Qiu, Z., Yao, T., Mei, T.: Learning spatio-temporal representation with pseudo-3D residual networks. In: 2017 IEEE International Conference on Computer Vision (ICCV). IEEE (2017)
Wang, Z.H., Liu, Z., Song, Y.Q., et al.: Densely connected deep U-Net for abdominal multi-organ segmentation. In: 2019 IEEE International Conference on Image Processing (ICIP). IEEE (2019)
Zhao, N., Tong, N., Ruan, D., Sheng, K.: Fully automated pancreas segmentation with two-stage 3D convolutional neural networks. In: Shen, D., Liu, T., Peters, T.M., Staib, L.H., Essert, C., Zhou, S., Yap, P.-T., Khan, A. (eds.) MICCAI 2019. LNCS, vol. 11765, pp. 201–209. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32245-8_23
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., Wojna, Z.: Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2818–2826.AWERTQ (2016)
Wu, Y., He, K.: Group normalization. Int. J. Comput. Vis. (2018)
Lee, C., Xie, S., Gallagher, P.W., et al.: Deeply-supervised nets. Int. Conf. Artif. Intell. Stat. 562–570 (2015)
Duta, I.C., Liu, L., Zhu, F., et al.: Pyramidal convolution: rethinking convolutional neural networks for visual recognition. arXiv preprint arXiv:2006.11538 (2020)
Yu, Q., Xie, L., Wang, Y., et al.: Recurrent Saliency Transformation Network: Incorporating Multi-Stage Visual Cues for Small Organ Segmentation. In: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition. IEEE (2018)
Zhou, X., Ito, T., Takayama, R., Wang, S., Hara, T., Fujita, H.: Three-dimensional CT image segmentation by combining 2D fully convolutional network with 3D majority voting. In: Carneiro, G., Mateus, D., Peter, L., Bradley, A., Tavares, J.M.R.S., Belagiannis, V., Papa, J.P., Nascimento, J.C., Loog, M., Lu, Z., Cardoso, J.S., Cornebise, J. (eds.) LABELS/DLMIA -2016. LNCS, vol. 10008, pp. 111–120. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46976-8_12
Roth, H.R., et al.: Deep learning and its application to medical image segmentation. Med. Imaging Technol. 36(2), 63–71 (2018)
Long, J., Evan, S., Trevor, D.: Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2015)
Yushkevich, P.A., Piven, J., Hazlett, H.C., et al.: User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31(3), 1116–1128 (2006)
Cheng, J., Liu, Y., Tang, X., Sheng, V.S., Li, M., et al.: DDOS attack detection via multi-scale convolutional neural network. Comput. Mater. Continua 62(3), 1317–1333 (2020)
Acknowledgment
This work was supported by Zhenjiang Key Deprogram “Fire Early Warning Technology Based on Multimodal Data Analysis” (SH2020011) Jiangsu Emergency Management Science and Technology Project “Research on Very Early Warning of Fire Based on Multi-modal Data Analysis and Multi-Intelligent Body Technology” (YJGL-TG-2020–8).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Wang, W. et al. (2021). Efficient 3D Pancreas Segmentation Using Two-Stage 3D Convolutional Neural Networks. In: Sun, X., Zhang, X., Xia, Z., Bertino, E. (eds) Artificial Intelligence and Security. ICAIS 2021. Lecture Notes in Computer Science(), vol 12736. Springer, Cham. https://doi.org/10.1007/978-3-030-78609-0_17
Download citation
DOI: https://doi.org/10.1007/978-3-030-78609-0_17
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-78608-3
Online ISBN: 978-3-030-78609-0
eBook Packages: Computer ScienceComputer Science (R0)