Abstract
Centromeres are chromosomal regions that are essential for the faithful transmission of genetic material through each cell division. They represent the chromosomal platform on which assembles a protein complex, the kinetochore, which mediates attachment to the mitotic spindle. In most organisms, centromeres assemble on large arrays of tandem satellite repeats, although their DNA sequences and organization are highly divergent among species. It has become evident that centromeres are not defined by underlying DNA sequences, but are instead epigenetically defined by the deposition of the centromere-specific histone H3 variant, CENP-A. In addition, and although long regarded as silent chromosomal loci, centromeres are in fact transcriptionally competent in most species, yet at low levels in normal somatic cells, but where the resulting transcripts participate in centromere architecture, identity, and function. In this chapter, we discuss the various roles proposed for centromere transcription and their transcripts, and the potential molecular mechanisms involved. We also discuss pathological cases in which unscheduled transcription of centromeric repeats or aberrant accumulation of their transcripts are pathological signatures of chromosomal instability diseases. In sum, tight regulation of centromeric satellite repeats transcription is critical for healthy development and tissue homeostasis, and thus prevents the emergence of disease states.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
7.1 Satellite DNA Underlies (Peri)centromeric Chromosomal Regions
Centromeres are chromosomal domains specialized in the faithful segregation of the genetic material between daughter cells at each cell division. In most higher eukaryotes, centromeres are made up of satellite DNA, i.e., large arrays of short, mostly A/T-rich, DNA sequences repeated in tandem over chromosomal domains that can range from hundreds of kilobases (Kb) to tens of megabases (Mb) on each centromere. Different families of satellite DNA define two distinct functional chromosomal domains (Choo 2001) with distinct chromatin landmarks (Karpen and Allshire 1997), i.e., the centromere per se and the juxta- or pericentromeric domains. The centromere is defined by the presence of unique nucleosomes, in which histone H3 is replaced by its variant called CENP-A in humans (Cse4 in budding yeast, Cnp1 in fission yeast, and CID/CenH3 in fruit flies) (Earnshaw and Rothfield 1985), interspersed with canonical nucleosomes. The CENP-A nucleosomal domain creates a platform for the assembly of a proteinaceous structure known as the kinetochore, that links chromosomes to mitotic spindle microtubules (Palmer et al. 1991; Fukagawa and Earnshaw 2014; Gambogi et al. 2020). CENP-A deposition is tightly regulated and mediated by a specific histone chaperone Holliday Junction Recognition Protein (HJURP) (Dunleavy et al. 2009; Foltz et al. 2009), although it remains unclear how HJURP directs CENP-A to its default location (Hoffmann and Fachinetti 2017). In juxta- or pericentromeric position, satellite repeats are enriched in repressive epigenetic marks which makes up the bulk of constitutive heterochromatin in mammals (Schueler and Sullivan 2006; Eymery et al. 2009b; Saksouk et al. 2015), and to which several functions have been assigned, including in sister chromatid cohesion at centromeres (Pidoux and Allshire 2005), maintenance of genome stability (Peters et al. 2001), and functional organization of the interphase nucleus (Wijchers et al. 2015; Muller et al. 2019; Francastel et al. 2000).
In contrast to the presence of CENP-A nucleosomes being the main and evolutionary conserved determinant of centromere identity, with very few exceptions in some insect lineages and kinetoplastids (Akiyoshi and Gull 2014; Drinnenberg et al. 2014; Navarro-Mendoza et al. 2019), the evolution of the underlying DNA sequences has been quite dynamic and gave rise to highly divergent centromere organization (Malik and Henikoff 2009). Phylogenetic analysis also found little evidence for satellite sequence conservation, which contrasts with the ancestral conserved structure of tandem repeats at telomeres (Meyne et al. 1989). Centromeres in different species display a wide variety of sequences, repeats organization, and chromosomal positions (Melters et al. 2013; Plohl et al. 2014), that can even diverge between chromosomes of the same species like in human and Drosophila (Bracewell et al. 2019; Sullivan and Sullivan 2020). Tandem repeats are highly prevalent at (peri)centromeres of most animal and plant genomes, and monocentric centromeres are the fundamental unit for chromosome inheritance in most species. However, chromosomes in certain insects lack a primary constriction and have adopted holocentric centromeres, i.e., in which the activity of the kinetochore extends over the whole chromosome arms, and therefore lacks a satellite repeats signature (Drinnenberg et al. 2014). There are also examples of atypical, yet functional, centromeres that spontaneously form on unique non-satellite sequences. Originally described in humans, the so-called neocentromeres are functionally and structurally similar to endogenous centromeres but lack the underlying repetitive sequences (Scott and Sullivan 2014). In the domestic horse, the discovery of satellite-less centromeres with variable positions among individuals (Giulotto et al. 2017) reinforced the idea that centromeres are defined, at least in part, by a centromere-specific histone variant, and not by the underlying DNA sequences.
Besides the abundance of repeats and association of centromeres with pericentromeric heterochromatin domains in most eukaryotes, a 17 base-pair (bp) motif is also highly conserved throughout species and called the CENP-B-box (B-Box). This sequence is the consensus binding site for the CENP-B protein, the only centromeric protein with sequence-specific DNA-binding activity (Masumoto et al. 1989). CENP-B protein is itself highly conserved, although its essential nature is debatable since certain centromeres lack a B-box in their satellite repeats, like on the human Y chromosome (Jain et al. 2018), and because CENP-B seems dispensable in the mouse (Kapoor et al. 1998). The development of human artificial chromosomes (HAC) has been instrumental in the determination of the minimal requirements for a functional centromere in terms of protein factors and DNA sequences (Bergmann et al. 2012). Many reports highlighted that both satellite DNA and CENP-B were necessary for de novo assembly of centromeres and HAC formation (Ohzeki et al. 2002; Masumoto et al. 2004), although alternative ways to establish a centromere during HAC formation have been reported (Logsdon et al. 2019), still questioning CENP-B requirement for establishment or maintenance of centromeres.
7.1.1 Examples of Centromere Organization Across Species
The budding yeast Saccharomyces cerevisiae and some relatives represent some sort of exceptions in the centromere world. In these organisms, a so-called “point centromere” is defined by short and unique sequences on which the centromere-specific nucleosome is positioned (Lechner and Ortiz 1996; Furuyama and Biggins 2007). This is in contrast with other eukaryotic organisms that feature regional centromeres assembled on kilo- to megabase-scale arrays of tandem repeats at the primary constriction site of the chromosome, and at which CENP-A nucleosomes are interspersed with canonical ones (Blower et al. 2002). The yeast Schizosaccharomyces pombe centromeres are composed of a 4–7 Kb-long central core element (ctr) flanked by centromere-specific innermost repeats (imr) sequences and pericentric outer repeats (otr), with an overall size range of 30–110 Kbs depending on the chromosome (Polizzi and Clarke 1991). In maize, centromeres are composed of CentC repeat of 156 bp, which form tandem arrays that span 180 Kb, separated by one or more copies of centromeric retrotransposable (CR) elements. Similar organization at centromeres is shared by the fruit fly Drosophila melanogaster, where the centromere is primarily composed of AATAT and TTCTC satellites, interspersed with complex A/T-rich repeats and mobile elements (Sun et al. 2003; Chang et al. 2019).
A model of choice for the study of centromere organization is the laboratory mouse Mus musculus due to its fairly homogeneous centromeres across all chromosomes (Kalitsis et al. 2006). The basic repeat unit in murine centromeres, called a minor satellite, is 120 bp-long and repeated in tandem over about 600 Kbs, which represents around 0.45% of the mouse genome. Murine chromosomes are telocentric, meaning that the centromere is nearly adjacent to the telomere of the short chromosome arm. On this short arm, minor satellite repeats are flanked by a retrotransposable DNA element, the truncated Long Interspersed Nucleotide Element 1 (tL1) and clusters of telocentric tandem (TLC) repeats, which share between 74% and 77% of homology with minor satellites, but lay in the opposite orientation (Kalitsis et al. 2006). On the long chromosome arm, the flanking pericentromeric domains are made up of tandem repeats of 234 bp-long major satellite repeat units over around 6 Mb and representing up to 3% of the mouse genome (Choo 1997; Kalitsis et al. 2006).
As opposed to homogeneous murine satellite repeats, each human centromere shows distinct polymorphisms in the number and sequence of α-satellite repeats (Aldrup-Macdonald and Sullivan 2014). Centromeric regions contain 171 bp-long α-satellite repeat units arranged in a tandem head-to-tail fashion, into higher-order repeat (HORs) units, themselves repeated in a largely uninterrupted fashion up to 5 Mb. The re-iteration of the HOR forms the centromeric α-satellite array, with occasional interruptions by transposable elements (She et al. 2004). In a given HOR, individual α-satellite repeats may only share 50–70% sequence similarity, whereas different HORs from the same chromosome share up to 98% of homology (Miga 2019). Neighboring pericentromeric regions account for about 4% to 5% of the human genome. They can be made of three types of satellite repeats: type I, which are formed by an alternation of 17 and 25 bp monomers and are restricted to chromosomes 2, 3, and acrocentric chromosomes. Type II and type III satellites are made of a 5 bp-long GGAAT repeat unit, found on all chromosomes, although unevenly distributed over several Mb. Notably, large blocks of heterochromatin in juxtacentromeric position on the long arm of chromosomes 1, and 16, or the long arm of chromosome 9, are composed of satellites type II or III, respectively (Vourc’h and Biamonti 2011).
All in all, the few unifying features in centromere organization across eukaryote kingdoms pose somewhat of a paradox given the essential nature of centromeres for the maintenance of genome integrity and the conserved functions and dynamics of kinetochores. Perhaps the most common characteristic of DNA sequences underlying centromeres is that they are transcriptionally competent in most of the species studied.
7.2 Transcription of Centromeric Repeats and Their Transcripts
At a time when recognition of the relatively pervasive aspect of genome transcription was in its infancy, the findings that (peri)centromeric satellite repeats could be transcribed have almost gone unnoticed. However, transcription at centromeric repeats was hinted at by the existence of centromeric transcripts in murine cells (Harel et al. 1968; Cohen et al. 1973) and in lung cells of the newt Taricha granulosa (Rieder 1978).
Nowadays, both the transcription of centromeric repeats and its products, the centromeric RNAs (cenRNA), are viewed as a conserved feature of centromeres in a broad range of organisms (Table 7.1), including yeast (Volpe et al. 2002; Choi et al. 2011; Ohkuni and Kitagawa 2011), plants (Topp et al. 2004; Du et al. 2010), beetles (Pezer and Ugarković 2008), Drosophila (Grewal and Elgin 2007; Rošić et al. 2014), amphibians (Varley et al. 1980; Diaz et al. 1981; Blower 2016), mouse (Rudert et al. 1995; Bouzinba-Segard et al. 2006; Ferri et al. 2009), and humans (Chan et al. 2012; Quénet and Dalal 2014; McNulty et al. 2017). Interestingly, work using a structurally dicentric chromosome that contains two α-satellite arrays demonstrated that RNA Polymerase II (RNA Pol II) localizes at active centromeres, i.e., at which the kinetochore assembles, but not at the inactive one (Chan et al. 2012), although another study reported that inactive arrays can also produce cenRNAs but just less stable than those originating from active arrays (McNulty et al. 2017).
In essence, in the absence of conserved centromeric DNA sequences across species, it is tempting to speculate that transcription through centromeric repeats or their derived transcripts may be functionally relevant to centromeres identity or function.
7.2.1 Regulation of Centromere Transcription/Transcripts Levels
Most of our knowledge of centromeric repeats transcription has been inferred from the existence of transcripts with centromeric sequences. Yet, the consequence of their highly repetitive and near-identical nature in some species is that they are mostly absent from reference genomes and specifically excluded from high-throughput sequencing analysis. Yet, transcripts with sequences of the identified centromeric repeat units can be found in various databases, including Expressed Sequence Tags (EST) databases. In addition, dedicated experimental testing of the levels of cenRNAs in a given tissue or at a specific developmental stage argued against the idea of simple transcription noise and even provided evidence for some level of transcriptional regulation. The challenge is rather to determine whether the observed differences in transcript abundance are the result of transcriptional or posttranscriptional control mechanisms.
In most species, the levels of cenRNAs appear to vary with particular developmental stages and with cell types, tissues, or organs. They have been detected in coleopteran insect species at all three developmental stages: larvae, pupae, and adults (Pezer and Ugarković 2008). In chicken and zebrafish, transcripts from an α-like satellite repeat are detected during early embryogenesis but are limited to the cardiac neural crest, the head, and the heart (Li and Kirby 2003). During mouse early development, pericentromeric major satellite RNAs (pericenRNAs) start being detected at the 2-cells stage and are required for the major reorganization of the nucleus that occurs at this stage, most notably characterized by the assembly of pericentromeric heterochromatin nuclear compartments, concomitantly with zygotic gene activation (Probst et al. 2010). In somatic cells, murine cen- and pericenRNAs accumulate with terminal differentiation, a process also accompanied by major spatial reorganization of constitutive heterochromatin compartments and changes in gene expression programs (Terranova et al. 2005; Bouzinba-Segard et al. 2006).
Transcription of centromeric repeats seems to be also regulated during the cell cycle. In cycling murine cells, cenRNAs begin to accumulate at the end of S phase and peak in the G2/M phase, just before the onset of mitosis (Ferri et al. 2009). This accumulation coincides with the late S phase when murine centromeres are being replicated (Müller and Almouzni 2017), although no formal demonstration that this would facilitate active transcription has been established. In human cells, the levels of cenRNAs do not change throughout the cell cycle (McNulty et al. 2017), although a recent study showed that their levels could fluctuate and peak in G2/M (Bury et al. 2020). In contrast, active RNA Pol II has been detected at human centromeric repeats in G1 (Quénet and Dalal 2014), when cenRNAs levels are low (Bury et al. 2020). More strikingly, elongating RNA Pol II was detected at mitotic centromeres in humans and mice (Chan et al. 2012). This is paradoxical since mitosis is regarded as a phase during which the bulk of the genome is transcriptionally silent (Christova and Oelgeschläger 2002), and this RNA Pol II localization could represent storage or bookmarking for further transcriptional activation when cells reenter the cell cycle. However, incorporation of fluorescent uridine-5′-triphosphate nucleotides (UTP) at the mitotic centromere suggested that human and murine centromeres are indeed actively transcribed during mitosis (Chan et al. 2012).
7.2.2 Mechanisms of Transcriptional Regulation
7.2.2.1 Transcriptional Machinery at Centromeric Repeats
The characteristic organization of most satellite DNA sequences is based on tandem repeats devoid of canonical promoter sequences, which led to the proposal that they could be transcribed by read-through from upstream genes or promoters of transposable elements, which is the case in maize (Topp et al. 2004). It should be noted that not all centromeres, like the human Y chromosome, may contain transposon sequences (Miga et al. 2014). In addition, a candidate TATA-box has been identified within human α-satellite sequences, as well as an SV40 enhancer-core sequence with spacing and orientation characteristic of RNA Pol II-transcribed genes (Vissel et al. 1992). The hypothesis that cryptic promoter elements are present within repeat sequences would not be surprising since, like any genomic sequence, centromeric repeats are stuffed with consensus binding sites for regulatory proteins. Some of these binding sites have long been known to serve as entry sites for the basal transcriptional machinery, like GATA sites, in place of a canonical TATA-box (Aird et al. 1994). Of note, consensus binding sites for GATA factors (WGATAR in which W indicates A/T and R indicates A/G) are frequently occurring in mammalian genomes, including at murine and human centromeric repeats, although occupancy by GATA-family members has not been described.
Whether cells have adapted to the fortuitous binding of various transcription factors to genomic loci essential for their survival is not known. Nevertheless, it may explain why centromere transcription appears to be regulated depending on cellular contexts, and hence, may rely on context-specific transcription factors. In addition, accumulation of cenRNAs of the size of a repeat unit in the mouse suggested that each repeat unit might contain a transcription start site (Bouzinba-Segard et al. 2006), although we cannot exclude that posttranscriptional processing of longer transcripts occurs, which is discussed below.
The question of the RNA polymerase(s) involved, and the means employed to regulate transcription of centromeric repeats, is also important. Centromeres of both budding and fission yeast are transcribed by RNA Pol II (Ohkuni and Kitagawa 2011; Sadeghi et al. 2014). In beetles, the presence of a cap structure and poly(A) tails in a subset of cenRNAs, termed PRAT, is also indicative of RNA Pol II-dependent transcription (Pezer and Ugarković 2008). Differential inhibition of RNA Pol I, II, and III and detection of active RNA Pol II at humans (Quénet and Dalal 2014; McNulty et al. 2017) and murine (Chan et al. 2012) centromeres indicated that RNA Pol II orchestrates the transcription of their centromeres.
In sum, RNA Pol II seems to be responsible for most part of the transcription of both point and regional centromeres, suggesting that transcription of this essential chromosomal domain has been conserved throughout evolution.
7.2.2.2 Transcription Factors
Compatible with the plethora of putative consensus binding sites for transcription factors in centromeric sequences, activators and repressors have been identified to control transcription of satellite repeats in various systems in a similar mode to the regulation of gene promoters.
In S. cerevisiae, the transcription factor Centromere-binding protein 1 (Cbf1) has been implicated as an activator of centromere transcription in an RNA Pol II-dependent manner (Ohkuni and Kitagawa 2011), although this is still debated. Indeed, other studies reported the upregulation of cenRNAs in cells lacking Cbf1, in association with chromosomal instability through the downregulation of the protein levels and mislocalization of CENP-A, HJURP, and components of the Chromosome Passenger Complex (CPC) (Ling and Yuen 2019; Chen et al. 2019). Along the same line, Htz1 (human homolog of H2A.Z) was identified as a transcriptional repressor since its deletion resulted in an upregulation of cenRNAs levels (Ling and Yuen 2019). Interestingly, the double invalidation of Cbf1 and Htz1 resulted in an additive effect on the upregulation of cenRNAs, suggesting that these two proteins operate in distinct pathways to repress centromere transcription (Ling and Yuen 2019). Noteworthy, Cbf1 is conserved among species with point centromeres, but not in eukaryotic species that have regional centromeres.
The Daxx-like motif-containing GATA factor Ams2 was actually one of the first transcription factors shown to be required for centromere function in S. pombe (Chen et al. 2003). Ams2 is a cell cycle-regulated factor that occupies centromeric chromatin in S phase where it is required for SpCENP-A deposition, although if this occurs through promoting transcription of cenRNAs has not been established. Again, whether a role for GATA factors is conserved among species has not yet been tested.
The only reported transcriptional regulator for transcriptional activation at murine and human centromeric repeats is the Zinc Finger and AT-Hook Domain Containing (ZFAT) protein, through binding to the ZFAT box, a short sequence present at centromeres of all chromosomes in mouse and human (Ishikura et al. 2020). In mammals, more is known about the transcription of the neighboring pericentromeric satellite repeats. In the mouse, transcriptional repressors YY1 (Shestakova et al. 2004), C/EBPα (Liu et al. 2007), and Ikaros (Brown et al. 1997; Cobb et al. 2000) appear to bind directly to major satellite sequences, although their link with transcriptional repression of these repeats has not been tested. In contrast, heat-shock transcription factor 1 (HSF1) has been shown to promote transcriptional activation of Sat III repeats in response to cellular stress in human cells (Jolly et al. 2004; Rizzi et al. 2004).
7.2.2.3 Histone Marks and DNA Methylation
In addition to occupancy by transcription factors, epigenetic modifications are likely to participate in the control of the transcription of centromeric repeats. In contrast to nearby pericentromeric heterochromatin, and in addition to CENP-A-containing nucleosomes, centromeres exhibit marks of euchromatin such as dimethylation of lysine 4 of histone H3 (H3K4me2) and lack of heterochromatin marks such as di- and tri-methylation of lysine 9 at histone H3 (H3K9me2 or me3) (Sullivan and Karpen 2004). In addition, a hypoacetylated state at centromeres seems to be conserved across eukaryotes (Wako et al. 2003; Sullivan and Karpen 2004; Choy et al. 2011). More specifically, hypoacetylated lysine 16 of histone H4 (H4K16) was shown to be required for kinetochore function and accurate chromosome segregation, although the link with transcriptional competence of centromeric repeats was not assessed (Choy et al. 2011). More recently, acetylated lysine 4 of histone H3 (H3K4ac) was described at centromeres, at which it is required for the centromeric localization of the chromatin reader Bromodomain-containing protein 4 (BRD4), which in turn recruits the RNA Pol II (Ishikura et al. 2020). At pericentromeric satellite repeats, SIRT6, a member of the Sirtuin family of deacetylases, has been implicated in the maintenance of their silent state through deacetylation of histone H3 at lysine 18 (H3K18ac) (Tasselli et al. 2016). Knockdown of SIRT6 caused an aberrant accumulation of pericenRNAs associated with mitotic errors through desilencing of pericentromeric repeats, which also lent support to the importance of heterochromatin maintenance in centromere function.
Additional important insights into the causal roles of histone modifications for transcription at centromeres came from the study of human artificial chromosomes (HACs) in which a CENP-B box was replaced by a tetracycline operator (tetO). Demethylation of H3K4me2, through targeting of the Lysine-Specific histone Demethylase 1A (LSD1) to the tetO sequence, induced a strong decrease in centromere transcription associated with impaired recruitment of HJURP, the CENP-A chaperone (Bergmann et al. 2011). This study provided a nice demonstration of a causal link between a specific activating histone mark, transcription of centromeric repeats, maintenance of CENP-A, and kinetochore functions.
Ubiquitination of histones is also important for the transcription of satellite repeats, with activating or repressive roles for ubiquitination of histone H2B or H2A (H2A-Ub; H2B-Ub), respectively (Zhu et al. 2011; Sadeghi et al. 2014). Loss of function of the Breast Cancer type 1 susceptibility protein (BRCA1) in cancer cells led to reduced H2A-Ub at pericentromeric satellite repeats, accompanied by their transcriptional derepression and loss of heterochromatin integrity (Zhu et al. 2011). Knockdown of Ring Finger Protein 20 (RNF20), the ubiquitin ligase responsible for H2B-Ub in S. pombe, led to reduced levels of transcription and nucleosome turnover at centromeres, associated with impaired centromere function. These data suggested that H2B-ub is essential for the maintenance of active centromeric chromatin (Sadeghi et al. 2014).
In contrast to their divergent sequence and structure, a common feature of mammalian satellite sequences is their methylated state at CpG dinucleotides, the main context for DNA methylation at least in mammals, which is also a major epigenetic mechanism to consider. The density of methylatable CpGs is higher at pericentromeres, which is consistent with their heterochromatin status, whereas centromeric repeat units in mice and humans contain only 2 to 3 CpGs per repeat unit. At repetitive elements, DNA methylation has been implicated in the inhibition of transposition and mitotic recombination between repeats, in part through maintaining these elements in a silent state (reviewed in Saksouk et al. 2014; Francastel and Magdinier 2019; Scelfo and Fachinetti 2019). However, our vision of the direct relationship between DNA methylation and transcriptional states of these regions may only be partial. For example, treatment of cells with the DNA demethylating agent 5-aza-2′-deoxycytidine (5AZA) led to increased levels of pericenRNAs in human cells (Eymery et al. 2009a) and of cenRNAs in murine cells (Bouzinba-Segard et al. 2006). However, it remains to be determined whether is it directly through demethylation of satellite repeats or through more indirect effects caused by 5AZA treatment, e.g., increased DNA damage also known to cause accumulation of Sat III pericenRNAs in humans (Valgardsdottir et al. 2008) or cenRNAs in murine cells (Hédouin et al. 2017). The identification of the DNA methyltransferases (DNMTs) responsible for de novo methylation (DNMT3A and DNMT3B) (Okano et al. 1999) or maintenance of methylation (DNMT1) (Bestor et al. 1988) was decisive in the dissection of more direct links. Notably, mouse minor satellites and human pericentromeric Sat II and Sat III repeats were identified as specific targets of the de novo methyltransferase DNMT3B (Okano et al. 1999; Xu et al. 1999). Human cells deficient for DNMT1 and DNMT3B are hypomethylated on Sat III repeats but do not accumulate pericenRNAs compared to wild-type cells (Eymery et al. 2009a). Similarly, hypomethylation of (peri)centromeric regions in murine embryonic stem cells (mESCs) deficient for Dnmt1 and Dnmt3a/b is not sufficient to promote their transcriptional activation (Lehnertz et al. 2003; Martens et al. 2005). However, in physiopathological cellular contexts further discussed below, loss of DNA methylation at (peri)centromeres has been associated with their transcriptional derepression, although it remains to be determined whether this is a direct consequence or a mere byproduct of the disease states.
It is possible that the methylated state at satellite repeats could influence the binding of transcriptional activators or repressors. It is important to note that two of the methylatable CpGs in human and murine centromeric repeat units reside within the CENP-B-box. Yet, the impact of CpG methylation on CENP-B binding and centromere architecture is still debated (Scelfo and Fachinetti 2019). DNA methylation was shown to prevent CENP-B binding to the CENP-B-box in vitro (Tanaka et al. 2005). Conversely, global demethylation using 5AZA treatment in cultured cells led to CENP-B spreading over demethylated repeats (Mitchell et al. 1996), although nearby pericentromeric repeats without a CENP-B-box are also demethylated in these conditions, making it difficult to conclude. Nevertheless, DNA methylation could be directly linked to the correct assembly of centromere architecture independently of the transcription of the repeats.
In sum, hypomethylation may create a favorable environment for transcriptional activation of satellite repeats, although it might not be sufficient. Hence, one has to consider that specific cellular contexts and their associated tissue- or context-specific transcription factors would be a prerequisite.
7.3 Functional Relevance of Centromeric Transcripts/Transcription
7.3.1 Centromeres Transcription and Chromatin Remodeling Processes
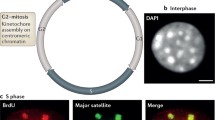
It appeared that low transcriptional activity is a characteristic of centromeric repeats in normal somatic cells. Several studies suggested that active transcription at centromeres would not only be compatible with but even required for centromere function. Indirect evidence came from the global inhibition of transcription by RNA Pol II inhibitors which led to compromised centromere function (Chan et al. 2012; Quénet and Dalal 2014). Transcription or nucleosome turnover at centromeres may be important for the dynamics of nucleosomes at centromeric repeats and deposition of CENP-A (Fig. 7.1). Notably, the complex Facilitates Chromatin Transcription (FACT) is localized to centromeres (Foltz et al. 2006) and is involved in CENP-A deposition via the recruitment of the chromatin remodeler Chromodomain Helicase DNA Binding Protein 1 (CHD1) in chickens (Okada et al. 2009). In mammals, a subunit of FACT, the Structure Specific Recognition Protein 1 (SSRP1), co-localized with RNA Pol II at centromeres during mitosis and was necessary for the efficient deposition of CENP-A in early G1 (Chan et al. 2012). Similarly, FACT-mediated transcription was also shown to be required for the de novo incorporation of CENP-A in Drosophila S cells (Chen et al. 2015). In this study, knockdown of FACT led to the loss of transcription at centromeres and reduced CENP-A loading. In recent years, a two-step process for Drosophila CENP-A loading was proposed (Bobkov et al. 2018). The first step was transcription-independent, during which CENP-A localized to centromeres through the Drosophila-specific chaperone Chromosome Alignment defect 1 (CAL1) (Chen et al. 2014), but a second step of active transcription was necessary for its stable incorporation into chromatin. In S. pombe mutants that are unable to restart stalled RNA Pol II at centromeres, CENP-A was still efficiently deposited, suggesting that halting RNA Pol II at centromeres may promote local chromatin remodeling events sufficient for CENP-A deposition (Catania et al. 2015). In a context where centromeric transcription-dependent chromatin remodeling is required for stable incorporation of its epigenetic determinant CENP-A, which also relies on the eviction of previously deposited H3/H3.3-placeholder nucleosomes (Dunleavy et al. 2011), the replication-independent histone chaperone and transcription elongation factor Spt6 (SUPT6H in human) was identified as a conserved CENP-A maintenance factor (Bobkov et al. 2020). Spt6 was shown to prevent loss of centromere identity in a transcription-dependent manner, through promoting the recycling of CENP-A and maintenance of parental CENP-A nucleosomes in both Drosophila and human cells (Bobkov et al. 2020).
Role of centromere transcription and transcripts in physiological conditions. Centromeric chromosomal domains are marked by a combination of nucleosomes containing histone H3 (gray) or CENP-A (orange) and are flanked by pericentromeric heterochromatin domains (green). RNA pol II-mediated transcription of centromeric repeats, together with chromatin remodelers such as the FACT complex, contribute to the dynamics of chromatin at centromeres and to the deposition of CENP-A. The derived centromeric transcripts serve as scaffolds or guides for the correct localization of centromeric proteins (e.g., CENP-C) and their associated complexes such as the chromosomal passenger complex (CPC), which includes Survivin, INCENP, and Aurora B kinase
Importantly, targeting of a strong trans-activation domain from Herpes simplex virus (VP16) to centromeres of HACs impaired the incorporation of newly synthesized CENP-A and led to the eviction of the parental one (Bergmann et al. 2011). Hence, even though transcriptional activation of centromeric repeats is necessary for centromere identity, increased transcription at this locus is incompatible with centromere function since it leads to loss of CENP-A at HAC centromeres (Bergmann et al. 2011).
To date, our knowledge of the immediate contribution of centromere transcription on centromere identity in different species still remains incomplete. Since the direct output from transcription at centromeres is the production of cenRNAs, whether long or short-lived, a question that arises is whether cenRNAs themselves could be implicated in the maintenance of centromere identity and function.
7.3.2 Functional Relevance of Centromeric Transcripts themselves
A growing body of evidence suggests that cenRNAs themselves may contribute to proper kinetochore assembly (Chen et al. 2003; Nakano et al. 2003; Topp et al. 2004; Ferri et al. 2009). Notably, cenRNAs are an integral part of the centromeric fraction (Ferri et al. 2009; McNulty et al. 2017; Kabeche et al. 2018), and coprecipitate with CENP-A in maize (Topp et al. 2004), mouse (Ferri et al. 2009), and humans (Chueh et al. 2009). Importantly, knockdown of human cenRNAs, without affecting transcription of the locus per se, led to impaired CENP-A deposition (Quénet and Dalal 2014). This finding suggested that correct loading of CENP-A does not only depend on active transcription at centromeres (see above), but also requires the transcripts themselves. This was further evidenced by the knockdown of cenRNAs in extracts of Xenopus oocytes, which led to decreased occupancy of CENP-A at centromeres (Grenfell et al. 2016).
Besides CENP-A deposition, several studies showed a direct association between cenRNAs and CENP-C in gel shift or immunoprecipitation experiments, probably through the CENP-C RNA binding domain. This was the case for example in maize (Du et al. 2010) and Drosophila (Rošić et al. 2014). RNase treatment of human cells induced the delocalization of CENP-C but not that of CENP-A (Wong et al. 2007). Similarly, inhibition of transcription in mitosis, a stage at which centromeres are transcribed by RNA Pol II, impaired the localization of CENP-C at centromeres (Chan et al. 2012).
Beyond their association with the constitutive components of the centromere, cenRNAs also interact with components of the CPC: Aurora B, INCENP, and Survivin in the G2/M phase in murine cells (Ferri et al. 2009). More specifically, the association of cenRNAs with the mitotic kinase Aurora B was necessary for Aurora B interaction with its partners Survivin and CENP-A and potentiated its kinase activity (Fig. 7.1). Since cenRNAs peak in G2/M, these data therefore suggested a role for cenRNAs in the timely recruitment or stabilization of the CPC components specifically at centromeres before the onset of mitosis. In turn, unscheduled accumulation of murine cenRNAs throughout the cell cycle led to ectopic localization of CPC proteins, mitotic abnormalities, and loss of cohesion between sister chromatids (Bouzinba-Segard et al. 2006). This interaction between cenRNAs and CPC proteins is conserved in Xenopus and humans, as seen with the knockdown of cenRNAs which impaired the recruitment of CPC at centromeres (Ideue et al. 2014; Blower 2016). Strikingly, the association of cenRNAs with Aurora B was also shown to be required for both telomerase activity and maintenance of telomere length in mESCs (Mallm and Rippe 2015). In the mouse, centromeres and telomeres being in close proximity, it is possible that cenRNAs may favor a local concentration of the kinase for shared functions on two essential chromosomal domains.
Together, these data emphasize that the fine-tuning of centromeric transcription/transcript levels is absolutely required, since too low or too high transcription or cenRNAs levels have deleterious consequences for centromere identity and function, and hence for normal cell growth and survival, and are emerging as new kinds of players in the development of disease as discussed in the following chapters.
7.3.3 Regulation of the Levels of (Peri)centromeric Transcripts Themselves
As mentioned above, the dynamic levels of cenRNAs according to cellular contexts may not only result from regulatory processes at the level of transcription but also at the level of the transcripts themselves, through fine-tuning of their stability or their processing. In S. cerevisiae, low levels of cenRNAs might be insured by their degradation by the exosome (Houseley et al. 2007; Ling and Yuen 2019). In many species, centromeric repeats are transcribed in both sense and antisense orientations (Topp et al. 2004; Li et al. 2008; Carone et al. 2009; Ideue et al. 2014), which would favor the formation of double-stranded RNAs (dsRNAs) that are substrates for further processing by the RNA interference (RNAi) machinery. The first example of the possibility that cenRNAs could be processed into smaller species was provided by the discovery of endogenous small interfering RNA (siRNAs) of centromeric origin in S. pombe (Reinhart and Bartel 2002). In addition, deletion of factors of the S. pombe RNAi machinery such as Dicer, the RNA-binding protein Argonaute (AGO), or the RNA-dependent RNA polymerase (RdRP), led to aberrant accumulation of cenRNAs and chromosome missegregation due to defective pericentromeric heterochromatin formation (Volpe et al. 2002). Similarly, in human–chicken hybrid cells (chicken DT40 cells carrying a human chromosome), ablation of Dicer led to mitotic defects and premature sister chromatid separation that was attributed to the loss of HP1 at pericentromeric heterochromatin and mislocalization of the cohesin complex (Fukagawa et al. 2004). In mouse embryonic stem cells (mESCs), Dicer deficiency also caused an accumulation of pericenRNAs, ranging from 40 nt to over 200 nt in size, i.e., not in the size range of siRNAs (Kanellopoulou et al. 2005; Murchison et al. 2005). Dicer has been involved in the repression of pericentromeric repeats in many species, but its depletion did not lead to mitotic defects in mESCs, although they exhibited differentiation and proliferation defects. In human cells, knockdown of Dicer or AGO2 resulted in chromosome lagging and increased levels of cenRNAs (Huang et al. 2015).
In fact, outside of the well-characterized S. pombe model, the literature is punctuated by opposing views of the role of Dicer/RNAi pathway in the regulation of the levels of satellite transcripts, with consequences for heterochromatin assembly and chromosomal stability. This is probably related to the failure to detect 25–30 nt RNA species, at least in the mouse (Kanellopoulou et al. 2005; Bouzinba-Segard et al. 2006). However, in mESCs, 150 nt-long cenRNAs, and smaller species but longer than siRNAs, have been detected and shown to rely on Dicer for their biogenesis (Kanellopoulou et al. 2005). Whether these transcripts play similar roles as (peri)centromeric siRNAs found in S. pombe is an interesting possibility. Of note, whereas exponentially growing somatic murine cells exhibit low levels of 2–4 Kb cenRNAs, the accumulation of 120 nt-long cenRNAs in physiopathological conditions recapitulated the same phenotypic defects observed in Dicer-deficient S. pombe that failed to produce centromeric siRNAs, including loss of sister chromatid cohesion, impaired centromere architecture and heterochromatin organization, associated with mitotic defects (Bouzinba-Segard et al. 2006). Although it is still not known whether these shorter cenRNAs result from cleavage through unknown mechanisms or are produced by multiple transcription initiation events, these data suggested that the absence of mature siRNAs or the accumulation of unprocessed longer RNA species have the same impact on the integrity of centromeric regions (Bouzinba-Segard et al. 2006).
The low levels of cenRNAs in normal conditions could indicate a short half-life caused by the rapid degradation of the transcripts through mechanisms exposed above or through posttranscriptional modifications shown to regulate the stability of transcripts (Nachtergaele and He 2017; Boo and Kim 2020). Among the myriad of known posttranscriptional RNA modifications, the adenosine to inosine (A to I) modification mediated by the adenosine deaminase acting on RNA (ADAR) machinery was shown to edit structured dsRNAs, with selectivity for certain internal loops and bulges rather than for a consensus sequence (Levanon et al. 2005). Just like the excitement of a connection between the RNAi machinery in keeping (peri)centromeric transcription in check in a broad range of eukaryotes in the early years of 2000 (Lippman and Martienssen 2004), the involvement of ADAR in the silencing of repetitive sequences in heterochromatin also attracted much interest a few years later (Fernandez et al. 2005). The A/T-rich and potentially dsRNAs produced from murine pericentromeric satellite repeats (Kanellopoulou et al. 2005) would make excellent substrates for the deamination of adenosine to inosine residues by ADAR. However, the search for such editing and for an immunolocalization of factors of the ADAR complex at (peri)centromeric domains remained unsuccessful (Lu and Gilbert 2008). Yet, it is interesting to note that RNA editing by ADAR is incompatible with the RNAi machinery (Scadden and Smith 2001). This is exemplified by the A to I conversion on microRNAs (miRNAs) derived from repetitive Long interspersed nuclear element 2 (LINE2), which blocks their cleavage by Dicer in human and mouse (Kawahara et al. 2007).
Thus, there is an exciting possibility that the dynamic size range of cenRNAs, with potentially distinct roles, could result from the fine-tuning of a balance between RNA modifications and RNA processing depending on phases of the cell cycle or cellular contexts.
7.4 Centromeric Transcripts: Cause or Consequence of Disease?
7.4.1 Accumulation of Satellite Transcripts in Various Types of Cellular Stress
Cells are constantly exposed to various environmental or endogenous stresses that may jeopardize their identity and viability. Exogenous sources of cellular stress include high temperatures [heat shock (HS)], DNA damaging chemicals, UV and ionizing radiation, hyperosmotic and oxidative stresses, whereas endogenous stress can originate from cellular metabolism or replication defects. In this context, a major challenge for the cells is therefore to safeguard their genome integrity. This is fundamental for their survival but also for the normal functioning of the whole organism and protection from the emergence of disease states. In that respect, cells have evolved sophisticated mechanisms that can trigger a rapid and adapted cellular response, including cell-cycle arrest, to allow time to repair DNA lesions or activate sets of genes to recover from stress, and therefore maintain their genomic integrity. There is now a large body of evidence showing that transcription of (peri)centromeric satellite repeats is rapidly induced in cells under various stress conditions, through the activity of transcription factors belonging to different stress-response pathways, thereby increasing the levels of resulting satellite transcripts as integral components of the stress response.
The accumulation of human pericenRNAs from Sat III of chromosome 9 was the first and best-characterized example of the accumulation of satellite transcripts in stress conditions. Originally described in response to HS (Jolly et al. 2004; Rizzi et al. 2004), it occurs in all of the above-cited types of stress conditions (Valgardsdottir et al. 2008). In response to HS, transient subnuclear organelles, called nuclear stress bodies (nSB), assemble on blocks of Sat III DNA repeats at which the Heat Shock Factor 1 (HSF1) binds, which in turn recruits the RNA Pol II and promotes transcriptional activation of the repeats (Biamonti and Vourc’h 2010). In stressed cells, nSBs are thought to contribute to the rapid and transient shutdown or reprogramming of gene expression programs required for the cells to recover from stress, through a Sat III transcripts-mediated trapping of a subset of splicing and transcription factors away from their site of action (Eymery et al. 2010). Pericentromeric heterochromatin is also central to the functional organization of the cell nucleus and to the maintenance of gene silencing through their positioning in the vicinity of subnuclear heterochromatin compartments (Francastel et al. 2000; Fisher and Merkenschlager 2002). Hence, transcriptional activation of pericentromeric Sat III sequences may also have a long-range impact on gene expression programs through the disorganization of these repressive nuclear compartments. It is interesting to note that activated transcription of satellite repeats in response to thermal perturbations appears to be a shared feature as it also occurs in beetles (Pezer and Ugarkovic 2012), Arabidopsis thaliana (Pecinka et al. 2010; Tittel-Elmer et al. 2010), and mouse (Hédouin et al. 2017). However, whether it triggers similar mechanisms in these organisms is not known. In fact, in the mouse, HS stimulates only a modest increase in cenRNAs levels but not that from pericentromeric repeats, whereas nSBs described in human cells do not form in murine cells. Hence, cellular responses to stress may vary between organisms although satellite transcripts appear to be central to the stress response.
In contrast to HS in murine cells, genotoxic stress led to a strong and rapid transcriptional activation of centromeric repeats, followed by local accumulation of cenRNAs at their site of transcription (Hédouin et al. 2017). Transcriptional activation, and not the transcripts themselves, has been causally linked to the loss of centromere identity characterized by the delocalization of CENP-A away from its default location. CENP-A delocalization, or eviction of CENP-A nucleosomes, was dependent on the chromatin remodeler FACT, pinpointing another function of FACT at centromeres in nucleosome destabilization at centromeres, and on the DNA Damage Response (DDR) effector ATM (Hédouin et al. 2017). Importantly, genotoxic stress-induced transcriptional activation of centromeric repeats had distinct functional consequences for cellular phenotypes depending on the integrity of the p53 checkpoint. Whereas immortalized cells continued to cycle, while accumulating micronuclei indicative of mitotic errors and centromere dysfunction, primary cells with normal p53 entered premature cell-cycle arrest and senescence (Hédouin et al. 2017). Hence, in the mouse, activated transcription at centromeric repeats provides a safeguard mechanism to prevent genomic instability in the context of persistent DNA damage signaling, through the disassembly of the core components of centromere identity and function. Whether this mechanisms is related to the acrocentric structure of murine chromosomes, i.e., with telomeres close to centromeric repeats, or to the mouse having very long telomeres so they enter senescence through mechanisms distinct than telomere shortening, is not known. However, together with the example of increased levels of Sat III pericenRNAs in heat-shocked cells, this illustrated the functional relevance for satellite transcripts in the stress response and protection of genome integrity.
7.4.2 Deregulation of Satellite Transcription/Transcripts in Cancer
Consistent with the above-mentioned links between transcription of satellite repeats and the triggering of safeguard mechanisms, it is not surprising that aberrant accumulation of transcripts from satellite repeats characterizes disease states with chromosomal instability like cancer, and that they actually represent good biomarkers of cancerous lesions (Eymery et al. 2009a; Ting et al. 2011; Zhu et al. 2011; Bersani et al. 2015; Tasselli et al. 2016; Hall et al. 2017). Yet, whether they are mere byproducts of disease phenotypes or act as drivers of disease, and through which mechanisms, are still open questions.
Macroarray-based approaches, designed to assess levels of transcripts from various repeated elements including satellite sequences, showed that the levels of satellite transcripts are higher in a variety of cancer cells compared to their normal healthy counterparts (Eymery et al. 2009a). Aberrant accumulation of satellite transcripts has also been reported in a wide range of primary epithelial tumors, both in humans and mice (Eymery et al. 2009a; Ting et al. 2011; Zhu et al. 2011). High-throughput sequencing has further highlighted that satellite transcripts actually represent up to 50% of transcriptional output in these tumors, which is in stark contrast to the low levels of these RNAs in normal cells (Ting et al. 2011).
An important open question remains as to the mechanisms that lead to the pathological transcriptional derepression of satellite sequences. Abnormal levels of satellite transcripts are often associated with the global hypomethylation that characterizes cancer cells, which in fact reflects reduced DNA methylation at repeated sequences owing to the large fraction of the genome they represent and their heavily methylated state in normal cells (Ross et al. 2010). The derepression of satellite transcripts was indeed shown to correlate with reduced DNA methylation of the underlying repeats (Ting et al. 2011; Unoki et al. 2020).
More hints into causal links between the aberrant accumulation of satellite transcripts and chromosomal instability came from gain-of-function experiments where cenRNAs were ectopically transcribed from expression vectors. In murine cells, ectopic expression of minor satellites from one repeat unit was sufficient to promote mitotic defects and alterations of nuclear organization typical of cancer cells (Bouzinba-Segard et al. 2006). Unscheduled accumulation of cenRNAs led to mitotic errors and disorganized centromere architecture through the trapping of centromeric protein complexes away from their default location (Bouzinba-Segard et al. 2006), providing a direct link between high levels of cenRNAs and centromere dysfunction. Likewise, ectopic expression or injection of satellite transcripts in cultured human or murine cells also led to mitotic errors (Zhu et al. 2011, 2018; Kishikawa et al. 2016, 2018), in correlation with the accumulation of foci of phosphorylated histone H2A.X (γ-H2A) that marks DNA double-strand breaks (DSBs) (Zhu et al. 2011). These data suggested that increased levels of satellite transcripts, and not transcriptional activation per se, is deleterious to the cells and leads to increased DNA mutation rates. They also put forward an interesting interplay between pathological high levels of satellite transcripts and DNA damage. In support of this hypothesis, there is the finding that these satellite transcripts accumulate strongly in breast cancer cells deficient for the BRCA1 gene (Zhu et al. 2011). Likewise, BRCA1 depletion has been causally linked to elevated levels of cenRNAs associated with impaired centromere architecture and chromosome missegregation (Di Paolo et al. 2014). BRCA1 is an important repair factor which, in normal conditions, occupies centromeric chromatin in interphase and throughout mitosis in normal cells (Pageau and Lawrence 2006; Di Paolo et al. 2014; Gupta et al. 2018), and may serve as a guardian of centromere integrity. Aberrant levels of cenRNAs, just like it has been described for kinetochore proteins (Bouzinba-Segard et al. 2006), were proposed to lead to BRCA1 delocalization away from centromeres and further exposure of this locus to the accumulation of unrepaired genotoxic insults (Zhu et al. 2018). Yet, the molecular mechanisms may not be that straightforward since BRCA1, besides its role in DNA damage repair, operates pleiotropic functions linked to the maintenance of chromosomal stability including at the replication fork, control of the cell cycle, and many other regulatory functions (Savage and Harkin 2015). BRCA1 was also recently shown to be an important determinant of the epigenetic states of centromeric and pericentromeric chromatin, through its ubiquitin ligase activity (Zhu et al. 2011). H2A ubiquitination at Lys 19 by BRCA1 provides a repressive mark at centromeric repeats important for their transcriptional repression. Hence, pathogenic variants of BRCA1 would promote DNA damage through the derepression of centromeric repeats in addition to, or instead of, promoting the accumulation of satellite transcripts. Along the same lines, centromeric targeting of VP16 for transcriptional activation of the underlying repeats in murine cells also promoted chromosomal instability (Zhu et al. 2018). Conversely, genotoxic stress using DSB inducers triggered the rapid transcriptional activation of murine centromeric repeats in a p53- and ATM-dependent dependent manner (Hédouin et al. 2017). In that case, transcriptional activation preceded, and was required for, eviction of CENP-A from centromeric chromatin, suggesting a direct link between activated transcription at centromeres and loss of centromere function and identity.
All these data suggested that our vision of the functional consequences of activated transcription of satellite repeats or accumulation of their related transcripts, and the actors at play, on centromere function and chromosomal stability is still only partial and may depend on organisms and cellular contexts. In the mouse, activated transcription of centromeric repeats and delocalization of CENP-A is associated with premature senescence in primary cells, whereas immortalized cells with impaired p53 checkpoint continue to cycle while accumulating mitotic errors and micronuclei, indicative of chromosomal instability (Hédouin et al. 2017). Thus, at least in the mouse, a functional p53 pathway is an important surveillance mechanism for centromere integrity, although the mechanisms remain unknown. Interestingly, p53-deficient mice ectopically expressing either human cenRNAs or murine pericenRNAs, were susceptible to tumor formation in mammary glands (Zhu et al. 2018). Hence, alterations to centromeric transcription may cooperate with oncogenic events or loss of tumor-suppressor function to promote oncogenesis.
Many questions remain unanswered as to the direct and reciprocal links between pathological hypomethylation of satellite repeats, their transcriptional derepression, the accumulation of DNA damage, and chromosomal instability. As cancer is a complex multifactorial disease, the current challenge is to dissect further and order these events.
7.4.3 Deregulation of Satellite Transcripts in the ICF Syndrome
A major breakthrough in the medical field came from the identification of inherited disorders of the epigenetic machinery, which provided interesting monogenic contexts and unsuspected players in a number of biological processes (Velasco and Francastel 2019). The first example of such developmental rare diseases was the Immunodeficiency with Centromeric instability and Facial anomalies (ICF) syndrome, a rare autosomal recessive immunological/neurological disorder with typical centromeric instability, including the presence of unusual multiradial chromosomal figures, decondensation, and rearrangement of (peri)centromeric regions (Ehrlich et al. 2006; Francastel and Magdinier 2019). At the molecular level, it is a remarkable case where these chromosomal alterations are caused by constitutive defects in DNA methylation, especially visible at heterochromatin blocks in juxtacentromeric position regions of chromosomes 1, 9, and 16 (Satellites type II and III) in all patients (Ehrlich et al. 2006). In a subset of patients, additional hypomethylation of centromeric α-satellite repeats suggested the genetic heterogeneity of the disease (Jiang et al. 2005; Toubiana et al. 2018).
Studies of the etiology of this rare disease have been instrumental in the identification of essential factors for the methylated state of (peri)centromeric repeats and the maintenance of their integrity. Hypomorphic mutations in the DNMT3B gene were the first identified genetic cause in about half of the patients (Xu et al. 1999), concomitantly implicated in de novo DNA methylation at centromeres in the mouse (Okano et al. 1999). The disease gained renewed interest when, in the reminder of patients, under the same diagnosis but with additional DNA methylation loss at centromeric repeats, exome sequencing identified mutations in factors with very few known functions and strikingly devoid of DNA methyltransferase (DNMT) activity (de Greef et al. 2011; Thijssen et al. 2015). These factors are transcription factors (ZBTB24, CDCA7) or chromatin remodeler of the SWI/SNF2 family (HELLS), the latter having already been shown to play a role in DNA methylation at murine centromeric repeats (Zhu et al. 2006). RNA interference performed in somatic cells, where DNA methylation profiles are already established, further demonstrated the requirement for ZBTB24, CDCA7, and HELLS in DNA methylation maintenance at murine centromeric repeats (Thijssen et al. 2015). These findings represented a major breakthrough in the knowledge of the determinants of DNA methylation at centromeric repeats. Yet, they raised again the question of the mechanisms that link hypomethylation of centromeric repeats to centromere loss of integrity, and the question of the contribution of non-DNMT ICF factors in DNA methylation and integrity of centromeres.
Independently of a putative role in DNA methylation pathways, a role for CDCA7 and HELLS in DNA repair pathways has been recently reported, reinforcing the idea of a link between DNA damage and centromere integrity (Burrage et al. 2012; Unoki et al. 2019). Notably, human embryonic kidney HEK-293T cells engineered to reproduce CDCA7 and HELLS mutations found in ICF patients exhibited a compromised nonhomologous end joining (NHEJ) DNA repair pathway (Unoki et al. 2019). Consistent with an aberrant accumulation of defects in DNA repair at centromeres, these cells accumulated micronuclei and suffered from abnormal chromosome segregation, while satellite repeats retained their methylated status. These engineered ICF cells, as well as cells from ICF patients, also exhibited increased transcription of (peri)centromeric repeats (Unoki et al. 2020). Given that genotoxic stress promotes a rapid transcriptional activation at centromeres (Hédouin et al. 2017), and along with the findings that satellite transcripts accumulate in breast cancer cells deficient for the DNA repair factor BRCA1 (Zhu et al. 2011), these data, therefore, suggested that DNA damage may trigger transcriptional activation at satellite repeats. An alternative, or concomitant, the mechanism could be that factors like CDCA7, HELLS, or BRCA1 may protect transcribed centromeric repeats from the accumulation of deleterious DNA:RNA hybrids (R-loops), just like BRCA1 does at transcriptional termination pause sites of actively transcribed genes (Hatchi et al. 2015). R-loops are dynamic and abundant structures that have been involved in a variety of physiological processes including chromosome segregation (Kabeche et al. 2018), whereas their unscheduled accumulation is also a source of DNA damage and genome instability (Costantino and Koshland 2018; Mishra et al. 2021). R-loops have been observed to accumulate at (peri)centromeres in engineered ICF cells and cells from ICF patients (Unoki et al. 2020). However, whether the transcriptional derepression and subsequent R-loop formation arise directly through DNA methylation loss or loss of function of ICF factors acting as “guardians” or transcriptional repressors of (peri)centromeric repeats, remains to be determined.
Like in cancer cells, transcriptional derepression or accumulation of the related satellite transcripts may represent intermediate steps between pathological hypomethylation of satellite repeats and chromosomal instability (Fig. 7.2). Importantly, the ICF syndrome leads to the premature death of the patients in early childhood from repeated infections, and despite a few reported cases where patients developed cancer, it is not clear whether pathological hypomethylation of centromeric repeats would favor later complications and further emergence of cancer. It also pointed out again that the impact of higher levels of satellite repeats transcripts on cellular phenotypes depends on the context in which it occurs.
Increased centromere transcription/transcripts levels: missing links between physiopathological DNA hypomethylation/DNA damage at centromeres and loss of centromere identity and function. Pathological hypomethylation, DNA damage at centromeres, or potentially impaired RNA processing or editing could promote: (a) unscheduled activated transcription of centromeric repeats, which in turn would lead to the formation of genotoxic R-loops; (b) aberrant accumulation of cenRNAs and trapping of kinetochore and DNA repair proteins away from centromeres; (c) the formation of double-stranded cenRNAs, known to trigger inflammatory responses. Although direct links between all these events remain to be formally dissected, the deregulation of centromeric transcription/transcripts ultimately lead to loss of centromere identity and integrity, as exemplified by multiradial chromosome figures, loss of sister chromatid cohesion, or recombination events between satellite repeats
A possibility that has not been evoked yet is that loss of DNA methylation at (peri)centromeric repeats and its associated abnormal levels of satellite RNAs may trigger surveillance mechanisms, which in fine would activate an interferon inflammatory response (Rajshekar et al. 2018). This has been nicely shown in a Zebrafish ICF model where one of the earliest in vivo consequences of ZBTB24 loss of function is a progressive loss of DNA methylation at pericentromeric regions associated with the derepression of sense and antisense pericenRNAs. This in turn triggered an interferon-dependent immune response mediated by the Melanoma Differentiation-Associated gene 5 (MDA5) and Mitochondrial AntiViral Signaling (MAVS) machinery, an antiviral surveillance mechanism that senses dsRNAs (Berke et al. 2013). Injection of sense and antisense pericenRNAs in Zebrafish embryos was also sufficient to stimulate the innate immunity (Rajshekar et al. 2018), implicating the accumulation of pericenRNAs as an important trigger of autoimmunity in a variety of diseases.
7.5 Conclusion
All of the data exposed in this chapter lend support to the essential nature of temporal control of the act of transcription through centromeric satellite repeats for the determination and correct functioning of this chromosomal region in most of the species studied so far. Transcription per se would facilitate the dynamic exchange of nucleosomes for the deposition of the key determinant of centromere identity, CENP-A, but would also favor a local concentration of the transcripts themselves for the timely recruitment of other centromere components. Yet, it is still unclear which molecular mechanisms and regulatory pathways are involved for a timely control in normal conditions, although we mentioned transcription factors acting in defined chromatin environments and RNA-based mechanisms for the regulation of the transcripts levels.
In turn, unscheduled transcription or aberrant levels of the transcripts have profound consequences for both centromere function and cell fate. We have seen that activated transcription of (peri)centromeric repeats or ectopic accumulation of the transcripts, i.e. elsewhere than at centromeres, under stress conditions is a mechanism adopted by many organisms to trigger rapid cellular responses for cells to recover from stress. This type of response is possible through the trapping of various regulatory factors away from their site of action and impairment of their associated functions, to favor genome repair or remodeling of gene expression programs. In turn, unscheduled transcription or accumulation of the transcripts coincides with disease states. In that case, they are not seen as safeguard mechanisms, which implicitly infer collaborative effects with disease conditions like defective checkpoints, oncogenic events, or even an inflammatory environment that ultimately alter cellular phenotypes. Pathological hypomethylation of satellite repeats like in cancer or ICF syndrome is a good candidate for uncontrolled transcription of satellite repeats, although we have seen that it is not necessarily sufficient and that opportunistic tissue- or context-specific factors may come into play. This might explain why all cancer cells do not necessarily exhibit increased transcription of satellite repeats, and why ICF patients do not have widespread alterations in all their tissues. Alternatively, or in addition to, defective RNA-based surveillance mechanisms might also contribute to the abnormal elevation of the levels of satellite transcripts.
In sum, the use of a wide range of model organisms and artificial centromeres allowed to identify a large number of centromere and kinetochore proteins, to address the relevance of DNA sequences for centromere identity, and to tackle the functional relevance of centromeres transcription/transcripts for centromere identity and function. Studies of the etiology of complex or monogenic human diseases further identified key determinants for centromere integrity and function, among which we can cite factors with DNA repair or chromatin remodeling activities, many of which could not be suspected before their implication in centromeric instability diseases. Yet, our vision of the intricate contribution of all the actors and mechanisms mentioned throughout this chapter still remains fragmentary and will require the development of targeted approaches, many of which are still missing in mammalian systems.
References
Aird WC, Parvin JD, Sharp PA, Rosenberg RD (1994) The interaction of GATA-binding proteins and basal transcription factors with GATA box-containing core promoters A model of tissue-specific gene expression. J Biol Chem 269:883–889
Akiyoshi B, Gull K (2014) Discovery of unconventional kinetochores in kinetoplastids. Cell 156:1247–1258
Aldrup-Macdonald ME, Sullivan BA (2014) The past, present, and future of human centromere genomics. Genes (Basel) 5:33–50
Bergmann JH, Rodríguez MG, Martins NMC, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LET, Earnshaw WC (2011) Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J 30:328–340
Bergmann JH, Martins NMC, Larionov V, Masumoto H, Earnshaw WC (2012) HACking the centromere chromatin code: insights from human artificial chromosomes. Chromosom Res 20:505–519
Berke IC, Li Y, Modis Y (2013) Structural basis of innate immune recognition of viral RNA. Cell Microbiol 15:386–394
Bersani F, Lee E, Kharchenko PV, Xu AW, Liu M, Xega K, MacKenzie OC, Brannigan BW, Wittner BS, Jung H et al (2015) Pericentromeric satellite repeat expansions through RNA-derived DNA intermediates in cancer. Proc Natl Acad Sci USA 112:15148–15153
Bestor T, Laudano A, Mattaliano R, Ingram V (1988) Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol 203:971–983
Biamonti G, Vourc’h C (2010) Nuclear stress bodies. Cold Spring Harb Perspect Biol 2:a000695
Blower MD (2016) Centromeric transcription regulates Aurora-B localization and activation. Cell Rep 15:1624–1633
Blower MD, Sullivan BA, Karpen GH (2002) Conserved organization of centromeric chromatin in flies and humans. Dev Cell 2:319–330
Bobkov GOM, Gilbert N, Heun P (2018) Centromere transcription allows CENP-A to transit from chromatin association to stable incorporation. J Cell Biol 217:1957–1972
Bobkov GOM, Huang A, van den Berg SJW, Mitra S, Anselm E, Lazou V, Schunter S, Feederle R, Imhof A, Lusser A et al (2020) Spt6 is a maintenance factor for centromeric CENP-A. Nat Commun 11:2919
Boo SH, Kim YK (2020) The emerging role of RNA modifications in the regulation of mRNA stability. Exp Mol Med 52:400–408
Bouzinba-Segard H, Guais A, Francastel C (2006) Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc Natl Acad Sci 103:8709–8714
Bracewell R, Chatla K, Nalley MJ, Bachtrog D (2019) Dynamic turnover of centromeres drives karyotype evolution in Drosophila. eLife 8
Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG (1997) Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell 91:845–854
Burrage J, Termanis A, Geissner A, Myant K, Gordon K, Stancheva I (2012) The SNF2 family ATPase LSH promotes phosphorylation of H2AX and efficient repair of DNA double-strand breaks in mammalian cells. J Cell Sci 125:5524–5534
Bury L, Moodie B, Ly J, McKay LS, Miga KH, Cheeseman IM (2020) Alpha-satellite RNA transcripts are repressed by centromere-nucleolus associations. eLife 9:e59770
Carone DM, Longo MS, Ferreri GC, Hall L, Harris M, Shook N, Bulazel KV, Carone BR, Obergfell C, O’Neill MJ et al (2009) A new class of retroviral and satellite encoded small RNAs emanates from mammalian centromeres. Chromosoma 118:113–125
Catania S, Pidoux AL, Allshire RC (2015) Sequence features and transcriptional stalling within centromere DNA promote establishment of CENP-A chromatin. PLoS Genet 11:e1004986
Chan FL, Marshall OJ, Saffery R, Kim BW, Earle E, Choo KHA, Wong LH (2012) Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc Natl Acad Sci USA 109:1979–1984
Chang C-H, Chavan A, Palladino J, Wei X, Martins NMC, Santinello B, Chen C-C, Erceg J, Beliveau BJ, Wu C-T et al (2019) Islands of retroelements are major components of Drosophila centromeres. PLoS Biol 17:e3000241
Chen ES, Saitoh S, Yanagida M, Takahashi K (2003) A cell cycle-regulated GATA factor promotes centromeric localization of CENP-A in fission yeast. Mol Cell 11:175–187
Chen C-C, Dechassa ML, Bettini E, Ledoux MB, Belisario C, Heun P, Luger K, Mellone BG (2014) CAL1 is the Drosophila CENP-A assembly factor. J Cell Biol 204:313–329
Chen C-C, Bowers S, Lipinszki Z, Palladino J, Trusiak S, Bettini E, Rosin L, Przewloka MR, Glover DM, O’Neill RJ et al (2015) Establishment of Centromeric chromatin by the CENP-A assembly factor CAL1 requires FACT-mediated transcription. Dev Cell 34:73–84
Chen C-F, Pohl TJ, Chan A, Slocum JS, Zakian VA (2019) Saccharomyces cerevisiae centromere RNA is negatively regulated by Cbf1 and its unscheduled synthesis impacts CenH3 binding. Genetics 213:465–479
Choi ES, Strålfors A, Castillo AG, Durand-Dubief M, Ekwall K, Allshire RC (2011) Identification of noncoding transcripts from within CENP-A chromatin at fission yeast centromeres. J Biol Chem 286:23600–23607
Choo KH (1997) Centromere DNA dynamics: latent centromeres and neocentromere formation. Am J Hum Genet 61:1225–1233
Choo KHA (2001) Domain organization at the centromere and neocentromere. Dev Cell 1:165–177
Choy JS, Acuña R, Au W-C, Basrai MA (2011) A role for histone H4K16 hypoacetylation in Saccharomyces cerevisiae kinetochore function. Genetics 1:11–21
Christova R, Oelgeschläger T (2002) Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nat Cell Biol 4:79–82
Chueh AC, Northrop EL, Brettingham-Moore KH, Choo KHA, Wong LH (2009) LINE retrotransposon RNA is an essential structural and functional epigenetic component of a core neocentromeric chromatin. PLoS Genet 5:e1000354
Cobb BS, Morales-Alcelay S, Kleiger G, Brown KE, Fisher AG, Smale ST (2000) Targeting of Ikaros to pericentromeric heterochromatin by direct DNA binding. Genes Dev 14:2146–2160
Cohen AK, Huh TY, Helleiner CW (1973) Transcription of satellite DNA in mouse L-cells. Can J Biochem 51:529–532
Costantino L, Koshland D (2018) Genome-wide map of R-loop-induced damage reveals how a subset of R-loops contributes to genomic instability. Mol Cell 71:487–497e3
de Greef JC, Wang J, Balog J, den Dunnen JT, Frants RR, Straasheijm KR, Aytekin C, van der Burg M, Duprez L, Ferster A et al (2011) Mutations in ZBTB24 are associated with immunodeficiency, centromeric instability, and facial anomalies syndrome type 2. Am J Hum Genet 88:796–804
Di Paolo A, Racca C, Calsou P, Larminat F (2014) Loss of BRCA1 impairs centromeric cohesion and triggers chromosomal instability. FASEB J 28:5250–5261
Diaz MO, Barsacchi-Pilone G, Mahon KA, Gall JG (1981) Transcripts from both strands of a satellite DNA occur on lampbrush chromosome loops of the newt Notophthalmus. Cell 24:649–659
Drinnenberg IA, de Young D, Henikoff S, Malik HS (2014) Recurrent loss of CenH3 is associated with independent transitions to holocentricity in insects. eLife 3:e03676
Du Y, Topp CN, Dawe RK (2010) DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet 6:e1000835
Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G (2009) HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137:485–497
Dunleavy EM, Almouzni G, Karpen GH (2011) H33 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G1 phase. Nucleus 2:146–157
Earnshaw WC, Rothfield N (1985) Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91:313–321
Ehrlich M, Jackson K, Weemaes C (2006) Immunodeficiency, centromeric region instability, facial anomalies syndrome (ICF). Orphanet J Rare Dis 1:1
Eymery HB, El Atifi-Borel M, Fourel G, Berger F, Vitte A-L, Van den Broeck A, Brambilla E, Fournier A, Callanan M et al (2009a) A transcriptomic analysis of human centromeric and pericentric sequences in normal and tumor cells. Nucleic Acids Res 37:6340–6354
Eymery A, Callanan M, Vourc’h C (2009b) The secret message of heterochromatin: new insights into the mechanisms and function of centromeric and pericentric repeat sequence transcription. Int J Dev Biol 53:259–268
Eymery A, Souchier C, Vourc’h C, Jolly C (2010) Heat shock factor 1 binds to and transcribes satellite II and III sequences at several pericentromeric regions in heat-shocked cells. Exp Cell Res 316:1845–1855
Fernandez HR, Kavi HH, Xie W, Birchler JA (2005) Heterochromatin: on the ADAR radar? Curr Biol 15:R132–R134
Ferri F, Bouzinba-Segard H, Velasco G, Hubé F, Francastel C (2009) Non-coding murine centromeric transcripts associate with and potentiate Aurora B kinase. Nucleic Acids Res 37:5071–5080
Fisher AG, Merkenschlager M (2002) Gene silencing, cell fate and nuclear organisation. Curr Opin Genet Dev 12:193–197
Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR 3rd, Cleveland DW (2006) The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol 8:458–469
Foltz DR, Jansen LET, Bailey AO, Yates JR, Bassett EA, Wood S, Black BE, Cleveland DW (2009) Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 137:472–484
Francastel C, Magdinier F (2019) DNA methylation in satellite repeats disorders. Essays Biochem 63:757–771
Francastel C, Schübeler D, Martin DIK, Groudine M (2000) Nuclear compartmentalization and gene activity. Nat Rev Mol Cell Biol 1:137–143
Fukagawa T, Earnshaw WC (2014) The centromere: chromatin foundation for the kinetochore machinery. Dev Cell 30:496–508
Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, Takami Y, Nakayama T, Oshimura M (2004) Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol 6:784–791
Furuyama S, Biggins S (2007) Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc Natl Acad Sci USA 104:14706–14711
Gambogi CW, Dawicki-McKenna JM, Logsdon GA, Black BE (2020) The unique kind of human artificial chromosome: bypassing the requirement for repetitive centromere DNA. Exp Cell Res 391:111978
Giulotto E, Raimondi E, Sullivan KF (2017) The unique DNA sequences underlying equine centromeres. Prog Mol Subcell Biol 56:337–354
Grenfell AW, Heald R, Strzelecka M (2016) Mitotic noncoding RNA processing promotes kinetochore and spindle assembly in Xenopus. J Cell Biol 214:133–141
Grewal SIS, Elgin SCR (2007) Transcription and RNA interference in the formation of heterochromatin. Nature 447:399–406
Gupta R, Somyajit K, Narita T, Maskey E, Stanlie A, Kremer M, Typas D, Lammers M, Mailand N, Nussenzweig A et al (2018) DNA repair network analysis reveals Shieldin as a key regulator of NHEJ and PARP inhibitor sensitivity. Cell 173:972–988e23
Hall LL, Byron M, Carone DM, Whitfield TW, Pouliot GP, Fischer A, Jones P, Lawrence JB (2017) Demethylated HSATII DNA and HSATII RNA foci sequester PRC1 and MeCP2 into cancer-specific nuclear bodies. Cell Rep 18:2943–2956
Harel J, Hanania N, Tapiero H, Harel L (1968) RNA replication by nuclear satellite DNA in different mouse cells. Biochem Biophys Res Commun 33:696–701
Hatchi E, Skourti-Stathaki K, Ventz S, Pinello L, Yen A, Kamieniarz-Gdula K, Dimitrov S, Pathania S, McKinney KM, Eaton ML et al (2015) BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol Cell 57:636–647
Hédouin S, Grillo G, Ivkovic I, Velasco G, Francastel C (2017) CENP-A chromatin disassembly in stressed and senescent murine cells. Sci Rep 7:42520
Hoffmann S, Fachinetti D (2017) A time out for CENP-A. Mol Cell Oncol 4:e1293596
Houseley J, Kotovic K, El Hage A, Tollervey D (2007) Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J 26:4996–5006
Huang C, Wang X, Liu X, Cao S, Shan G (2015) RNAi pathway participates in chromosome segregation in mammalian cells. Cell Discov 1:15029
Ideue T, Cho Y, Nishimura K, Tani T (2014) Involvement of satellite I noncoding RNA in regulation of chromosome segregation. Genes Cells 19:528–538
Ishikura S, Nakabayashi K, Nagai M, Tsunoda T, Shirasawa S (2020) ZFAT binds to centromeres to control noncoding RNA transcription through the KAT2B-H4K8ac-BRD4 axis. Nucleic Acids Res 48:10848–10866
Jain M, Olsen HE, Turner DJ, Stoddart D, Bulazel KV, Paten B, Haussler D, Willard HF, Akeson M, Miga KH (2018) Linear assembly of a human centromere on the Y chromosome. Nat Biotechnol 36:321–323
Jiang YL, Rigolet M, Bourc'his D, Nigon F, Bokesoy I, Fryns JP, Hultén M, Jonveaux P, Maraschio P, Mégarbané A, Moncla A, Viegas-Péquignot E (2005) DNMT3B mutations and DNA methylation defect define two types of ICF syndrome. Hum Mutat 25:56–63
Jolly C, Metz A, Govin J, Vigneron M, Turner BM, Khochbin S, Vourc’h C (2004) Stress-induced transcription of satellite III repeats. J Cell Biol 164:25–33
Kabeche L, Nguyen HD, Buisson R, Zou L (2018) A mitosis-specific and R loop-driven ATR pathway promotes faithful chromosome segregation. Science 359:108–114
Kalitsis P, Griffiths B, Choo KHA (2006) Mouse telocentric sequences reveal a high rate of homogenization and possible role in Robertsonian translocation. Proc Natl Acad Sci USA 103:8786–8791
Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K (2005) Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19:489–501
Kapoor M, Montes de Oca Luna R, Liu G, Lozano G, Cummings C, Mancini M, Ouspenski I, Brinkley BR, May GS (1998) The cenpB gene is not essential in mice. Chromosoma 107:570–576
Karpen GH, Allshire RC (1997) The case for epigenetic effects on centromere identity and function. Trends Genet 13:489–496
Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K (2007) RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep 8:763–769
Kishikawa T, Otsuka M, Yoshikawa T, Ohno M, Yamamoto K, Yamamoto N, Kotani A, Koike K (2016) Quantitation of circulating satellite RNAs in pancreatic cancer patients. JCI Insight 1:e86646
Kishikawa T, Otsuka M, Suzuki T, Seimiya T, Sekiba K, Ishibashi R, Tanaka E, Ohno M, Yamagami M, Koike K (2018) Satellite RNA increases DNA damage and accelerates tumor formation in mouse models of pancreatic cancer. Mol Cancer Res 16:1255–1262
Lechner J, Ortiz J (1996) The Saccharomyces cerevisiae kinetochore. FEBS Lett 389:70–74
Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AHFM (2003) Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol 13:1192–1200. https://doi.org/10.1016/s0960-9822(03)00432-9
Levanon EY, Hallegger M, Kinar Y, Shemesh R, Djinovic-Carugo K, Rechavi G, Jantsch MF, Eisenberg E (2005) Evolutionarily conserved human targets of adenosine to inosine RNA editing. Nucleic Acids Res 33:1162–1168
Li Y-X, Kirby ML (2003) Coordinated and conserved expression of alphoid repeat and alphoid repeat-tagged coding sequences. Dev Dyn 228:72–81
Li F, Sonbuchner L, Kyes SA, Epp C, Deitsch KW (2008) Nuclear non-coding RNAs are transcribed from the centromeres of plasmodium falciparum and are associated with centromeric chromatin. J Biol Chem 283:5692–5698
Ling YH, Yuen KWY (2019) Centromeric non-coding RNA as a hidden epigenetic factor of the point centromere. Curr Genet 65:1165–1171
Lippman Z, Martienssen R (2004) The role of RNA interference in heterochromatic silencing. Nature 431:364–370
Liu X, Wu B, Szary J, Kofoed EM, Schaufele F (2007) Functional sequestration of transcription factor activity by repetitive DNA. J Biol Chem 282:20868–20876
Logsdon GA, Gambogi CW, Liskovykh MA, Barrey EJ, Larionov V, Miga KH, Heun P, Black BE (2019) Human artificial chromosomes that bypass centromeric DNA. Cell 178:624–639e19
Lu J, Gilbert DM (2008) Cell cycle regulated transcription of heterochromatin in mammals vs fission yeast: functional conservation or coincidence? Cell Cycle 7:1907–1910
Malik HS, Henikoff S (2009) Major evolutionary transitions in centromere complexity. Cell 138:1067–1082
Mallm J-P, Rippe K (2015) Aurora kinase B regulates telomerase activity via a centromeric RNA in stem cells. Cell Rep 11:1667–1678
Martens JH, O’Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T (2005) The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J 24:800–812
Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T (1989) A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol 109:1963–1973
Masumoto H, Nakano M, Ohzeki J-I (2004) The role of CENP-B and alpha-satellite DNA: de novo assembly and epigenetic maintenance of human centromeres. Chromosom Res 12:543–556
McNulty SM, Sullivan LL, Sullivan BA (2017) Human centromeres produce chromosome-specific and Array-specific alpha satellite transcripts that are complexed with CENP-A and CENP-C. Dev Cell 42:226–240e6
Melters DP, Bradnam KR, Young HA, Telis N, May MR, Ruby J, Sebra R, Peluso P, Eid J, Rank D et al (2013) Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol 14:R10
Meyne J, Ratliff RL, MoYzIs RK (1989) Conservation of the human telomere sequence (TTAGGG) among vertebrates. Proc Natl Acad Sci USA 5
Miga KH (2019) Centromeric satellite DNAs: hidden sequence variation in the human population. Genes (Basel) 10:352
Miga KH, Newton Y, Jain M, Altemose N, Willard HF, Kent WJ (2014) Centromere reference models for human chromosomes X and Y satellite arrays. Genome Res 24:697–707
Mishra PK, Chakraborty A, Yeh E, Feng W, Bloom KS, Basrai MA (2021) R-loops at centromeric chromatin contribute to defects in kinetochore integrity and chromosomal instability in budding yeast. MBoC 32:74–89
Mitchell AR, Jeppesen P, Nicol L, Morrison H, Kipling D (1996) Epigenetic control of mammalian centromere protein binding: does DNA methylation have a role? J Cell Sci 109:2199–2206
Müller S, Almouzni G (2017) Chromatin dynamics during the cell cycle at centromeres. Nat Rev Genet 18:192–208
Muller H, Gil J, Drinnenberg IA (2019) The impact of centromeres on spatial genome architecture. Trends Genet 35:565–578
Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ (2005) Characterization of dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA 102:12135–12140
Nachtergaele S, He C (2017) The emerging biology of RNA post-transcriptional modifications. RNA Biol 14:156–163
Nakano M, Okamoto Y, Ohzeki J, Masumoto H (2003) Epigenetic assembly of centromeric chromatin at ectopic alpha-satellite sites on human chromosomes. J Cell Sci 116:4021–4034
Navarro-Mendoza MI, Pérez-Arques C, Panchal S, Nicolás FE, Mondo SJ, Ganguly P, Pangilinan J, Grigoriev IV, Heitman J, Sanyal K et al (2019) Early diverging fungus Mucor circinelloides lacks centromeric histone CENP-A and displays a mosaic of point and regional centromeres. Curr Biol 29:3791–3802e6
Ohkuni K, Kitagawa K (2011) Endogenous transcription at the centromere facilitates centromere activity in budding yeast. Curr Biol 21:1695–1703
Ohkuni K, Kitagawa K (2012) Role of transcription at centromeres in budding yeast. Transcription 3:193–197
Ohzeki J, Nakano M, Okada T, Masumoto H (2002) CENP-B box is required for de novo centromere chromatin assembly on human alphoid. DNA J Cell Biol 159:765–775
Okada M, Okawa K, Isobe T, Fukagawa T (2009) CENP-H-containing complex facilitates centromere deposition of CENP-A in cooperation with FACT and CHD1. Mol Biol Cell 20:3986–3995
Okano M, Bell DW, Haber DA, Li E (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247–257
Pageau GJ, Lawrence JB (2006) BRCA1 foci in normal S-phase nuclei are linked to interphase centromeres and replication of pericentric heterochromatin. J Cell Biol 175:693–701
Palmer DK, O’Day K, Trong HL, Charbonneau H, Margolis RL (1991) Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc Natl Acad Sci USA 88:3734–3738
Pecinka A, Dinh HQ, Baubec T, Rosa M, Lettner N, Mittelsten Scheid O (2010) Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell 22:3118–3129
Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schöfer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A et al (2001) Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107:323–337
Pezer Z, Ugarković D (2008) RNA Pol II promotes transcription of centromeric satellite DNA in beetles. PLoS One 3:e1594
Pezer Z, Ugarkovic D (2012) Satellite DNA-associated siRNAs as mediators of heat shock response in insects. RNA Biol 9:587–595
Pidoux AL, Allshire RC (2005) The role of heterochromatin in centromere function. Philos Trans R Soc Lond Ser B Biol Sci 360:569–579
Plohl M, Meštrović N, Mravinac B (2014) Centromere identity from the DNA point of view. Chromosoma 123:313–325
Polizzi C, Clarke L (1991) The chromatin structure of centromeres from fission yeast: differentiation of the central core that correlates with function. J Cell Biol 112:191–201
Probst AV, Okamoto I, Casanova M, El Marjou F, Le Baccon P, Almouzni G (2010) A strand-specific burst in transcription of pericentric satellites is required for chromocenter formation and early mouse development. Dev Cell 19:625–638
Quénet D, Dalal Y (2014) A long non-coding RNA is required for targeting centromeric protein A to the human centromere. eLife 3:e03254
Rajshekar S, Yao J, Arnold PK, Payne SG, Zhang Y, Bowman TV, Schmitz RJ, Edwards JR, Goll M (2018) Pericentromeric hypomethylation elicits an interferon response in an animal model of ICF syndrome. eLife 7
Reinhart BJ, Bartel DP (2002) Small RNAs correspond to centromere heterochromatic repeats. Science 297:1831
Rieder CL (1978) Effect of elevated temperatures on spindle microtubules and chromosome movements in cultured newt lung cells. Cytobios 18:201–233
Rizzi N, Denegri M, Chiodi I, Corioni M, Valgardsdottir R, Cobianchi F, Riva S, Biamonti G (2004) Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol Biol Cell 15:543–551
Rošić S, Köhler F, Erhardt S (2014) Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J Cell Biol 207:335–349
Ross JP, Rand KN, Molloy PL (2010) Hypomethylation of repeated DNA sequences in cancer. Epigenomics 2:245–269
Rudert F, Bronner S, Garnier JM, Dollé P (1995) Transcripts from opposite strands of gamma satellite DNA are differentially expressed during mouse development. Mamm Genome 6:76–83
Sadeghi L, Siggens L, Svensson JP, Ekwall K (2014) Centromeric histone H2B monoubiquitination promotes noncoding transcription and chromatin integrity. Nat Struct Mol Biol 21:236–243
Saksouk N, Barth TK, Ziegler-Birling C, Olova N, Nowak A, Rey E, Mateos-Langerak J, Urbach S, Reik W, Torres-Padilla M-E et al (2014) Redundant mechanisms to form silent chromatin at pericentromeric regions rely on BEND3 and DNA methylation. Mol Cell 56:580–594
Saksouk N, Simboeck E, Déjardin J (2015) Constitutive heterochromatin formation and transcription in mammals. Epigenetics Chromatin 8:3
Savage KI, Harkin DP (2015) BRCA1, a “complex” protein involved in the maintenance of genomic stability. FEBS J 282:630–646
Scadden AD, Smith CW (2001) RNAi is antagonized by A-->I hyper-editing. EMBO Rep 2:1107–1111
Scelfo A, Fachinetti D (2019) Keeping the centromere under control: a promising role for DNA methylation. Cell 8
Schueler MG, Sullivan BA (2006) Structural and functional dynamics of human centromeric chromatin. Annu Rev Genomics Hum Genet 7:301–313
Scott KC, Sullivan BA (2014) Neocentromeres: a place for everything and everything in its place. Trends Genet 30:66–74
She X, Horvath JE, Jiang Z, Liu G, Furey TS, Christ L, Clark R, Graves T, Gulden CL, Alkan C et al (2004) The structure and evolution of centromeric transition regions within the human genome. Nature 430:857–864
Shestakova EA, Mansuroglu Z, Mokrani H, Ghinea N, Bonnefoy E (2004) Transcription factor YY1 associates with pericentromeric gamma-satellite DNA in cycling but not in quiescent (G0) cells. Nucleic Acids Res 32:4390–4399
Sullivan BA, Karpen GH (2004) Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol 11:1076–1083
Sullivan LL, Sullivan BA (2020) Genomic and functional variation of human centromeres. Exp Cell Res 389:111896
Sun X, Le HD, Wahlstrom JM, Karpen GH (2003) Sequence analysis of a functional Drosophila centromere. Genome Res 13:182–194
Tanaka Y, Kurumizaka H, Yokoyama S (2005) CpG methylation of the CENP-B box reduces human CENP-B binding. FEBS J 272:282–289
Tasselli L, Xi Y, Zheng W, Tennen RI, Odrowaz Z, Simeoni F, Li W, Chua KF (2016) SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat Struct Mol Biol 23:434–440
Terranova R, Sauer S, Merkenschlager M, Fisher AG (2005) The reorganisation of constitutive heterochromatin in differentiating muscle requires HDAC activity. Exp Cell Res 310:344–356
Thijssen PE, Ito Y, Grillo G, Wang J, Velasco G, Nitta H, Unoki M, Yoshihara M, Suyama M, Sun Y et al (2015) Mutations in CDCA7 and HELLS cause immunodeficiency-centromeric instability-facial anomalies syndrome. Nat Commun 6:7870
Ting DT, Lipson D, Paul S, Brannigan BW, Akhavanfard S, Coffman EJ, Contino G, Deshpande V, Iafrate AJ, Letovsky S et al (2011) Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science 331:593–596
Tittel-Elmer M, Bucher E, Broger L, Mathieu O, Paszkowski J, Vaillant I (2010) Stress-induced activation of heterochromatic transcription. PLoS Genet 6:e1001175
Topp CN, Zhong CX, Dawe RK (2004) Centromere-encoded RNAs are integral components of the maize kinetochore. Proc Natl Acad Sci USA 101:15986–15991
Toubiana S, Velasco G, Chityat A, Kaindl AM, Hershtig N, Tzur-Gilat A, Francastel C, Selig S (2018) Subtelomeric methylation distinguishes between subtypes of immunodeficiency, centromeric instability and facial anomalies syndrome. Hum Mol Genet 27:3568–3581
Unoki M, Funabiki H, Velasco G, Francastel C, Sasaki H (2019) CDCA7 and HELLS mutations undermine nonhomologous end joining in centromeric instability syndrome. J Clin Invest 129:78–92
Unoki M, Sharif J, Saito Y, Velasco G, Francastel C, Koseki H, Sasaki H (2020) CDCA7 and HELLS suppress DNA:RNA hybrid-associated DNA damage at pericentromeric repeats. Sci Rep 10:17865
Valgardsdottir R, Chiodi I, Giordano M, Rossi A, Bazzini S, Ghigna C, Riva S, Biamonti G (2008) Transcription of satellite III non-coding RNAs is a general stress response in human cells. Nucleic Acids Res 36:423–434
Varley JM, Macgregor HC, Erba HP (1980) Satellite DNA is transcribed on lampbrush chromosomes. Nature 283:686–688
Velasco G, Francastel C (2019) Genetics meets DNA methylation in rare diseases. Clin Genet 95:210–220
Vissel B, Nagy A, Choo KH (1992) A satellite III sequence shared by human chromosomes. 13, 14, and 21 that is contiguous with alpha satellite DNA. Cytogenet Cell Genet 61:81–86
Volpe TA, Kidner C, Hall IM, Teng G, Grewal SIS, Martienssen RA (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297:1833–1837
Vourc’h C, Biamonti G (2011) Transcription of satellite DNAs in mammals. Prog Mol Subcell Biol 51:95–118
Wako T, Houben A, Furushima-Shimogawara R, Belyaev ND, Fukui K (2003) Centromere-specific acetylation of histone H4 in barley detected through three-dimensional microscopy. Plant Mol Biol 51:533–541
Wijchers PJ, Geeven G, Eyres M, Bergsma AJ, Janssen M, Verstegen M, Zhu Y, Schell Y, Vermeulen C, de Wit E et al (2015) Characterization and dynamics of pericentromere-associated domains in mice. Genome Res 25:958–969
Wong LH, Brettingham-Moore KH, Chan L, Quach JM, Anderson MA, Northrop EL, Hannan R, Saffery R, Shaw ML, Williams E et al (2007) Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res 17:1146–1160
Xu GL, Bestor TH, Bourc’his D, Hsieh CL, Tommerup N, Bugge M, Hulten M, Qu X, Russo JJ, Viegas-Péquignot E (1999) Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402:187–191
Zhu H, Geiman TM, Xi S, Jiang Q, Schmidtmann A, Chen T, Li E, Muegge K (2006) Lsh is involved in de novo methylation of DNA. EMBO J 25:335–345
Zhu Q, Pao GM, Huynh AM, Suh H, Tonnu N, Nederlof PM, Gage FH, Verma IM (2011) BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature 477:179–184
Zhu Q, Hoong N, Aslanian A, Hara T, Benner C, Heinz S, Miga KH, Ke E, Verma S, Soroczynski J et al (2018) Heterochromatin-encoded satellite RNAs induce breast Cancer. Mol Cell 70:842–853e7
Acknowledgments
We are grateful to Ekaterina Boyarchuk for the critical reading of the manuscript and Florent Hubé for the help with figures.
We thank our tutelles University de Paris, Centre National de la Recherche Scientifique (CNRS) and Institut National de la Recherche Médicale (INSERM). Work in C.F.’s lab is supported by research grants from Agence Nationale pour la Recherche (grant ANR-19-CE12-0022), Ligue Nationale Contre le Cancer, Fondation ARC and Fondation Maladies Rares. P.M. is a recipient of a fellowship from the French Ministry of Research through the Ecole Universitaire Génétique et Epigénétique Nouvelle Ecole (EUR GENE).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Mihìc, P., Hédouin, S., Francastel, C. (2021). Centromeres Transcription and Transcripts for Better and for Worse. In: Ugarković, Ð. (eds) Satellite DNAs in Physiology and Evolution. Progress in Molecular and Subcellular Biology, vol 60. Springer, Cham. https://doi.org/10.1007/978-3-030-74889-0_7
Download citation
DOI: https://doi.org/10.1007/978-3-030-74889-0_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-74888-3
Online ISBN: 978-3-030-74889-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)