Abstract
The brain tumor segmentation task aims to classify tissue into the whole tumor (WT), tumor core (TC) and enhancing tumor (ET) classes using multimodel MRI images. Quantitative analysis of brain tumors is critical for clinical decision making. While manual segmentation is tedious, time-consuming, and subjective, this task is at the same time very challenging to automatic segmentation methods. Thanks to the powerful learning ability, convolutional neural networks (CNNs), mainly fully convolutional networks, have shown promising brain tumor segmentation. This paper further boosts the performance of brain tumor segmentation by proposing hyperdense inception 3D UNet (HI-Net), which captures multi-scale information by stacking factorization of 3D weighted convolutional layers in the residual inception block. We use hyper dense connections among factorized convolutional layers to extract more contexual information, with the help of features reusability. We use a dice loss function to cope with class imbalances. We validate the proposed architecture on the multi-modal brain tumor segmentation challenges (BRATS) 2020 testing dataset. Preliminary results on the BRATS 2020 testing set show that achieved by our proposed approach, the dice (DSC) scores of ET, WT, and TC are 0.79457, 0.87494, and 0.83712, respectively.
S. Qamar and P. Ahmad—Equal contribution.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Primary and secondary are two types of brain tumors. Primary brain tumors originate from brain cells, whereas secondary tumors metastasize into the brain from other organs. Gliomas are primary brain tumors. Gliomas can be further sub-divided into two parts: low-grade (LGG) and high-grade (HGG). High-grade gliomas are an aggressive type of malignant brain tumor that proliferates, usually requires surgery and radiotherapy, and has a poor survival prognosis. Magnetic resonance imaging (MRI) is a critical diagnostic tool for brain tumor analysis, monitoring, and surgery planning. Usually, several complimentary 3D MRI modalities - such as T1, T1 with contrast agent (T1c), T2, and fluid attenuation inversion recover (FLAIR) are required to emphasize different tissue properties and areas of tumor spread. For example, the contrast agent, usually gadolinium, emphasizes hyperactive tumor subregions in T1c MRI modality.
Deep learning techniques, especially CNNs, are prevalent for the automatic segmentation of brain tumors. CNN can learn from examples and demonstrate state-of-the-art segmentation accuracy both in 2D natural images and in 3D medical image modalities. The information of segmentation provides an accurate, reproducible solution for further tumor analysis and monitoring. Multi-modal brain tumor segmentation challenge (BRATS) aims to evaluate state-of-the-art methods for the segmentation of brain tumors by providing a 3D MRI dataset with ground truth labels annotated by physicians [1,2,3,4, 14]. A 3D UNet is a popular CNN architecture for automatic brain tumor segmentation [8]. The multi-scale contextual information of the encoder-decoder sub-networks is effective for the accurate brain tumor segmentation task. Several variations of the encoder-decoder architectures were proposed for MICCAI BraTS 2018 and 2019 competitions. The potential of several deep architectures [12, 13, 17] and their ensembling procedures for brain tumor segmentation was discussed by a top-performing method [11] for MICCAI BRATS 2017 competition. Wang et al. [18] proposed architectures with factorized weighted layers to save the GPU memory and the computational time. At the same time, the majority of these architectures used either the bigger input sizes [16] or cascaded training [10] or novel pre-processing [7] and post-processing strategies [9] to improve the segmentation accuracy. In contrast, few architectures demonstrate the important memory consumption of 3D convolutional layers. Chen et al. [5] used an important concept in which each weighted layer was split into three branches in a parallel fashion, each with a different orthogonal view, namely axial, sagittal, and coronal. However, more complex combinations exist between features within and in-between different orthogonal views, which can significantly increase the learning representation [6]. Inspired by the S3D UNet architecture [5, 19], we propose a variant encoder-decoder based architecture for the brain tumor segmentation. The key contributions of our study are as follows:
-
A novel hyperdense inception 3D UNet (HI-Net) architecture is proposed by stacking factorization of 3D weighted convolutional layers in the residual inception block.
-
In each residual inception block, hyper-dense connections are used in-between different orthogonal views to learn more complex feature representation.
-
Our network achieves state-of-the-art performance as compared to other recent methods.
2 Proposed Method
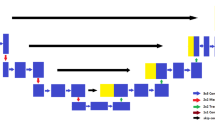
Figure 1 shows the proposed HI-Net architecture for brain tumor segmentation. The network’s left side works as an encoder to extract the features of different levels, and the right component of the network acts as a decoder to aggregate the features and the segmentation mask. The modified residual inception blocks of the encoder-decoder sub-networks have two 3D convolutional layers, and each layer has followed the structure of Fig. 2(b). In contrast, traditional residual inception blocks are shown in Fig. 2(a). This study employed inter-connections of dense connections within and in-between different orthogonal views to learn more complex feature representation. In the stage of encoding, the encoder extracts feature at multiple scales and create fine-to-coarse feature maps. Fine feature maps contain low-level features but more spatial information, while coarse feature maps provide the opposite. Skip connection is used to combine coarse and fine feature maps for accurate segmentation. Unlike standard residual UNet, the encoder sub-network uses a self-repetition procedure on multiple levels to generate semantic maps for fine feature maps and thus select relevant regions in the fine feature maps to concatenate with the coarse feature maps.
Proposed HI-Net architecture. The element-wise addition operations (\(+\) symbol with the oval shape) are employed to design the proposed architecture. The modified residual inception blocks (violet), known as hyperdense residual inception blocks, are used in the encoder-decoder paths. The length of the encoder path is longer than the decoder part by performing repetitions on several levels. The maximum repetition is 4 on the last level of the encoder part to draw the semantic information from the lowest input resolution. Finally, the softmax activation is performed for the outcomes. (Color figure online)
3 Implementation Details
3.1 Dataset
The BRATS 2020 [1,2,3,4, 14] training dataset included 369 cases (293 HGG and 76 LGG), each with four rigidly aligned 3D MRI modalities (T1, T1c, T2, and FLAIR), resampled to \(1\times 1\times 1\) mm isotropic resolution and skull-stripped. The input image size is \(240\times 240\times 155\). The data were collected from 19 institutions, using various MRI scanners. Annotations include 3 tumor subregions: WT, TC, and ET. Two additional datasets without the ground truth labels are provided for validation and testing. These datasets required participants to upload the segmentation masks to the organizers’ server for evaluations. In validation (125 cases) and testing (166) datasets, each subject includes the same four modalities of brain MRI scans but no ground truth. In our experiment, the training set is applied to optimize the trainable parameters in the network. The validation and testing sets are utilized to evaluate the performance of the trained network.
3.2 Experiments
The network is implemented by Keras and trained on Tesla V100–SXM2 32 GB GPU card with a batch size of 1. Adam optimizer with an initial learning rate \(3\times 10{^{-5}}\) is employed to optimize the parameters. The learning rate is reduced by 0.5 per 30 epochs. The network is trained for 350 epochs. During network training, augmentation techniques such as random rotations and mirroring are employed. The size of the input during the training of the network is \(128\times 128\times 128\). The multi-label dice loss function [15] addressed the class imbalance problem. Equation 1 shows the mathematical representation of loss function.
where \(P_{(j, d)}\) and \(T_{(j, d)}\) are the prediction obtained by softmax activation and ground truth at voxel j for class d, respectively. D is the total number of classes.
3.3 Evaluation Metrics
Multiple criteria are computed as performance metrics to quantify the segmentation result. Dice coefficient (DSC) is the most frequently used metric for evaluating medical image segmentation. It measures the overlap between the segmentation and ground truth with a value between 0 and 1. The higher the Dice score, the better the segmentation performance. Sensitivity and specificity are also commonly used statistical measures. The sensitivity called true positive rate is defined as the proportion of positives that are correctly predicted. It measures the portion of tumor regions in the ground truth that is also predicted as tumor regions by the segmentation method. The specificity, called true negative rate, is defined as the proportion of correctly predicted negatives. It measures the portion of normal tissue regions in the ground truth that is also predicted as normal tissue regions by the segmentation method.
3.4 Results
The performance of our proposed architecture is evaluated on training, validation, and the testing sets provided by BRATS 2020. Table 1 presents the quantitative analysis of the proposed work. We have secured mean DSC scores of ET, WT, and TC as 0.74191, 0.90673, and 0.84293, respectively, on the validation dataset, while 0.80009, 0.92967, and 0.90963 on the training dataset. At the same time, our proposed approach obtained mean DSC scores of ET, WT, and TC as 0.79457, 0.87494, and 0.83712, respectively, on the testing dataset. In Table 1, sensitivity and specificity are also presented on training, validation, and the testing datasets. Table 2 shows the comparable study of proposed work with the baseline work [5]. Our proposed HI-Net achieves higher scores for each tumor than the baseline work. Furthermore, ablation studies are conducted to assess the modified residual inception blocks’ influence with and without the inter-connected dense connections. The influence of these connections on DSCs of ET, WT, and TC is shown in Table 2. To provide qualitative results of our method, three-segmented images from training data are shown in Fig 3. In summary, modified inception blocks significantly improve the DSCs of ET, WT, and TC against the baseline inception blocks.
4 Conclusion
We proposed a HI-Net architecture for brain tumor segmentation. Each 3D convolution is splitted into three parallel branches in the residual inception block, each with different orthogonal views, namely axial, sagittal and coronal. We also proposed hyperdense connections among factorized convolutional layers to extract more contextual information. The HI-Net architecture secures high DSC scores for all types of tumors. This network has been evaluated on the BRATS 2020 Challenge testing dataset and achieved average DSC scores of 0.79457, 0.87494, and 0.83712 for the segmentation of ET, WT, and TC, respectively. Compared with the performance of the validation dataset, the scores on the testing set are higher. In the future, we will work to enhance the robustness of the network to improve the segmentation performance by using some post-processing methods such as a fully connected conditional random field (CRF).
References
Bakas, S., et al.: Segmentation labels and radiomic features for the pre-operative scans of the TCGA-GBM collection. The Cancer Imaging Archive (2017) (2017)
Bakas, S., et al.: Segmentation labels and radiomic features for the pre-operative scans of the TCGA-LGG collection. Cancer Imaging Archive 286 (2017)
Bakas, S., et al.: Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Scientific Data 4, 170117 (2017). https://doi.org/10.1038/sdata.2017.117 10.0.4.14/sdata.2017.117
Bakas, S., et al.: Identifying the Best Machine Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and Overall Survival Prediction in the BRATS Challenge. CoRR abs/1811.0 (2018), http://arxiv.org/abs/1811.02629
Chen, W., Liu, B., Peng, S., Sun, J., Qiao, X.: S3D-UNet: separable 3D U-Net for brain tumor segmentation. In: Crimi, A., Bakas, S., Kuijf, H., Keyvan, F., Reyes, M., van Walsum, T. (eds.) BrainLes 2018. LNCS, vol. 11384, pp. 358–368. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-11726-9_32
Dolz, J., Gopinath, K., Yuan, J., Lombaert, H., Desrosiers, C., Ayed, I.B.: HyperDense-Net: a hyper-densely connected CNN for multi-modal image segmentation. CoRR abs/1804.0 (2018), http://arxiv.org/abs/1804.02967
Feng, X., Tustison, N., Meyer, C.: Brain tumor segmentation using an ensemble of 3D U-Nets and overall survival prediction using radiomic features. In: Crimi, A., Bakas, S., Kuijf, H., Keyvan, F., Reyes, M., van Walsum, T. (eds.) BrainLes 2018. LNCS, vol. 11384, pp. 279–288. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-11726-9_25
Isensee, F., Kickingereder, P., Wick, W., Bendszus, M., Maier-Hein, K.H.: Brain Tumor Segmentation and Radiomics Survival Prediction: Contribution to the BRATS 2017 Challenge. CoRR abs/1802.1 (2018), http://arxiv.org/abs/1802.10508
Isensee, F., Kickingereder, P., Wick, W., Bendszus, M., Maier-Hein, K.H.: No New-Net. In: Crimi, A., Bakas, S., Kuijf, H., Keyvan, F., Reyes, M., van Walsum, T. (eds.) BrainLes 2018. LNCS, vol. 11384, pp. 234–244. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-11726-9_21
Jiang, Z., Ding, C., Liu, M., Tao, D.: Two-stage cascaded U-Net: 1st place solution to BraTS challenge 2019 segmentation task. In: Crimi, A., Bakas, S. (eds.) BrainLes 2019. LNCS, vol. 11992, pp. 231–241. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-46640-4_22
Kamnitsas, K., et al.: Ensembles of Multiple Models and Architectures for Robust Brain Tumour Segmentation. CoRR abs/1711.0 (2017), http://arxiv.org/abs/1711.01468
Kamnitsas, K., Ledig, C., Newcombe, V.F.J., Simpson, J.P., Kane, A.D., Menon, D.K., Rueckert, D., Glocker, B.: Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 36, 61–78 (2017)
Long, J., Shelhamer, E., Darrell, T.: Fully Convolutional Networks for Semantic Segmentation. CoRR abs/1411.4 (2014), http://arxiv.org/abs/1411.4038
Menze, B.H., et al.: The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 34(10), 1993–2024 (2015). https://doi.org/10.1109/TMI.2014.2377694
Milletari, F., Navab, N., Ahmadi, S.A.: V-Net: fully convolutional neural networks for volumetric medical image segmentation. CoRR abs/1606.0 (2016), http://arxiv.org/abs/1606.04797
Myronenko, A.: 3D MRI brain tumor segmentation using autoencoder regularization. CoRR abs/1810.1 (2018), http://arxiv.org/abs/1810.11654
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. CoRR abs/1505.0 (2015), http://arxiv.org/abs/1505.04597
Wang, G., Li, W., Ourselin, S., Vercauteren, T.: Automatic Brain Tumor Segmentation using Cascaded Anisotropic Convolutional Neural Networks. CoRR abs/1709.0 (2017), http://arxiv.org/abs/1709.00382
Xie, S., Sun, C., Huang, J., Tu, Z., Murphy, K.: Rethinking Spatiotemporal Feature Learning For Video Understanding. CoRR abs/1712.0 (2017), http://arxiv.org/abs/1712.04851
Acknowledgment
This work is supported by the National Natural Science Foundation of China under Grant No. 91959108.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Qamar, S., Ahmad, P., Shen, L. (2021). HI-Net: Hyperdense Inception 3D UNet for Brain Tumor Segmentation. In: Crimi, A., Bakas, S. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2020. Lecture Notes in Computer Science(), vol 12659. Springer, Cham. https://doi.org/10.1007/978-3-030-72087-2_5
Download citation
DOI: https://doi.org/10.1007/978-3-030-72087-2_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-72086-5
Online ISBN: 978-3-030-72087-2
eBook Packages: Computer ScienceComputer Science (R0)