Abstract
Detecting suspicious lesions in MRI imaging is a critical task in preventing deaths from cancer. Deep learning systems have produced remarkable accuracy for the task of detecting lesions in MRI images. Although these systems show remarkable performance, they often ignore an indispensable component which is interpretability. Interpretability is essential for many deep learning applications in medicine because of ethical, monetary, and legal factors. Interpretation also builds a necessary degree of trust and transparency between the doctor, patient, and system. This work proposes a framework for the interpretation of medical deep learning systems. The proposed approach is based on the idea that it is advantageous to use different interpretation techniques to show multiple views of reasoning behind the classification. This work demonstrates deep learning interpretations for various patient data modalities using the proposed Multiple Views of Interpretation for Deep Learning framework.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
There have been recent advances using deep learning techniques, such as convolutional neural networks [1], to detect prostate cancer from MRI images with impressive performance. [2] showed deep learning can detect prostate cancer with accuracy suitable to be integrated into a clinical environment. These results demonstrate the great potential for using deep learning to aid medical practitioners. However, these advances often ignore an indispensable component of such systems which is interpretability.
Although deep learning models can produce accurate cancer classification, they are often treated as black-box models that lack interpretability and transparency of their inner working [3]. The models provide an accurate classification but do not demonstrate how they arrived at the decision. If such systems are to be implemented into medical settings, integrating interpretability is an essential, often overlooked component. Interpretability is needed for various reasons. First, there are legal and ethical requirements along with laws and regulations that are required for deep learning cancer detection systems to be implemented in a clinical setting. An example of a regulation is the European Union’s General Data Protection Regulation (GDPR) requiring organizations that use patient data for classifications and recommendations to provide on-demand explanations [4]. The inability to provide such explanations on demand may result in large penalties for the organizations involved. Thus, there are monetary incentives associated with interpretable deep learning models. Beyond ethical and legal issues, clinicians and patients need to be able to trust the classifications provided by these systems. Interpretation attempts to show the reasoning behind the model’s classification thus building a degree of trust between the system, clinician, and patient. Theoretically, this will reduce the number of misdiagnosed cases that would be a possible consequence of non-interpretable systems. Finally, interpretable deep learning systems will provide the clinician with practical features as a second-order effect. Examples of these practical features are the ability to provide segmentation of a medical region of interest (ROI) [5] and the localization of lesions [6].
Interpretation methods can be categorized as post-hoc and ad-hoc. Post-hoc refers to interpretation after the classification is made whereas ad-hoc refers to engineering interpretation into the deep learning system. This work will largely focus on post-hoc approaches. There are various types of interpretation techniques that highlight different aspects of classification for the same sample. Some of them highlight the localization of a lesion and others highlight the size or area of a tumor or cyst. This paper will shed light on the importance of interpretation for medical deep learning systems, the current state of interpretation for deep learning within a medical context, and will propose a viable approach for medical deep learning interpretation moving forward. The main contributions of this paper are (1) showing that the integration of multiple interpretation techniques produce a new quality and delivers greater insight into the model’s classification opposed to using a single method (2) establish an evaluation methodology for measuring visual interpretation performance (3) demonstrate that Grad-CAM can precisely localize prostate lesions in T2W and ADC MRI images.
2 Related Work
2.1 Classification of Prostate Cancer and Lesions
Deep learning has been widely applied to the classification of medical conditions ranging from diabetes to cancer. There are attempts to use deep learning techniques to detect cancer, some of which produce remarkable performance. [7] demonstrate a transfer learning approach to detect prostate lesions using MRI images. The study implements the InceptionV3 and VGG16 models which were both initialized with imagenet weights for the task of detecting prostate lesions using the PROSTATEx dataset [8] Transfer learning is used in many cancer detection systems because of the advantage from initializing a network with pre-trained parameters. Ensemble learning techniques were implemented to improve the area under the curve. Their results range from 0.82 to 0.91 for AUROC. [9] implement a three-dimensional convolutional neural network for the task of classifying clinically significant lesions. The study reports an AUROC of 0.80 and argues that with the 3D network, spatial information is captured thus producing a model with greater insight into 3D medical volumes. [10] proposed a method called XmasNet. Their work performs novel data augmentation using three-dimensional rotation and slicing, in order to incorporate the three-dimensional information of the lesion volume. The study reports an AUC of 0.92. [11] propose a fully automated approach to prostate lesion detection using MRI images reporting an AUROC of 0.84 (Fig. 1).
2.2 Post-Hoc Interpretation for Deep Learning in Medicine
Post-hoc interpretation attempts to provide reasoning after the classification is made as opposed to engineering interpretation into the deep learning system. These post-hoc visual interpretation techniques generally either use perturbation forward propagation, backward propagation, or gradient-based visual explanation methods. Perturbation forward propagation make perturbations to individual inputs or neurons and observe the impact on later neurons in the network. Backward propagation is the opposite. Instead of propagating forward through the network, a signal is propagated from the output neuron(s) back to the input neurons. Gradient-based methods propose taking the gradient of an output variable with respect to input variables to calculate which input variables change the outcome the most. [12] examine post-hoc interpretation for the classification of melanoma in histology slides. Their work trains a ResNet50 and VGG19 using transfer learning to for the classification of melanoma. Class activation map (CAM) [13] is used to provide a post-hoc visual explanation. [14] design a neural network to perform analysis on frames collected from an endoscopic examination taken from a video stream. This uses Grad-CAM and guided Grad-CAM as gradient-based post-hoc interpretation techniques to explain findings in these frames. [15] show post-hoc interpretability using LIME for clinical data. [16] show post-hoc interpretability using LIME for acute kidney injury in cardiac surgery patients. Their work uses LIME to attempt to explain the onset of the condition. [17] created a deep learning system to classification hypertension and then used LIME to explain these classifications. [18] developed a model to classifications diabetic retinopathy progression in individual patients. They then use SHAP to give insight into the model’s decisions.
Emerging literature has highlighted the ability to use these interpretation techniques to localize lesions or other medical regions of interest. [19] use a modified version of Grad-CAM, coined pyramid gradient-based class activation mapping (PG-CAM), to localize meningioma. They report a 23% increase from vanilla Grad-CAM for the localization of brain tumors. [20] propose high-resolution CAM (HR-CAM) which aggregates feature maps together. They localize ependymomas using this technique. [21] use saliency maps to segment lesions in dermoscopy images with a DICE coefficient of 0.858. To the best of our knowledge, there are not any studies that localize prostate lesions from MRI images using interpretation techniques.
3 Multiple Views of Interpretations for Deep Learning
Interpretation methods (i.e. Grad-Cam, LIME, SHAP, saliency maps) show a certain aspect of the reasoning behind a deep learning model’s classification. Each interpretation method by itself provides some method-specific information. For example, grad-CAM highlights an area of interest whereas LIME highlights which clinical features contribute the most to the classification. Saliency maps show the important structure and clusters of important individual pixels. These individual interpretation techniques can be considered individual parts of a larger system. By combining these methods together, a higher quality interpretation is produced.
Using a multiple interpretation approach is advantageous for multiple reasons. First, the clinician and patient will receive more insight into the model’s classification using a combination of methods as opposed to a sole method. Second, you gain a higher degree of confidence using an approach which includes multiple interpretations. If the different methods are in unison, then the consistency delivers a degree of confidence in the interpretation and classification. Third, it is possible for interpretations can be fooled and produce misleading interpretations. Using multiple interpretations approach, theoretically, you can uncover issues with the classification because of a lack of consistency between interpretations. Lastly, you can provide an interpretation for many different data modalities (i.e. images, genetic information, clinical information, patient history) which is essential for healthcare applications.
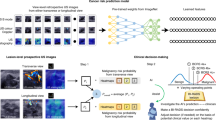
This work presents a framework for interpretation, Multiple Views of Interpretation for Deep Learning, for prostate lesion detection and interpretation (Fig. 2). Combining multiple interpretation methods will increase transparency and give a multifaceted view into how the model arrives at the classification thus providing a holistic interpretation.
3.1 Deep Learning Model
In this paper, convolution neural network (CNN) based on the VGG16 [22] was implemented with some improvements. InceptionV3, VGG16, VGG19, ResNet50, MobileNet, and WideResNet were all implemented and compared to select the optimal model. Each network’s hyper-parameters were tuned using grid search. After tuning, the VGG16 produced the highest performance. Two extra convolutional layers followed by max-pooling were concatenated to the end for increased classification performance. The network is designed to classify individual slices with a lesion from slices without a lesion. Clinical features are concatenated to the fully connected layers to incorporate patient records. The hyper-parameters after tuning were: 18 layers, a weight decay of 0.00001, a learning rate of 0.001, the ADAM optimizer, the binary cross-entropy loss function, a softmax activation as the final layer, and the network was initialized with imagenet weights (Fig. 3).
3.2 Interpretation Techniques for Prostate Lesion Detection
Combining interpretation techniques produces a more transparent system by giving a holistic view of the model’s decision. Each technique gives unique insight, therefore, to get a comprehensive interpretation, each technique is needed as individual parts of a larger system. For this work, these techniques are split into two categories: image data interpretation and clinical data interpretation. Image data refers to MRI imaging in this work and clinical data refers to patient characteristics including weight, age, height, and body mass index (BMI). The techniques for image interpretation are focused solely on providing explanations for image data thus do not take into account clinical information. The second category is clinical data interpretation which takes into account clinical information but does not provide an explanation for image data. This work shows Grad-CAM [23] and saliency maps [24, 25] as the techniques for image data. SHAP [26] and LIME [27] as the techniques for clinical data. Each technique will be explained in detail below.
Gradient-weighted Class Activation Mapping (Grad-CAM) uses the gradients flowing into a convolutional layer to produce a map that highlights important regions in the image for the classification of a class.
Grad-CAM calculates the gradients of an individual class score, c, with respect to feature map, A.
Neuron importance is calculated by globally average pooling the gradients.
A weighted combination of forward activation maps is followed by a ReLU to obtain the final activation map. The advantage of Grad-CAM is it takes into account feature maps thus showing how well your model learns quality features. This is important when training a model for clinical diagnosis because you can examine if your model is learning interpretable features using Grad-CAM. A disadvantage is the heatmaps can be unclear.
Saliency maps show the individual pixels that contribute the most to the class score. This is useful for showing the structures and clusters of pixels that contribute the most to the class score. Mathematically, saliency maps calculate the partial derivative of the class score with respect to an individual pixel at a specific pixel.
It repeats this process for each pixel in the image and assigns each pixel a numeric value. This value represents the contribution to the class score for each pixel. You will notice in the results, the highlighting of the structure as well as pixel clustering around ‘important’ areas in the image. The advantage of saliency maps is they highlight individual pixels that are important thus providing a precise interpretation. The disadvantage is individual pixels may not matter as much as clusters of pixels (i.e. feature maps).
Local Interpretable Model-agnostic Explanations (LIME) creates an interpretable model locally around a classification. It produces a bar graph showing the contribution of each feature from the patient records. Each bar shows the direction and magnitude of contribution.
The explanation produced by LIME is obtained by the following:
Where f is a machine learning model, g is the explanation defined as a model, pix(z) is used as a proximity measure between an instance z to x, so as to define locality around x, and omega(g) is a measure of complexity. The advantages of LIME are it produces a clear and concise graph that is easily interpretable. The disadvantage is the method does not support multi-modal input.
SHAP (SHapley Additive exPlanations is a unified, model-agnostic approach to interpretation based on game theoretically optimal Shapley Values. The way SHAP calculates feature importance is as follows.
Where g is the explanation model, z is the coalition vector, M is the maximum coalition size, and phi is the feature attribution for feature j. For global importance, we average the absolute Shapley values per feature across the data as shown below. The advantage of SHAP is that it provides a model-agnostic, personalized global and local interpretation.
4 Dataset
The dataset used for this work was the PROSTATEx dataset [28] from the SPIE-AAPM-NCI PROSTATEx challenge. The PROSTATEx dataset consists of 330 lesions from 204 patients. The dataset provides DICOM coordinates for the centroid of the prostate lesions. The lesions in the dataset were labeled as clinically significant or not clinically significant depending on their pi-rads score. The dataset provides T2W transaxial, T2W sagittal, T2W coronal, ADC, and BVAL image modalities. This study includes T2W and ADC images. Six patients were excluded from this work due to poor image quality. This results in 199 patients and 322 lesions. The test set for both classification and interpretation includes 103 images of lesions and 103 images without lesions.
The data preprocessing steps are as follows: The T2W images are downsized from \(350\times 350\) to \(224\times 224\) pixels. The ADC images are downsized from \(120\times 80\) to \(50\times 50\) pixels. All images are then converted to RGB images. The pixel values are normalized using z-scoring. Then data augmentation is carried out using shearing, rotation, and translating data augmentation techniques. The lesion centroid coordinates are then converted to the resized coordinate frame using the following formulas:
5 Experimental Results
This section is split into four sub-sections: classification results, image interpretation results, clinical data interpretation results, and Multiple Views for Interpretation for Deep Learning results. In Sect. 5.1, lesion classification results are introduced after tuning the parameters of methodology with ProstateX data set. The image interpretation shows the different visualizations used to gain insight into the classifications. This part also demonstrates the precision of the localization of prostate lesions using image interpretation technique (Grad-Cam) in Sect. 5.2. In Sect. 5.3, clinical data interpretation results demonstrate local and global clinical data interpretation with LIME and SHAP interpretation techniques. Lastly, the advantages of using multiple interpretation techniques are demonstrated in Sect. 5.4.
5.1 Classification Result
The classification results are shown in Table 1. These results demonstrate that engineering interpretation into deep learning can still produce models with classification performance. As can we see on Table 1, our work (VGG Net) has almost similar result when we compare with previous works for accuracy. Although XmasNet has better values, our results are close enough to go second step which are interpretation techniques. These results also show that true positives and false positives are captured using this approach. False negatives and false positives are likely to be correctly filtered out and classified correctly. It provides a credible model with results comparable to models in relevant literature to demonstrate the interpretation techniques.
5.2 Evaluation of Grad-Cam Precision Results
Grad-CAM highlights the area of the image that contributes most to the classification. An additional finding is this highlights the lesion centroid in MRI images with high precision. To measure interpretation quality for Grad-CAM, the assumption is made that a credible interpretation would highlight the lesion centroid as the most important area. That is, if the slice contained a lesion in the ground truth. We propose the following performance measures for interpretation for image data: the distance between centroids, false positives, false positive, correctly localized, and incorrectly localized.
Incorrectly localized interpretations are measured as the number of samples that produce a heatmap that does not accurately highlight lesion, see Fig. 6. The heatmap is considered correctly localized if the centroid of the lesions falls within the radius of the heatmap. False Positives are measured as the number of cases that produce a heatmap given a slice without a lesion, see Fig. 5. False Negatives are measured as the number of cases that do not show a heatmap given an input slice that contains a lesion, see Fig. 4.
The interpretation is considered correctly localized if the coordinates of the lesion centroid are located within the radius of the heat map. If this does not hold true then it is considered incorrectly localized. The threshold is the radius of the heat map which varies from 5 pixels to 16 pixels. Table 2 shows the errors for Grad-CAM visualizations in terms of false positives, false negatives, and incorrect localization. Examples of false negatives, false positives, and incorrect localization are shown in Figs. 4, 5, and 6 respectively.
Distance is calculated between the centroid of the lesion and the geometric center of the heatmap using the distance formula shown below:
103 images were analyzed and the precision of lesion localization was calculated using Grad-CAM. T2W images were \(224\times 224\) pixels and ADC images were \(50\times 50\) pixels. Figure 7 and Fig. 8 show the precision of Grad-Cam for T2W and ADC images respectively. The interpretation results for Grad-CAM, measured as a localization task, for image data interpretation are a mean distance of 6.93 for T2W images and a mean distance of 16.3 for ADC images. These results do not only show that the interpretation is clear for clinicians, but also that this method can precisely localize lesion location.
Grad-CAM is useful when you want the interpretation to highlight high-level (i.e. human interpretable) features. Examples of this would be tumor classification and neuroimaging studies. This method is also useful because of its ability to localize a region of interest such as a prostate lesion. This method does not work as well if you want to highlight the boundaries precisely.
Figure 7 and Fig. 8 show how T2W and ADC images distribute precision of Grad-Cam in percentage and pixel-wise. Based on these figures, the average sample is more likely to be localized accurately in T2W images opposed to ADC images. The inaccuracy in ADC images can be contributed to significantly lower image resolution (Table 3).
Saliency maps show the importance of the structure within the image. They also show which pixels are important for the classification. Thus with saliency maps, you can examine where the clusters of pixels are. These clusters are the areas the model considers most important. This is how saliency maps differ from Grad-CAM. Grad-CAM focuses on feature maps whereas saliency maps focus on individual pixels. If the cluster of pixels is in the same region as the Grad-CAM heatmap that shows consistency between methods and instills confidence in the classification. This provides a sense of trust for the interpretation that the area of the image contributes most to the classification.
Saliency maps show the structure of the image well. Also, they are useful if you want to outline an object within the image. For example, if you want to extract tumor shape or orientation. Saliency maps work well if you want to visualize which pixels contribute the most to the classification. You can gain insight into the important regions by examining where the clusters of pixels are. This does not work well if you want to produce a clear, concise interpretation because this method is often unclear.
5.3 Interpretation Results for Clinical Data
In previous sections, we mentioned visual results with interpretation techniques. In this part, non-visual interpretation techniques which are LIME and SHAP were tested for patient record. For LIME, the model shows BMI and age are most important for the personalized interpretation. The SHAP values representing global feature importance are consistent with this showing that BMI and age the most important features globally. This shows consistency between the local and global methods thus providing a sense of trust that the interpretation is accurate. These results show which clinical features contribute most to the classification.
LIME is useful if you want individualized interpretation. An advantage of LIME is the graphs it provides. They show not only magnitude, but also direction of feature importance. They also are color coded and organized in a clear and concise manner. If you want to examine global feature importance it is better to not use LIME (Figs. 9 and 10).
SHAP is useful if you want to examine global feature importance along with local feature importance. SHAP is also useful because of it’s model-agnostic feature. It can show reasoning behind the classification regardless of what type of model you are using. SHAP also provides straightforward interpretation using SHAP value. Future work can include more variables for these interpretation methods such as patient history, genetic information, additional patient characteristics, and anything else the clinician deems appropriate (Fig. 11).
5.4 Multiple Views for Interpretation for Deep Learning Results
In this section, two cases are shown for end-to-end personalized interpretations using the Multiple Views for Interpretation for Deep Learning Results framework. This shows what regions of the images contribute to the classification, how the structure influences classification, what cluster of pixels are important, what global features matter the most across the entire cohort, and what clinical information contributes to the system’s decision. This shows that using multiple interpretation techniques provides more complete reasoning behind the prediction as opposed to using a single technique. Using Multiple Views for Interpretation for Deep Learning, we show: (1) what areas of the image contribute most the prediction (2) the importance of the structure of the prostate and surrounding area (3) what individual pixels, and clusters of pixels, contribute the most (4) what features from the patient’s medical records contribute the most (5) localization of the lesion and (6) what medical record features contribute the most globally across the cohort. This provides a more complete explanation than an individual technique. This combines the strength of each individual method to create a greater whole. There is consistency between the different image interpretation methods. This shows that the area where the heatmap is, and the pixels are clustered, is the most important region of the image for the classification. Since both interpretation techniques are consistent, there is a degree of trust in the reasoning the model gives for the classification. For clinical data interpretation, the local interpretations are consistent with global interpretations. This provides confidence the neural network is using sound reasoning to make life-critical classifications. In Figs. 12, 13, 14 and 15, each example is tested using image and patient records from the PROSTATEx dataset. Three patient were selected and tested with our framework. The first patient has a age, weight, BMI, and height of 58 years, 70 kg, 23 kg/m2 and 176 cm. The second patient has a age, weight, BMI, and height of 75 years, 80 kg, 28 kg/m\(^2\), and 200 cm.
Based on Fig. 12 and 13, the heatmap shows the area of the image that contributes the most to the class score. This heatmap highlights the lesion centroid. Saliency map shows the individual pixels that contribute the most to the class score. You can see a cluster of pixels at the lesion centroid. You can also see how it highlights the edges, shapes, and structure of the prostate area. SHAP shows global feature importance.
LIME shows the clinical information features that contribute the most to the class score. For this patient, his bmi and age contribute the most. The bmi is low so even though the network says the age contributes towards malignant, the bmi is low enough to give confidence he is healthy.
Based on Fig. 14 and 15, the heatmap shows the area of the image that contributes the most to the class score. This heatmap highlights the lesion centroid. Saliency map shows the individual pixels that contribute the most to the class score. You can see a cluster of pixels at the lesion centroid. You can also see how it highlights the edges, shapes, and structure of the prostate area. SHAP shows global feature importance. LIME shows the clinical information features that contribute the most to the class score. For this patient, his bmi and age contribute the most. The bmi and age are both high so the network considers this patient malignant overall.
6 Discussion and Future Work
This work proposes Multiple Views for Interpretation for Deep Learning which is an interpretation framework for deep learning medical systems. We utilize a deep convolution neural network for the task of image classification. This is used to demonstrates classification performance greater than, or on par with, similar prostate cancer classification models in the literature. Clinical information is concatenated to the fully connected layers of the CNN. Thus, we use image data (i.e. MRI images) and clinical data (i.e. patient information). This model is then used to show that multiple interpretation techniques gives greater insight compared to a single interpretation method. The network and interpretation methods are trained and tested on the PROSTATEx dataset because of the number of images and availability of patient information and lesion centroid location. The framework is extendable to other data modalities such as clinical notes or genetic data. The four methods included are Grad-CAM, saliency maps, SHAP, and LIME. Using Grad-CAM we show, not only that one can gain insight into the model’s classifications, but also precisely localize lesion location using this technique. Saliency maps show the importance of the structure and individual pixels that contribute most to the class score. The results from Grad-CAM are consistent with clusters of pixels from saliency maps showing agreement between image interpretation methods. For clinical data interpretation, LIME is used to show personalized classification using patient information such as weight, age, height, and BMI. SHAP is used to show global feature importance which can be used when examining LIME’s personalized interpretation to determine if the classification is reasonable. These techniques are then integrated to provide a multifaceted approach to deep learning interpretation within a medical context. An interesting additional finding is using Grad-CAM we were able to accurately localize lesions in slices that contained a lesion. This work shows and analyzes different approaches to handle interpretability, one of the problems that come along with computer-aided diagnosis systems. Using multiple interpretations, the number of incorrect diagnoses influenced by deep learning systems can be reduced. The legal issues that come along with these systems will be mitigated. The systems will be more successful and ethical in practice. This makes using these systems more ethical because the reasoning will enable these systems to work alongside clinicians as opposed to carrying out their tasks. Lastly, they will ensure a degree of trust and credibility by showing the reasoning behind the model’s decision. Most importantly, more work needs to be done to validate interpretation approaches in clinical settings and testing the generalizability of interpretation methods. Future work should also include working with clinicians to tailor interpretation methods to suit their specific needs. This future work also should study the integrity of such systems. If deep learning cancer detection systems are going to be implemented into clinical settings, it is of utmost importance that we trust the classifications. An approach to this is to implement multiple integrated post-hoc interpretability into these systems to provide a holistic interpretation of the model’s decision. The clinical validation of this hypothesis is an important future direction.
References
LeCun, Y., Haffner, P., Bottou, L., Bengio, Y.: Object recognition with gradient-based learning. Shape, Contour and Grouping in Computer Vision. LNCS, vol. 1681, pp. 319–345. Springer, Heidelberg (1999). https://doi.org/10.1007/3-540-46805-6_19
Antonelli, M., Johnston, E.W., Dikaios, N., et al.: Machine learning classifiers can classification Gleason pattern 4 prostate cancer with greater accuracy than experienced radiologists. Eur. Radiol. 29, 4754–4764 (2019). https://doi.org/10.1007/s00330-019-06244-2
Lipton, Z.C.: The mythos of model interpretability. CoRR, abs/1606.03490 (2016). http://arxiv.org/abs/1606.03490
Hoofnagle, C.J., van der Sloot, B., Borgesius, F.Z.: The European Union general data protection regulation: what it is and what it means. Inf. Commun. Technol. Law 28(1), 65–98 (2019). https://doi.org/10.1080/13600834.2019.1573501
Canalini, L., Pollastri, F., Bolelli, F., Cancilla, M., Allegretti, S., Grana, C.: Skin lesion segmentation ensemble with diverse training strategies. In: Vento, M., Percannella, G. (eds.) CAIP 2019. LNCS, vol. 11678, pp. 89–101. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-29888-3_8
Yoon, H.J., et al.: A lesion-based convolutional neural network improves endoscopic detection and depth classification of early gastric cancer. J. Clin. Med. 8(9), 1310 (2019). https://doi.org/10.3390/jcm8091310
Chen, Q., Hu, S., Long, P., Lu, F., Shi, Y., Li, Y.: A transfer learning approach for malignant prostate lesion detection on multiparametric MRI. https://doi.org/10.1177/1533033819858363
Litjens, G., Debats, O., Barentsz, J., Karssemeijer, N., Huisman, H.: Computer-aided detection of prostate cancer in MRI. IEEE Trans. Med. Imaging 33, 1083–1092 (2014). https://doi.org/10.1109/TMI.2014.2303821
Mehrtash, A., et al.: Classification of clinical significance of MRI prostate findings using 3D convolutional neural networks. In: Proceedings of SPIE-the International Society for Optical Engineering, vol. 10134, p. 101342A (2017). https://doi.org/10.1117/12.2277123
Liu, S., Zheng, H., Feng, Y., Li, W.: Prostate cancer diagnosis using deep learning with 3D multiparametric MRI. In: Armato, S.G., Petrick, N.A. (eds.) Medical Imaging (2017)
Wang, X., Yang, W., Weinreb, J., et al.: Searching for prostate cancer by fully automated magnetic resonance imaging classification: deep learning versus non-deep learning. Sci. Rep. 7, 15415 (2017)
Xie, P., Zuo, K., Zhang, Y., Li, F., Yin, M., Lu, K.: Interpretable classification from skin cancer histology slides using deep learning: a retrospective multicenter study (2019)
Zhou, B., Khosla, A., Lapedriza, A., Oliva, A., Torralba, A.: Learning DeepFeatures for discriminative localization. In: CVPR (2016)
Hicks, S., et al.: Dissecting deep neural networks for better medical image classification and classification understanding. In: 2018 IEEE 31st International Symposium on Computer-Based Medical Systems (CBMS), Karlstad, pp. 363–368 (2018)
Zhang, Z., et al.: Opening the black box of neural networks: methods for interpreting neural network models in clinical applications. Ann. Transl. Med. 6(11), 216 (2018). https://doi.org/10.21037/atm.2018.05.32
Da Cruz, H.F., et al.: Classification of acute kidney injury in cardiac surgery patients: interpretation using local interpretable model-agnostic explanations. In: HEALTHINF (2019)
Elshawi, R., Al-Mallah, M.H., Sakr, S.: On the interpretability of machine learning-based model for predicting hypertension. BMC Med. Inform. Decis. Mak. 19, 146 (2019). https://doi.org/10.1186/s12911-019-0874-0
Arcadu, F., Benmansour, F., Maunz, A., et al.: Deep learning algorithm predicts diabetic retinopathy progression in individual patients. NPJ Digit. Med. 2, 92 (2019)
Lee, S., et al.: Robust tumor localization with pyramid Grad-CAM. arXiv abs/1805.11393 (2018)
Shinde, S., Chougule, T., Saini, J., Ingalhalikar, M.: HR-CAM: precise localization of pathology using multi-level learning in CNNs. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11767, pp. 298–306. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32251-9_33
Jahanifar, M., et al.: Segmentation of lesions in dermoscopy images using saliency map and contour propagation. arXiv (2017)
Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition. In: ICLR (2015)
Selvaraju, R.R., et al.: Grad-CAM: visual explanations from deep networks via gradient-based localization. Int. J. Comput. Vis. 128(2), 336–359 (2019)
Armato III, S.G., Petrick, N.A.: Computer-aided diagnosis. In: SPIE Proceedings, vol. 10134. International Society for Optics and Photonics, Bellingham (2017). 1013428
K. Simonyan, A. Vedaldi, and A. Zisserman. Deep inside convolutional networks: Visualising image classification models and saliency maps. CoRR, abs/1312.6034, 2013. 3
Lundberg, S.M., Lee, S.I.: A unified approach to interpreting model classifications. In: Proceedings of the Advances in Neural Information Processing Systems, pp. 4768–4777 (2017)
Ribeiro, M.T., Singh, S., Guestrin, C.: Why should i trust you?: Explaining the classifications of any classifier. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp. 1135–1144. ACM (2016)
Litjens, G., Debats, O., Barentsz, J., Karssemeijer, N., Huisman, H.: PROSTATEx challenge data. The Cancer Imaging Archive (2017)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Gulum, M.A., Trombley, C.M., Kantardzic, M. (2021). Multiple Interpretations Improve Deep Learning Transparency for Prostate Lesion Detection. In: Gadepally, V., et al. Heterogeneous Data Management, Polystores, and Analytics for Healthcare. DMAH Poly 2020 2020. Lecture Notes in Computer Science(), vol 12633. Springer, Cham. https://doi.org/10.1007/978-3-030-71055-2_11
Download citation
DOI: https://doi.org/10.1007/978-3-030-71055-2_11
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-71054-5
Online ISBN: 978-3-030-71055-2
eBook Packages: Computer ScienceComputer Science (R0)