Abstract
Microarray gene technology solves the problem of obtaining gene expression data. It is a significant part for current research to obtain effective information from omics genes quickly. Feature selection is an important step of data preprocessing, and it is one of the key factors affecting the capability of algorithm information extraction. Since single feature selection method causes the deviation of feature subsets, we introduce ensemble learning to solve the problem of clusters redundancy. We propose a new method called Multi-Cluster minimum Redundancy (MCmR). Firstly, features are clustered by L1-normth. And then, redundant features among clusters are removed according to the mRMR algorithm. Finally, it can be sorted by the calculation results of each feature MCFS_score in the features subset. By this process, the feature with higher score can be used as the output result. The concept lattice constructed by MCmR reduces redundant concepts while maintaining its structure and improve the efficiency of data analysis. We verify the valid of MCmR on multiple disease gene datasets, and its ACC in Prostate_Tumor, Lung_cancer, Breast_cancer and Leukemia datasets reached 95.4, 94.9, 96.0 and 95.8 respectively.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
The development of microarray gene technology has enabled researchers to obtain a large amount of gene expression data. These samples have the characteristics of small-scale samples at high dimension [1]. At the same time, there are a number of unrelated genes in the obtained data. Therefore, when mining deep information, it is the key that selecting a valid method to obtain accurate sets of pathogenic genes for analyzing gene expression data.
Formal Concept Analysis (FCA), proposed by R. Wille in 1980s, is an effective tool for data analysis. It essentially reflects the association between objects and attributes (samples and features), and embodies relationship between instantiation and generalization by Hasse. FCA is applied to gene expression data to mine deep information. The existed methods have extended and applied concept lattices in many ways.
In 2009, Mehdi proposed two algorithms based on inter-ordinal scaling and pattern structures, and the pattern-structures algorithm calculates interval algebra by adjusting standard algorithm [2]. In 2010, Dexing Wang introduced association rules to reduce concept lattice on biological information data [3]. In 2011, Benjamin J found gene sets that reflected the strong relationship among multiple diseases in the gene expression data of similar diseases by constructing concept lattice [4]. In 2018, Hongxiang Tang used the data structural dependence to construct concept lattice and mined interesting information in genetic data [5]. At present, there are many methods can get better results. However, in the constructed concept lattice, interesting concepts containing important information and redundant concepts caused by redundant genes are difficult to select.
Feature selection, as an important data preprocessing process, can select important feature genes in biological data to alleviate high-dimensional problems while removing irrelevant feature genes [6]. In genetic data, a sample often contains tens of thousands gene expression values, most of which cannot account for the disease. Therefore, feature selection selects the smallest number of features from the original data to contain as much information as possible [7]. The concept lattice constructed in this way achieves the purpose of reducing redundant concepts and improving the valid and accuracy of data expression. There are many feature selection methods. Single feature selection methods may make bias of classification and ignore interesting genes.
Chong Du proposed an integrated feature selection method. This model improves the accuracy of single selection method in gene expression. Maghsoudloo established a hybrid feature selection framework for extracting crucial genes of asthma and other lung diseases as biomarkers [8]. Gang Fang used feature selection of ensemble learning methods to find genes related to stroke patients and predict acute stroke in the dataset [9, 10].
The contributions of this paper are listed as follows:
-
1.
This paper proposed an integrated feature selection method called Multi-Cluster minimum Redundancy (MCmR). It improves redundant features of multiple clusters and reduces the interference of redundant genes on feature selection. This method can obtain excellent feature subsets in multiple clusters.
-
2.
This paper uses Prostate_Tumor, Lung_cancer, Breast_cancer and Leukemia to verify the valid of MCmR. The experimental results show that it obtains an excellent subset of features in all datasets. Compared with other methods, our method gets better results on feature selection set.
In what follows, Sect. 2 introduces the definition and concepts of related methods. Section 3 shows the experiment and results on datasets. Section 4 summarizes this paper.
2 Related Work
2.1 Formal Concept Analysis
FCA takes concepts as basic elements, and each node represents a concept. As the core of data analysis, the concept lattice constructs the conceptual hierarchy structure in the formal context out of the partial order relationship among concepts [1, 2]. As a visualization method of concept lattice, Hasse intuitively describes the information of data.
Definition 1: Let a triple \(K = (G,A,I)\) composed of object set \(G\), attribute set \(A\) and binary relation set \(I\) between objects and attributes constitute a formal context collection, where \(G = (g_{1} ,g_{2} ,...,g_{n} )\), \(A = \{ a_{1} ,a_{2} ,...,a_{m} \}\), \(I \subseteq G \times M\) [4]. In \(I\), for any \(g\) in \(G\) and any \(a\) in \(A\) satisfy “\((g,a) \in K\)” or “\(gIa\)”, it means that the object \(g\) has attribute \(a\), which is represented by 1 (otherwise, it is represented by 0).
Definition 2: Set a tuple \((X,M)\) extracted from the formal context \(K = (G,A,I)\) satisfies \(X^{\prime} = M\) and \(M^{\prime} = X\), and can be called a formal concept (abbreviated as concept, \(C\)). Where, \(X \in G\), \(M \in A\). \(X\) is called the extension of the concept, which is a set of objects shared by all attributes of the concept, and \(M\), as the set of the attributes shared by all objects of the concept, is called the intension of the concept [11]. Where, \(X^{\prime}\) and \(M^{\prime}\) satisfy the following equation:
Definition 3: Suppose two concepts \((X_{1} ,M_{1} )\) and \((X_{2} ,M_{2} )\) in the formal context, named \(C_{1}\) and \(C_{{2}}\). If the relationship conform to \(X_{1} \subseteq X_{2}\) (equivalent to \(M_{2} \subseteq M_{1}\)), \((X_{1} ,M_{1} )\) is called a sub-concept of \((X_{2} ,M_{2} )\), and \((X_{2} ,M_{2} )\) is called a super-concept of \((X_{1} ,M_{1} )\), denoted as \(C_{1} \le C_{2}\) [5, 12]. This relationship between concepts construct complete concept lattice, which called partial order. Formulated as follows (see Fig. 1) (Table 1):
2.2 Feature Selection Method
It achieves the purpose of removing redundant features by selecting feature subsets with strong resolution capacity from the high-dimensional raw data. The selected features retain information of raw data as much as possible [7]. Applying the feature selection method to gene data can obtain a subset of disease feature genes, and remove redundant genes. Feature selection are divided into supervised and unsupervised algorithms [13]. Unsupervised method become popular since it does not need to obtain labels in advance.
MCFS
Multi-Cluster Features Selection (MCFS) method spectral embedding for cluster analysis achieves the purpose of reducing dimensionality by constructing graph, defining the weight matrix (such as heat kernel weighting) and mapping feature. Eigenvalues and eigenvectors of the Laplacian matrix \(L\) will be calculated by the following formula [10]:
\(D\) is diagonal matrix, and \(D_{ii} = \sum\nolimits_{j} {W_{ij} }\). Thus, Laplacian \(L = D - W\). \(Y{\text{ = [y}}_{{1}} {\text{,y}}_{{2}} {,}...{\text{,y}}_{{\text{k}}} {]}\) and \(y_{k}\) is the eigenvector corresponding to the smallest \(k\) non-zero eigenvalues [14].
Sparse coefficient vectors \(\{ a_{k} \}_{k = 1}^{K} \in {\mathbb{R}}^{M}\) measure the significance of each dimension and the capability of each feature to distinguish different clusters, which can be calculated by LARs algorithm to optimize L1-normth [15]. We obtain \(k\) sparse coefficient vectors by solving the L1-normth of every \(y_{k}\) in \(Y\).
Definition 4: For each feature \(j\), \(MCFS(j) = \mathop {\max }\limits_{k} |a_{k,j} |\). It calculates the MCFS_score, and selects the top \(d\) features according to descending order [10].
mRMR
The maximum correlation minimum redundancy (mRMR) method uses mutual information for feature selection [16]. The correlation between features and target categories is represented by mutual information \(I(f_{i} ;c)\). Redundancy between features is represented by mutual information \(I(f_{i} ;f_{j} )\) [6]. The feature subset selected by mRMR has maximum correlation and minimum redundancy.
where \(R(S) = \frac{1}{{|S|^{2} }}\sum\limits_{{f_{i} ,f_{j} \in S}} {I(f_{i} ;f_{j} )}\) represents the redundancy of all features in set \(S\), and \(D(S,c) = \frac{1}{|S|}\sum\limits_{{f_{i} \in S}} {I(f_{i} ;c)}\) represents the correlation between all features in \(S\) and category \(c\).
Assuming that \(n\) features are already in the subset \(S_{n}\), selecting the next feature from set \(\{ S - S_{n} \}\) according to the following formula [17]:
2.3 Multi-cluster Minimum Redundancy
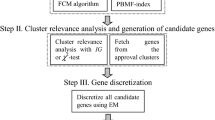
MCFS uses correlation within the cluster to select features, calculates MCFS_score for the selected features, and gets the first \(d\) features. This method only performs better when the clusters is less than 50, and it is not detected whether there is redundancy between features of clusters before calculating the score. The mRMR performs feature redundancy detection between clusters and eliminates features with higher redundancy. Thus, it can be used for further screening of feature selection to achieve its purpose. Therefore, we propose an integrated feature selection method named MCmR, which maintains its advantages in large scale clusters [17]. MCmR makes full use of effective information (see Fig. 2). This proposed feature selection algorithm effectively reduces the formal context and concept lattice [18].
The MCmR is described as follows:
-
1.
The inputting data will be performed by spectral cluster. It will get diagram matrix \(D\) from weights matrix \(W\), and gain set \(Y{ = [}y_{1} {,}y_{2} {,}...{,}y_{k} {]}\) by \(Ly = \lambda Dy\).
-
2.
Get \(k\) sparse coefficient vectors by solving the L1-regularized regression of each \(y_{k}\) in \(Y\).
-
3.
Set threshold of mRMR to filter the features with higher redundancy by \(mRMR = \mathop {\max }\limits_{S} [D(S) - R(S)]\).
-
4.
The MCFS_scores of selected features will be calculated and sorted in descending order. Thus, the top \(d\) features will be selected.
-
5.
Output Feature subset \(S\).
MCmR can not only gets more information feature genes cluster with less number, but also maintain accuracy when cluster is increases. MCmR makes up the shortcoming of single feature selection method in large number cluster, and improves the accuracy of feature selection by multiple selection with feature sets [19].
3 Experiment and Results
3.1 Datasets
This paper verified the valid of MCmR on Prostate_Tumor, Lung_Cancer, Breast Cancer and Leukemia. The detail informations of datasets are shown in Table 2.
3.2 Experiment
Evaluation Metrics
During the experiment, we take the Normalized Mutual Information (NMI) metric and Accuracy as evaluation metrics [14]. Comparing the cluster label calculated by algorithm and raw set can measure the cluster performance. Assuming two cluster labels set \(L\) and \(L^{^{\prime}}\), which respectively include the label provided by data and algorithm. The normalized mutual information metric \(MI(L,L^{^{\prime}} )\) is defined as follows:
where, \(P(i)\) is the probability that a feature picked at random falls into class \(L_{i}\). \(P(i,j)\) is the probability that a feature picked at random falls into both class \(L_{i}\) and \(L_{j}^{^{\prime}}\).
The NMI is as follows:
where, \(H(L)\) and \(H(L^{^{\prime}} )\) represent the entropies of \(L\) and \(L^{^{\prime}}\) respectively. The evaluation criterion ranges from 0 to 1. It is similar between A and B when the value is bigger. NMI = 1 if the two sets of clusters are identical, and NMI = 0 if the two sets are independent [10].
Results
In this part, we compare performance of MCmR with MCFS, mRMR and Relief for various cluster number on Prostate_Tumor (see Fig. 3).
In Fig. 3, it shows that the performance of different algorithms on 30, 50, 70 and 90 clusters. Picture (a) shown that MCmR performs as well as MCFS when the features are less than 50. Picture (b) and (c) show that the performance of MCmR is improving gradually, and always higher than MCFS when cluster is 50 and 70. MCFS gets best results when cluster is 70. When cluster is more than 90, the experiment performance become worse. Picture (d) shows that MCmR gets the best performance when cluster is 90. Too many clusters lead to more redundant genes. On country, it results in filtering out disease-related genes. Comparing with 70 and 110 clusters, MCmR gets better results on Prostate_Tumor when cluster is 90. The quantity of features also affects experimental results. It is shown from picture (a–d), MCmR gets the best performance when feature is 130. The quantity of feature d is half of the features set satisfied threshold of mRMR. Then, we compare the experimental effects in different features numbers, and verify the best d.
In Table 3, it shows that MCmR gets the best results on Prostate_Tumor. This prove our new method is valid. Compared with MCFS, mRMR and Relief, MCmR is improved by 4.1, 12 and 10.8 respectively.
In Table 4, it is easy to know that the ACC of Relief and mRMR are close which reaches 82.0 and 82.4 on average. The effect of MCFS is better than those two, which reaches 89.8 on average. The highest ACC belongs to MCmR that it is 95.5 on average. It shows the valid and superiority of MCmR in feature selection.
In Table 5, we use MCmR to construct a concept lattice in each dataset. Compared with MCFS, MCmR has less concept in concept lattice. It can be seen from the table that MCmR performs differently in different datasets. Among them, it performed best in Lung_cancer, with a concept reduction of 10.1%. Using feature selection to remove redundant genes from high-dimensional gene expression data and obtain feature genes. It can reduce the formal context and concepts in concept lattice (see Fig. 4).
The experimental results show that MCmR improves the accuracy of related gene selection and reduces redundant genes compared with other methods.
4 Conclusion
Aiming at the problem of poor feature selection effect in case of a large number of clusters, this paper proposes an integrated feature selection method MCmR to improve the capability of feature selection on genetic data. We focus on the extraction of feature genes by clustering to reduce attribute genes concept. Our experiments show that MCmR improved the accuracy of data classification. And it also reduces concept of concept lattices at the same time. However, this method has high computational overhead. Thus, we try to reduce it through improve framework of MCmR. For future work, we will pay attention to find a way to reduce computational overhead.
References
Kaytoue-Uberall, M., Duplessis, S., Napoli, A.: Using formal concept analysis for the extraction of groups of co-expressed genes. In: Le Thi, H.A., Bouvry, P., Pham Dinh, T. (eds.) Modelling, Computation and Optimization in Information Systems and Management Sciences. MCO 2008. Communications in Computer and Information Science, vol. 14, pp. 439–449. Springer, Heidelberg (2008). https://doi.org/10.1007/978-3-540-87477-5_47
Kaytoue M., Duplessis S., Kuznetsov S.O., Napoli A. (2009) Two FCA-based methods for mining gene expression data. In: Ferré, S., Rudolph, S. (eds.) Formal Concept Analysis. ICFCA 2009. Lecture Notes in Computer Science, vol. 5548, pp. 251–266. Springer, Heidelberg. https://doi.org/10.1007/978-3-642-01815-2_19
Wang, D., Cui, L., Wang, Y., Yuan, H., Zhang, J.: Association Rule mining based on concept lattice in bioinformatics research. In: 2010 International Conference on Biomedical Engineering and Computer Science, pp. 1–4. IEEE, April 2010
Keller, B.J., Eichinger, F., Kretzler, M.: Formal concept analysis of disease similarity. AMIA Summits Transl. Sci. Proc. 2012, 42 (2012)
Tang, H., Xia, F., Wang, S.: Information structures in a lattice-valued information system. Soft. Comput. 22(24), 8059–8075 (2018). https://doi.org/10.1007/s00500-018-3097-x
Chong, D.U., Chang Yin, Z.H.O.U., Yue, L.I., et al.: Application of ensemble feature selection in gene expression data. J. Shandong Univ. Sci. Technol. (Nat. Sci.) 38(1), 85–90 (2019)
Lei, Y., Wu, Z.: Time series classification based on statistical features. EURASIP J. Wireless Commun. Netw. 2020(1), 1–13 (2020). https://doi.org/10.1186/s13638-020-1661-4
Maghsoudloo, M., Jamalkandi, S.A., Najafi, A., Masoudi-Nejad, A.: An efficient hybrid feature selection method to identify potential biomarkers in common chronic lung inflammatory diseases. Genomics 112, 3284–3293 (2020)
Fang, G., Liu, W., Wang, L.: A Machine Learning Approach to Select Features Important to Stroke Prognosis. Comput. Biol. Chem. 88, 107316 (2020)
Cai, D., Zhang, C., He, X.: Unsupervised feature selection for multi-cluster data. In: Proceedings of the 16th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp. 333–342. July 2010
Hao, F., Min, G., Pei, Z., Park, D.S., Yang, L.T.: $ K $-clique community detection in social networks based on formal concept analysis. IEEE Syst. J. 11(1), 250–259 (2015)
Henriques, R., Madeira, S.C.: Pattern-based biclustering with constraints for gene expression data analysis. In: Pereira, F., Machado, P., Costa, E., Cardoso, A. (eds.) Progress in Artificial Intelligence. EPIA 2015. Lecture Notes in Computer Science, vol 9273. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-23485-4_34
Xie, J.Y., Ding, L.J., Wang, M.Z.: Spectral clustering based unsupervised feature selection algorithm. Ruan Jian Xue Bao/J. Softw. 31(4), 1009–1024 (2020)
He, X., Cai, D., Niyogi, P.: Laplacian score for feature selection. In: Advances in Neural Information Processing Systems, pp. 507–514 (2006)
Efron, B., Hastie, T., Johnstone, I., Tibshirani, R.: Least angle regression. Ann. Stat. 32(2), 407–499 (2004)
Zhang, Y., Ding, C., Li, T.: Gene selection algorithm by combining reliefF and mRMR. BMC Genomics 9(S2), S27 (2008). https://doi.org/10.1186/1471-2164-9-S2-S27
Shao, M., Liu, M., Guo, L.: Vector-based attribute reduction method for formal contexts. Fundamenta Informaticae 126(4), 397–414 (2013)
Mundra, P.A., Rajapakse, J.C.: SVM-RFE with MRMR filter for gene selection. IEEE Trans. Nanobiosci. 9(1), 31–37 (2009)
Liu, D., Hua, G., Viola, P., Chen, T.: Integrated feature selection and higher-order spatial feature extraction for object categorization. In: 2008 IEEE Conference on Computer Vision and Pattern Recognition, pp. 1–8. IEEE, June 2008
Funding
This work is partly supported by the Undergraduate Education Reform Project in Shandong Province (no. Z2018S022).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 ICST Institute for Computer Sciences, Social Informatics and Telecommunications Engineering
About this paper
Cite this paper
Wang, Z., Lei, Y., Zhang, L. (2021). A Concept Lattice Method for Eliminating Redundant Features. In: Qi, L., Khosravi, M.R., Xu, X., Zhang, Y., Menon, V.G. (eds) Cloud Computing. CloudComp 2020. Lecture Notes of the Institute for Computer Sciences, Social Informatics and Telecommunications Engineering, vol 363. Springer, Cham. https://doi.org/10.1007/978-3-030-69992-5_4
Download citation
DOI: https://doi.org/10.1007/978-3-030-69992-5_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-69991-8
Online ISBN: 978-3-030-69992-5
eBook Packages: Computer ScienceComputer Science (R0)