Abstract
The EndoTect challenge at the International Conference on Pattern Recognition 2020 aims to motivate the development of algorithms that aid medical experts in finding anomalies that commonly occur in the gastrointestinal tract. Using HyperKvasir, a large dataset containing images taken from several endoscopies, the participants competed in three tasks. Each task focuses on a specific requirement for making it useful in a real-world medical scenario. The tasks are (i) high classification performance in terms of prediction accuracy, (ii) efficient classification measured by the number of images classified per second, and (iii) pixel-level segmentation of specific anomalies. Hopefully, this can motivate different computer science researchers to help benchmark a crucial component of a future computer-aided diagnosis system, which in turn, could potentially save human lives.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
The human digestive system is prone to suffer from many different diseases and abnormalities throughout a human lifetime. Some of these may be life-threatening and pose a severe risk to a patient’s health and well-being. In most cases, if the detection of lethal disease is done early enough, it can be treated with a high chance of being fully healed. Therefore, it is important that all lesions are identified and reported during a routine investigation of the gastrointestinal (GI) tract. Currently, the gold-standard in performing these investigations is through video endoscopies, which is a procedure involving a small camera attached to a tube that is inserted either orally or rectally. However, there is one major downside to this procedure. The method is highly dependent on the skills and experience of the person operating the endoscope, which in turn results in a high operator variation and performance [18, 28, 47]. This is one of the reasons for high miss-rates when measuring polyp detection performance, with some miss-rates being as high as 20% [25]. Polyps are small mushroom-like growths that appear on the inner-lining of the GI wall and are the leading cause to colorectal cancer.
Automated detection of GI anomalies has been a research topic for at least two decades, and in the last few years, there have been various AI-based solutions have been proposed using both hand-crafted features and representation learning methods (such as neural networks). However, even though there are many approaches for detecting [1, 4, 7, 13, 32, 33, 35, 37, 42, 44, 45, 48] and segmenting [14, 23, 24] GI findings, even some targeting real-time analysis [2, 39, 40], there is room for improvement. One popular way of benchmarking and improving the state-of-the-art in machine learning is through publicly hosted challenges that motivate researchers to contribute to a use-case they otherwise would not work on. For GI automatic image and video analysis, there have been several such challenges hosted the last few years [3, 19, 38, 41], with each bringing new insights into the current state of the field.
This year, we present three different tasks for participants to complete. The tasks are as follows: (i) The detection task which aims for high classification accuracy among 23 different classes, (ii) the efficient detection task which targets real-time performance for the same 23 classes of the detection task, and (iii) the segmentation task that aims to segment polyps in GI images. To participate, the teams had to solve at least one of the provided tasks. Overall, six teams participated, where all participants, in one way or another, utilize deep neural networks to solve the provided tasks. The results vary between teams, but most are able to achieve satisfactory scores in terms of what is suitable for use in clinics [36].
We see this as an opportunity to aid medical doctors by helping them detect lesions through automatic frame analysis done live during endoscopy examinations. The pattern recognition community has a lot of knowledge that could assist in this task, making this challenge a perfect fit for the International Conference on Pattern Recognition (ICPR). The work done in this competition, detecting and segmenting medical findings in the GI tract, has the potential of making a real societal impact, as it directly affects the quality of care that healthcare professionals can provide.
2 Dataset Details
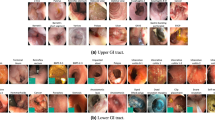
For this challenge, we provided the participants with a development dataset that was to be used to train their algorithms. This year, we provided HyperKvasir [6], which is a large GI dataset consisting of labeled and unlabeled images taken from several different GI endoscopies. The dataset is split into four distinct parts; Labeled image data, unlabeled image data, segmented image data, and annotated video data. In total, the dataset contains 110, 079 images (see Fig. 1 for examples) and 374 videos where it captures anatomical landmarks, pathological findings, and normal findings. The result is more than one million images and video frames altogether.
For the detection and efficient detection tasks, participants used the 23 classes provided in the labeled part of the dataset to develop their algorithms. The number of images per class is not balanced, which is a general challenge in the medical field due to the fact that some findings occur more often than others. This adds an additional challenge for researchers since methods applied to the data should also be able to learn from a small amount of training data. The participants could also use the unlabeled part of the dataset to further improve their algorithm by using, for example, a student-teacher approach or the pseudo labels provided in the HyperKvasir GitHub repositoryFootnote 1.
For the segmentation task, we provide the original image, a segmentation mask, and a bounding box for 1,000 images containing polyps. An example is shown in Fig. 2, where we see six samples taken from the segmentation dataset. For the image masks, the white pixels depict the area of the image containing a polyp, while the black background pixels do not. The bounding box is defined as the outermost pixels of the found polyp.
3 Tasks
With the end-goal of helping medical experts detect more lesions, we present three different tasks that each target a different requirement for in-clinic use. In the following, we give a detailed description of each task and describe how each was evaluated using the appropriate metrics. The script used to evaluate each task is on GitHubFootnote 2.
3.1 Detection Task
The detection task stems from the requirement of the high detection accuracy needed to be viable for use in a clinical setting. Participants are asked to develop algorithms that achieve high classification scores on the 23 different classes present in the labeled part of the development dataset (further described in Sect. 2). Submissions to this task was a comma-separated values (CSV) file, where each line contained the filename of the predicted image in the test dataset, the predicted label, and a confidence score ranging from 0 to 1 for the predicted label.
For this detection task, we use several standard metrics commonly used to evaluate classification tasks. We collect all true and false positives and negatives, and we then calculate metrics such as precision, recall/sensitivity, specificity, F1, and Matthews correlation coefficient (MCC) for multi-classification (also called \(R_k\) statistic for multiclass classification). The officially reported metric for evaluating this task is the MCC, which will also be the metric used to rank the submissions.
3.2 Efficient Detection Task
The efficient detection task focuses on the real-time analysis needed to deliver instant feedback to doctors performing endoscopies. To satisfy this requirement, the algorithm must achieve good classification scores while also being able to classify images as fast as they are put on screen, which is approximately 30 frames per second. For the efficient detection task, we asked participants to submit a Docker [31] image so that we can evaluate the speed and efficiency of the proposed algorithm on the same hardware. The Docker image was set up to produce a submission file similar to the one described for the detection task, but in addition to the aforementioned value entries, the classification processing time was also appended to the end of each row. All submissions submitted to this task were run on what could be considered consumer-grade hardware, that is, a computer running Arch Linux with an Intel Core i9-10900K processor, an Nvidia GeForce 1080 Ti graphics processing unit (GPU), and 32 gigabytes of RAM.
As one could generally achieve higher processing speeds with an algorithm with lower prediction accuracy, the evaluation used a combination of the MCC classification score and the number of frames processed per second. The focus here is on the “speed” aspect of the algorithm, so the only requirement from a classification standpoint is that it exceeds a set MCC threshold so that it is still viable for in-clinic use. We set the threshold of \(85\%\) as it is considered standard for automatic detection systems for colonoscopies [36].
3.3 Segmentation Task
In the segmentation task, we asked participants to use the segmented images provided in the dataset to generate segmentation masks of polyps automatically. Polyps are clumps of cells that form on the mucosal wall of the GI tract and come in a variety of shapes and sizes. Polyps are among the most critical findings in an endoscopy procedure as they are a precursor to different cancer types, including colorectal cancer, which is one of the most lethal cancer types worldwide [22]. The motivation behind this task is rooted in the requirement for not only detecting that a frame contains a polyp, but also showing where it is so that it can be properly removed. A typical example of a segmented polyp is shown in Fig. 2.
For the evaluation of this task, we use the standard metrics commonly used to evaluate segmentation tasks. This includes precision, recall, the Dice coefficient, and the Intersection over Union (IoU, also known as the Jaccard index). The metric which will be used to rank submissions will be the IoU. To calculate the metrics, we use the implementation provided by the Python library scikit-learn [34].
4 Participants
This year, we received 26 registrations, of which six submitted results. Each participating team was allowed to submit as many runs to each task as they wished. In the following, we give a short summary of each participant’s approach. A more detailed description of each approach can be found in the teams’ corresponding challenge papers.
4.1 Team DeepBlueAI
Team DeepBlueAI participated in the detection and segmentation tasks. For the detection task, they trained a series of (CNNs), of which the best performing approach is an ensemble network consisting of a ResNet-50 [15] with batch normalization and an EfficientNet B7 [43]. For the segmentation task, they used two different approaches, namely instance and semantic segmentation. The instance segmentation approach used the Mask Scoring R-CNN [21] with ResNeXt-101 [49] as the backbone. As for the semantic segmentation, they used DeepLab V3 plus [9] with multi-scale training. More information on the specific implementation for both tasks can be found in [30].
4.2 Team Spearheads
Team Spearheads participated in all three tasks, where two runs were submitted to the detection and efficient detection tasks, and one run to the segmentation task. For the detection and efficient detection task, they used a Tiny Darknet modelFootnote 3, which was trained using an augmented version of the provided development dataset. For the segmentation task, they used a standard UNet architecture trained on the provided segmentation dataset, which was expanded using augmentation by Augmentor [5]. More information about team Spearheads approach can be found in [11].
4.3 Team NKT
Team NKT participated in the segmentation task, where they submitted one run. Their approach used a novel CNN-based architecture, which they named Dual Decoder Attention Network (DDANet). The architecture uses a single encoder network together with multiple decoders that use a combination of residual learning [16] and squeeze and excitation networks [20]. A more detailed explanation of the approach can be found in [46].
4.4 Team aggcmab
Team aggcmab participated in the detection and segmentation tasks, for which they submitted one run to each. For the detection task, aggcmab used a ResNet-50x1 with a BiT-M [27] backbone trained with a hierarchical loss function. For the segmentation task, they use a double encoder-decoder network with a dual path network [10] for the encoders and a Feature-Pyramid [29] for the decoders. More information on the specifics of team aggcmab’s approach can be found in [12].
4.5 Team FAST-NU-DS
Team FAST-NU-DS participated in the detection task, where they submitted three runs. Their approach used bagging with 11 DenseNet169 models, where the final classification was made through hard majority voting. More information on the method can be found in [26].
4.6 Team howard
Team howard participated in all three tasks, where they submitted one run to each. For the detection and efficient detection task, they used a CNN based on the ResNet152 [15] architecture trained with a hybrid loss. During training, they also applied some data augmentation, namely, contrast augmentation, color shift, brightness augmentation, flipping, perspective transformation, and blur. For the segmentation task, their solution is based on Cascade Mask R-CNN [8]. More information about their solution can be found in [17].
5 Results and Discussion
Tables 1, 2, and 3 show the results for all tasks in the challenge. Looking at the results for the detection task (Table 1), we see that team howard achieved the best result with their use of ResNet-152 together with a custom hybrid loss. They achieved an MCC score of 0.903, 0.043 ahead of DeepBlueAI, who came in second place. For the efficient detection task (Table 2), only two teams participated, but also here, team howard achieved the best average frames per second (FPS) while also keeping the classification performance high. None of the teams reached the target MCC threshold of 85%, but team howard achieved an MCC of 0.765 at an FPS of 129, far above the real-time requirement. Thus, maybe some speed can be traded for a more complex model, achieving a slightly higher MCC while still reaching a real-time speed of 30 FPS. A common trend in this task was using neural networks with less parameters, like MobileNet or Tiny Darknet, to achieve a higher FPS. For the segmentation task (Table 3), team aggcmab achieved the highest IoU with their double encoder-decoder network approach. They reached an IoU score of 0.871, which is quite close to the runner up score of 0.861 submitted by team DeepBlueAI. Overall, the results prove that deep learning works well for analyzing GI image data and confirms the potential of computer-assisted detection and segmentation of GI anomalies, but they also suggest that there is still some room for improvement.
From an organizational perspective, the challenge went smoothly, without any significant hiccups or sudden difficulties. Docker submissions seem to work well, but may require some extra effort from the participants, which may explain why we only got two submissions to the efficient detection task. The difficulty level of the tasks appears to be quite balanced as the different teams achieved a variety of scores. Next year, we plan to hold the challenge again, but this time with an extended evaluation dataset and an additional task for efficient segmentation.
6 Conclusion
This paper described the EndoTect 2020 challenge, which asked participants to build algorithms that automatically detect different findings commonly found in the GI tract. The challenge consisted of three distinct tasks, where participants were given a large open dataset composed of videos from real endoscopies. We believe that computer scientists can make a real impact on the field of medicine, and the results presented in this paper show that we are at the point where machine learning algorithms have much potential in helping doctors detect more diseases.
References
Alammari, A., Islam, A.R., Oh, J., Tavanapong, W., Wong, J., De Groen, P.C.: Classification of ulcerative colitis severity in colonoscopy videos using CNN. In: Proceedings of the ACM International Conference on Information Management and Engineering (ACM ICIME), pp. 139–144 (2017). https://doi.org/10.1145/3149572.3149613
Angermann, Q., et al.: Towards real-time polyp detection in colonoscopy videos: adapting still frame-based methodologies for video sequences analysis. In: Cardoso, M.J., et al. (eds.) CARE/CLIP -2017. LNCS, vol. 10550, pp. 29–41. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-67543-5_3
Bernal, J., Aymeric, H.: MICCAI endoscopic vision challenge polyp detection and segmentation (2017). https://endovissub2017-giana.grand-challenge.org/home/. Accessed 11 Dec 2017
Bernal, J., et al.: Polyp detection benchmark in colonoscopy videos using GTCreator: a novel fully configurable tool for easy and fast annotation of image databases. In: Proceedings of Computer Assisted Radiology and Surgery (CARS) (2018). https://hal.archives-ouvertes.fr/hal-01846141
Bloice, M.D., Roth, P.M., Holzinger, A.: Biomedical image augmentation using Augmentor. Bioinformatics (Oxford Engl.) 35(21), 4522–4524 (2019). https://doi.org/10.1093/bioinformatics/btz259
Borgli, H., et al.: HyperKvasir, a comprehensive multi-class image and video dataset for gastrointestinal endoscopy. Sci. Data 7 (2020). https://doi.org/10.1038/s41597-020-00622-y. Article no. 283
Bychkov, D., et al.: Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci. Rep. 8(1), 3395 (2018). https://doi.org/10.1038/s41598-018-21758-3
Cai, Z., Vasconcelos, N.: Cascade R-CNN: high quality object detection and instance segmentation. IEEE Trans. Pattern Anal. Mach. Intell. (2019)
Chen, L.C., Zhu, Y., Papandreou, G., Schroff, F., Adam, H.: Encoder-decoder with atrous separable convolution for semantic image segmentation. arXiv:1802.02611 (2018)
Chen, Y., Li, J., Xiao, H., Jin, X., Yan, S., Feng, J.: Dual path networks. In: Proceedings of the Annual Conference on Advances in Neural Information Processing Systems (NeurIPS), pp. 4467–4475 (2017)
Dutta, A., Bhattacharjee, R.K., Barbhuiya, F.A.: Efficient detection of lesions during endoscopy. In: Proceedings of the ICPR 2020 Workshops and Challenges. LNCS. Springer (2020)
Galdran, A., Carneiro, G., Ballester, M.A.G.: A hierarchical multi-task approach to gastrointestinal image analysis. In: Proceedings of the ICPR 2020 Workshops and Challenges. LNCS. Springer (2020)
Ghatwary, N.M., Ye, X., Zolgharni, M.: Esophageal abnormality detection using DenseNet based faster R-CNN with gabor features. IEEE Access 7, 84374–84385 (2019). https://doi.org/10.1109/ACCESS.2019.2925585
Guo, Y., Bernal, J., Matuszewski, B.J.: Polyp segmentation with fully convolutional deep neural networks–extended evaluation study. J. Imaging 6(7), 69 (2020)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 770–778 (2016). https://doi.org/10.1109/CVPR.2016.90
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 770–778 (2016)
He, Q., Bano, S., Stoyanov, D., Zuo1, S.: Hybrid loss with network trimming for disease recognition in digestive endoscopy. In: Proceedings of the ICPR 2020 Workshops and Challenges. LNCS. Springer (2020)
Hewett, D.G., Kahi, C.J., Rex, D.K.: Efficacy and effectiveness of colonoscopy: how do we bridge the gap? Gastrointest. Endosc. Clin. 20(4), 673–684 (2010). https://doi.org/10.1016/j.giec.2010.07.011
Hicks, S., et al.: ACM multimedia BioMedia 2019 grand challenge overview. In: Proceedings of the ACM International Conference on Multimedia (ACM MM), pp. 2563–2567 (2019). https://doi.org/10.1145/3343031.3356058
Hu, J., Shen, L., Sun, G.: Squeeze-and-excitation networks. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 7132–7141 (2018). https://doi.org/10.1109/CVPR.2018.00745
Huang, Z., Huang, L., Gong, Y., Huang, C., Wang, X.: Mask scoring R-CNN. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 6402–6411 (2019). https://doi.org/10.1109/CVPR.2019.00657
International Agency for Research on Cancer - WHO: Cancer fact sheets (2019). https://gco.iarc.fr/today/fact-sheets-cancers. Accessed 16 Dec 2019
Jha, D., Riegler, M., Johansen, D., Halvorsen, P., Johansen, H.: DoubleU-Net: a deep convolutional neural network for medical image segmentation. In: Proceeding of the International Symposium on Computer Based Medical Systems (CBMS) (2020)
Jha, D., et al.: ResUNet++: an advanced architecture for medical image segmentation. In: Proceedings of the International Symposium on Multimedia (ISM), pp. 225–230 (2019). https://doi.org/10.1109/ISM46123.2019.00049
Kaminski, M.F., et al.: Quality indicators for colonoscopy and the risk of interval cancer. N. Engl. J. Med. 362(19), 1795–1803 (2010). https://doi.org/10.1056/NEJMoa0907667
Khan, Z., Tahir, M.A., Memon, S.: Medical diagnostic by data bagging for various instances of neural network. In: Proceedings of the ICPR 2020 Workshops and Challenges. LNCS. Springer (2020)
Kolesnikov, A., et al.: Big Transfer (BiT): general visual representation learning. arXiv preprint arXiv:1912.11370, June 2019
Lee, S.H., et al.: Endoscopic experience improves interobserver agreement in the grading of esophagitis by Los Angeles classification: conventional endoscopy and optimal band image system. Gut Liver 8(2), 154 (2014). https://doi.org/10.5009/gnl.2014.8.2.154
Lin, T.Y., Dollár, P., Girshick, R., He, K., Hariharan, B., Belongie, S.: Feature pyramid networks for object detection. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 2117–2125 (2017)
Luo, Z., Che, L., He, J.: A hierarchical multi-task approach to gastrointestinal image analysis. In: Proceedings of the ICPR 2020 Workshops and Challenges. LNCS. Springer (2020)
Merkel, D.: Docker: lightweight Linux containers for consistent development and deployment. Linux J. 2014(239) (2014)
Min, M., Su, S., He, W., Bi, Y., Ma, Z., Liu, Y.: Computer-aided diagnosis of colorectal polyps using linked color imaging colonoscopy to predict histology. Sci. Rep. 9(1), 2881 (2019). https://doi.org/10.1038/s41598-019-39416-7
Mori, Y., et al.: Real-time use of artificial intelligence in identification of diminutive polyps during colonoscopy: a prospective study. Ann. Intern. Med. 169(6), 357–366 (2018). https://doi.org/10.7326/M18-0249
Pedregosa, F., et al.: Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011)
Pogorelov, K., et al.: A holistic multimedia system for gastrointestinal tract disease detection. In: Proceedings of the ACM on Multimedia Systems Conference (MMSYS), pp. 112–123 (2017). https://doi.org/10.1145/3193740
Pogorelov, K., et al.: Deep learning and hand-crafted feature based approaches for polyp detection in medical videos. In: Proceedings of the IEEE International Symposium on Computer-Based Medical Systems (CBMS). IEEE (2018)
Pogorelov, K., et al.: Efficient disease detection in gastrointestinal videos-global features versus neural networks. Multimedia Tools Appl. 76(21), 22493–22525 (2017). https://doi.org/10.1007/s11042-017-4989-y
Pogorelov, K., et al.: Medico multimedia task at mediaeval 2018. In: Proceeding of the MediaEval Benchmarking Initiative for Multimedia Evaluation Workshop (MediaEval) (2018)
Pogorelov, K., et al.: GPU-accelerated real-time gastrointestinal diseases detection. In: Proceedings of the International Symposium on Computer-Based Medical Systems (CBMS), pp. 185–190. IEEE (2016). https://doi.org/10.1109/CBMS.2016.63
Riegler, M., et al.: EIR - efficient computer aided diagnosis framework for gastrointestinal endoscopies. In: Proceedings of the IEEE International Workshop on Content-Based Multimedia Indexing (CBMI), pp. 1–6 (2016). https://doi.org/10.1109/CBMI.2016.7500257
Riegler, M., et al.: Multimedia for medicine: the medico task at MediaEval 2017. In: Proceeding of the MediaEval Benchmarking Initiative for Multimedia Evaluation Workshop (MediaEval) (2017)
Silva, J., Histace, A., Romain, O., Dray, X., Granado, B.: Toward embedded detection of polyps in WCE images for early diagnosis of colorectal cancer. Int. J. Comput. Assist. Radiol. Surg. 9(2), 283–293 (2014). https://doi.org/10.1007/s11548-013-0926-3
Tan, M., Le, Q.V.: EfficientNet: rethinking model scaling for convolutional neural networks. In: Proceedings of the International Conference on Machine Learning, pp. 6105–6114 (2019)
Thambawita, V., et al.: The medico-task 2018: disease detection in the gastrointestinal tract using global features and deep learning. In: Proceeding of the MediaEval Benchmarking Initiative for Multimedia Evaluation Workshop (MediaEval) (2018)
Thambawita, V.L., et al.: An extensive study on cross-dataset bias and evaluation metrics interpretation for machine learning applied to gastrointestinal tract abnormality classification. ACM Trans. Comput. Healthcare 1 (2020)
Tomar, N.K., Jha, D., Ali, S., Johansen, H.D.J.D., Riegler, M.A., Halvorsen, P.: DDANet: dual decoder attention network for automatic polyp segmentation. In: Proceedings of the ICPR 2020 Workshops and Challenges. LNCS. Springer (2020)
Van Doorn, S.C., et al.: Polyp morphology: an interobserver evaluation for the Paris classification among international experts. Am. J. Gastroenterol. 110(1), 180–187 (2015). https://doi.org/10.1038/ajg.2014.326
Wang, Y., Tavanapong, W., Wong, J., Oh, J.H., De Groen, P.C.: Polyp-Alert: near real-time feedback during colonoscopy. Comput. Methods Programs Biomed. 120(3), 164–179 (2015). https://doi.org/10.1016/j.cmpb.2015.04.002
Xie, S., Girshick, R., Dollár, P., Tu, Z., He, K.: Aggregated residual transformations for deep neural networks. arXiv preprint arXiv:1611.05431 (2016)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Hicks, S.A., Jha, D., Thambawita, V., Halvorsen, P., Hammer, H.L., Riegler, M.A. (2021). The EndoTect 2020 Challenge: Evaluation and Comparison of Classification, Segmentation and Inference Time for Endoscopy. In: Del Bimbo, A., et al. Pattern Recognition. ICPR International Workshops and Challenges. ICPR 2021. Lecture Notes in Computer Science(), vol 12668. Springer, Cham. https://doi.org/10.1007/978-3-030-68793-9_18
Download citation
DOI: https://doi.org/10.1007/978-3-030-68793-9_18
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-68792-2
Online ISBN: 978-3-030-68793-9
eBook Packages: Computer ScienceComputer Science (R0)