Abstract
Strong experimental evidence from studies in human donor retinas and animal models supports the idea that the retinal pathology associated with age-related macular degeneration (AMD) involves mitochondrial dysfunction and consequent altered retinal metabolism. This chapter provides a brief overview of mitochondrial structure and function, summarizes evidence for mitochondrial defects in AMD, and highlights the potential ramifications of these defects on retinal health and function. Discussion of mitochondrial haplogroups and their association with AMD brings to light how mitochondrial genetics can influence disease outcome. As one of the most metabolically active tissues in the human body, there is strong evidence that disruption in key metabolic pathways contributes to AMD pathology. The section on retinal metabolism reviews cell-specific metabolic differences and how the metabolic interdependence of each retinal cell type creates a unique ecosystem that is disrupted in the diseased retina. The final discussion includes strategies for therapeutic interventions that target key mitochondrial pathways as a treatment for AMD.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
10.1 Introduction to Mitochondria
10.1.1 Mitochondrial Origins
Every cell contains two types of DNA, the nuclear DNA, which is inherited from both mother and father, along with the small circular mitochondrial DNA (mtDNA) that is inherited only from the mother. The “Endosymbiotic Theory” explains the existence of these very different types of DNA co-existing within cells. In early evolution, free-living, aerobic bacteria were incorporated into the early, ancestral eukaryotic cells. Over time, this symbiotic relationship resulted in increased energy production that allowed progression from single-cell eukaryotes to multicellular organisms, tissues, and the diversity of species found worldwide. Evidence supporting this theory includes similarities between bacterial and mitochondrial (MT) patterns of gene arrangements, small subunit ribosomal RNAs (rRNA), and protein data [1,2,3].
10.1.2 MT Distribution and Content
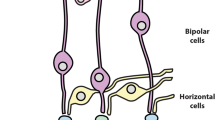
The retina is one of the most metabolically active tissues in the body due, in part, to the high concentration of MT present in nearly all cells. The MT within different retinal cell types are localized toward the sources of oxygen, which in the human retina are the choriocapillaris and the inner retinal vasculature. In the retinal pigment epithelium (RPE), MT are clustered along the basal border of the cell, in close proximity to the choriocapillaris (Fig. 10.1). Photoreceptors are nourished by both the choriocapillaris as well as the inner retinal vasculature. Reflecting the multiple sources of oxygen, photoreceptor MT are located at two sites—densely packed within the inner segments and at the synaptic terminals. Müller cells, which span the entire length of the neural retina and thus also derive oxygen from both sources, have MT that are evenly distributed throughout the cell [4]. MT in the retinal ganglion cells (RGC) are present in the soma where mtDNA replication occurs and are abundant along the unmyelinated axons where they move bidirectionally along microtubule tracks [5]. MT accumulate just anterior to the lamina cribrosa, but are drastically reduced once the axons exit the eye and form the myelinated optic nerve.
Mitochondrial localization. (a) Schematic of retinal cells showing location of mitochondria (labeled blue) within each cell type. Position of the two major blood supplies (choriocapillaris and the inner retinal vasculature) is indicated in red. (b) Mouse retinal section stained with anti-TOMM20 (green) to indicate position of mitochondria, anti-Ezrin (red) to indicate the RPE apical membrane, and DAPI (blue) for staining nuclei (Illustration by S. Atilano; Micrograph provided by John Ash and Emily Brown)
In addition to the cell-specific distribution of MT, the number of MT within a cell can vary from 100 to several thousand depending upon the cell’s energy requirement. Additionally, MT content is very dynamic and can be adjusted when energy demands change by either making more MT or eliminating MT via the processes of biogenesis and mitophagy, respectively. The master regulator of MT biogenesis is PGC-1α, a co-activator of transcription factors (NRF-1, PPARα, mtTFAM) that upregulate genes involved in MT biogenesis. Mitophagy is a specialized version of autophagy involving the selective degradation of MT that are either not needed for the current cellular energy requirements or are damaged. MT degradation involves multiple steps starting with the segregation of unwanted or damaged MT from healthy segments. The healthy MT can then fuse with other healthy MT thereby mixing contents and forming ever-changing MT networks (Fig. 10.2). This dynamic process of fission to remove damaged MT segments and fusion of healthy MT is essential for maintaining a population of functional MT. The damaged MT are surrounded by a double membrane structure called an autophagosome, which then fuses with lysosomes. Lysosomal enzymes then digest the contents of the phagosome. Thus, in the healthy cell, there is continual MT turnover involving coordination and balance between both biogenesis and mitophagy.
Mitophagy eliminates damaged MT. Fission of MT networks allows for segregation of damaged MT (star, green) from healthy MT (yellow). Healthy MT can then fuse with other healthy MT to replenish the dynamic MT network. The damaged MT are encapsulated by a double-walled membrane, forming the phagosome, which fuses with lysosomes. The lysosomal enzymes digest mitochondria in the phagosome (Illustration by S. Atilano)
There is good evidence suggesting the processes involved in MT turnover are adversely affected with AMD. Analysis of RPE MT in human donor eyes using electron microscopy showed there was a significant decrease in MT number and size in donors with AMD compared with nondiseased age-matched controls [6]. Lower MT numbers are consistent with defects in MT biogenesis and the smaller MT suggests defects in MT fusion. Autophagy defects, which will have a negative impact on mitophagy, have also been reported in human donors with AMD. In one study, immunohistochemical staining of human retinal sections showed p62 accumulated in the macula of donors with AMD [7]. Elevated p62, which is normally degraded via autophagy, is an indicator of decreased autophagic flux. Data from a study in primary cultures of RPE directly showed autophagic flux is reduced in RPE from donors with AMD [8].
10.1.3 MT Structure and Function
Each MT consists of two separate membranes (outer and inner) that divide discrete compartments (Intermembrane Space and Matrix), each containing different proteins with specific functions (Fig. 10.3). The outer membrane is permeable to small molecules (e.g., oxygen, calcium, sugars) that pass through the lipid bilayer. Channels within the outer mitochondrial membrane contain porin proteins (also known as voltage-dependent anion channels, VDAC) that allow passage of ions and molecules less than 5000 Daltons, such as ADP and ATP. The vast majority of proteins that reside in the MT (~1500 proteins) are encoded by the nuclear genome, produced in the cytosol, and imported through the outer MT membrane via the “translocase of the outer membrane” (TOM) complex into the Intermembrane Space [3, 9]. Also embedded in the outer membrane are the proteins involved in MT fission (Fis1/2) and fusion (Mitofusin). Proteins involved in regulating apoptosis (Bcl-2, Bax/Bak) bind to the outer membrane but are not integrated into the membrane.
Mitochondrial structure and function. Schematic shows gross mitochondrial structures, including the Inner and Outer membranes that separate the Intermembrane Space and the Matrix. Also depicted are the Complexes (I–V) that are part of the electron transport chain and the energy pathways that feed into these Complexes (Acetyl CoA, TCA cycle, NADH). Hydrogen ions (designated as H) that accumulate in the intermembrane space flow back into the matrix through Complex V, driving the phosphorylation of ADP to form ATP. Boxes outside the MT summarize key functions of energy production, regulation of apoptosis, ROS production and signaling to and from the nucleus via anterograde/retrograde signaling. E = reactive oxygen species; mtDNA = mitochondrial DNA; ROS = reactive oxygen species; VDAC = voltage dependent anion channel; ANT = adenine nucleotide translocase; MnSOD = manganese superoxide dismutase; GPX = glutathione peroxides; AIF Endonuclease = apoptosis-inducing factor; APAf-1 = apoptotic protease activating factor. (Modified from www.mitomap.org, and Nicholis and Minczuk, Exp Gerontol. 2014. 56:175-81. Illustration by S. Atilano)
The Intermembrane Space is the site of the “ion motive force,” a term that refers to the accumulation of hydrogen ions into this compartment as a consequence of these ions being pumped from the matrix by protein Complexes (I, III, IV) of the electron transport chain (ETC). The exit of these hydrogen ions through the ATP synthase, also known as Complex V of the ETC, back into the matrix is what generates ATP. The Intermembrane Space is also the site of multiple proteins involved in initiating apoptosis, such as cytochrome c, which is sequestered in this compartment by its interaction with cardiolipin, a MT-specific lipid of the inner membrane. Other regulators of apoptosis include apoptosis inducing factor, high-temperature requirement protein/Omi, and Smac/diablo. Release of these proteins from the Intermembrane Space into the cytosol initiates a cascade of events that culminates in apoptosis and cell death. Other proteins present in the Intermembrane Space are involved in the import and folding of proteins that reside in this compartment, as well as the antioxidant, superoxide dismutase, which helps to detoxify reactive oxygen species generated by the ETC.
The inner membrane, with similar lipid content as bacterial membranes, has high cardiolipin content and is selectively permeable, allowing specific molecules (e.g., oxygen, carbon dioxide, and water) into the matrix [10]. Cardiolipin is a negatively charged lipid that drives transport of positively-charged molecules across the outer membrane and is required for maintaining function of the ETC proteins. The surface area of the inner membrane is increased significantly by numerous invaginations called cristae that contain embedded protein respiratory complexes associated with the ETC (Complexes I, III, IV) and production of ATP (Complex V). Multiple proteins are involved in maintaining cristae integrity, including mitofilin, optic atrophy 1 (OPA1) and ChChd3. Maintenance of cristae integrity is essential for optimal energy production, which requires that the ETC proteins are localized in close proximity for effective transfer of electrons.
Data from human donor eyes support the idea that MT architecture is disrupted with AMD. In a study using electron microscopy to analyze RPE MT, Feher and colleagues reported significant disruptions in MT membranes and cristae in donors with AMD compared to age-matched controls [6]. In a proteomic analysis of RPE, increased mitofilin was reported in donors at more advanced stages of AMD [11]. Since mitofilin helps to maintain cristae integrity, the authors suggested the increase could be a compensatory response to help rescue and stabilize the degenerating cristae.
Also embedded in the inner membrane is the “translocase of the inner membrane” (TIM) complex that, working in conjunction with TOM, is responsible for importing nuclear-encoded proteins into the matrix. Multiple heat shock proteins (mtHSP60, mtHSP70, HSP90) assist with the import and folding of resident MT proteins and are localized both in the Intermembrane Space and Matrix. Other inner membrane proteins that work in concert with outer membrane proteins include the adenine nucleotide translocator (ANT), which associates with VDAC in the outer membrane and forms the MT permeability transition pore that opens to allow escape of cytochrome c and other proteins involved in initiating apoptosis. ANT is also involved in moving ADP and ATP into and out of the MT. The inner membrane protein OPA1 interacts with mitofusion in the outer membrane to initiate MT fusion. Mutations in the OPA1 gene have been associated with optic atrophy type 1, which is a dominantly inherited optic neuropathy resulting in progressive loss of visual acuity, often leading to legal blindness [12].
Proteomic analysis of RPE from human donors with AMD showed decreased content of several heat shock proteins that are critical for the import of MT resident proteins. Identified proteins include mtHSP60 and in two separate proteomic studies of the RPE and isolated MT, a decrease in mtHSP70 was observed [11, 13]. These two heat shock proteins assist TOM and TIM with the import and refolding of nuclear encoded proteins and therefore, a reduction in these two proteins in AMD suggests potential defects in this critical process in diseased RPE.
Recent analysis of the MT matrix has identified 495 proteins that reside in this central compartment [14]. The processes ongoing in the matrix and some of the associated proteins include: mtDNA replication and transcription (TFAM, TFB2M, POLRmt), protein translation (RNA polymerase, ribosomes, Tufm, Rnase P), protein quality control (Lon and ClpXP proteases, chaperones, methionine sulfoxide reductase), and energy production (ETC Complexes). In addition to proteins, the matrix also contains mtDNA (discussed in greater detail in Sect. 10.2) and all the machinery required to produce the 13 proteins that are encoded by the MT genome. These 13 proteins, which are part of the ~100 subunits that make up the multisubunit Complexes of the ETC, are absolutely required for a functional ETC. Many MT diseases caused by mutations in either mtDNA or in the nuclear-encoded genes of ETC subunits have ophthalmic involvement [12]. For example, Leber Hereditary Optic Neuropathy (LHON) is maternally-inherited and caused by specific point mutations in subunits of Complex I that are encoded by the MT genome.
The MT have been referred to as the “powerhouse of the cell” because all mechanisms for generating energy (ATP), except for glycolysis, are conducted within the MT matrix. The citric acid cycle (TCA) and β-oxidation, two energy-producing pathways that use glucose and fatty acids as substrates, produce acetyl-CoA (Fig. 10.3). This product is used to generate the energy substrates NADH and FADH2, which are utilized by the ETC in a process known as oxidative phosphorylation. The sequential transfer of electrons between each ETC Complex (I, III, IV) culminating in the reduction of oxygen to water provides the energy to pump hydrogen ions from the matrix into the Intermembrane Space. Release of hydrogen ions through Complex V (ATP synthase) into the matrix drives the phosphorylation of ADP to form ATP. Complex II, also known as succinate dehydrogenase, is the only MT enzyme that participates in both the TCA and oxidative phosphorylation. Complex II transfers electrons that are produced by the oxidation of succinate to fumarate (TCA) to Complex III (oxidative phosphorylation).
Strong experimental evidence supports the hypothesis that RPE MT function is detrimentally altered with AMD. Direct measurement of MT function in primary RPE cultures showed that RPE from human donors with AMD exhibited reduced MT respiration [15] and ATP production [8, 15]. Data from a proteomic analysis of isolated RPE MT from human donors with AMD reported decreased content for several protein subunits of the ETC [11]. Three of the subunits with lower content are part of Complex V, which could result in decreased ATP production. Additionally, Complex V is involved in stabilizing MT morphology and maintaining membrane potential. A loss of these functions would be detrimental to cell viability. Further discussion of how failures in retinal metabolism contribute to AMD pathology will be included in Sect. 10.3.
While the most familiar role of the MT is in energy production, this organelle performs a number of additional functions that are essential for cell viability. As previously mentioned, MT regulate apoptosis and subsequent cell death through the release of various proteins (cytochrome c, AIF endonuclease) that are normally sequestered in the MT. They are the site of heme and steroid biosynthesis, and provide calcium buffering for the cell when cytosolic calcium levels increase over normal levels. Calcium is freely permeable through the outer membrane, but requires the aid of the calcium uniporter and Na/Ca exchanger in order to be shuttled into and out of the matrix. Not only does the MT provide buffering from excess calcium, its influx into the MT matrix plays an important role in regulating MT energy production, for example, through the activation of isocitrate dehydrogenase, one of the key regulatory enzymes of the TCA. In neurons, concurrent calcium increases in the cytosol and MT act to synchronize neuronal activity with MT bioenergetics [16].
MT also serve as a signaling platform, communicating changes in the MT environment and cellular energy demands to and from the nucleus in a process referred to anterograde/retrograde signaling (Fig. 10.3). Reactive oxygen species (ROS), which are produced as a normal by-product of respiration, serve as signaling molecules between the MT and the nucleus. Communication between the MT and nucleus is also induced by changes in the MT electrochemical gradient, the unfolded protein response, and fluctuating levels of ATP or calcium. These varied signals initiate changes in gene expression in both the nucleus and MT to accommodate changing energy demands and cellular conditions. For example, changes in cellular ROS content can initiate signaling through redox-sensitive transcription factors, such as Nrf2 and NfKB, which are proteins that utilize the oxidation of cysteine residues as the signal for activation and respond by translocating into the nucleus and upregulating gene expression to counteract the oxidative stress. Examples of redox-regulated transcription factors include AP-1, p53, estrogen and glucocorticoid receptors, Hif-1, and Nrf-2.
Under normal conditions, these multiple signaling mechanisms play an essential role in maintaining cell health. Abnormal conditions, for example, excessive amounts of MT calcium or high ROS, can disrupt signaling and cell homeostasis. In particular, excessive ROS can cause oxidative damage to lipids, proteins, and DNA. Oxidation of lipids can disrupt membrane integrity and increase membrane permeability to previously excluded molecules. Lipid oxidation can also change the fluidity of membranes in a way that could adversely affect the function of transmembrane proteins, like ETC Complexes, whose function requires a specific lipid environment. Oxidation of the MT-specific lipid cardiolipin in the MT inner membrane disrupts its interaction with cytochrome c, which is tethered to the membrane by cardiolipin. Once cytochrome c is released into the cytosol, it activates apoptosis and causes cell death. Oxidative cleavage of lipids can also generate 4-hydroxynonenal (HNE) and carboxyethylpyrrole (CEP), which can form adducts on DNA and covalently modify lysine residues on proteins. CEP is a specific oxidative fragment of docosahexaenoic acid, which is found in neural tissue and is particularly abundant in the outer retina. Increased CEP-adducted protein has been found in drusen, AMD patient plasma, and AMD donor photoreceptors [17, 18]. AMD-like lesions were observed in mice after immunization with CEP-adducted albumin, suggesting that this specific oxidation product may be toxic to retinal tissue [19]. Abundant HNE adducts have also been reported in retinal proteins, although HNE was not significantly increased with AMD [20]. However, the enzymes used for detoxifying HNE were significantly elevated in retinas from AMD donors suggesting that the retina had mounted a compensatory response to this specific type of lipid oxidation product.
In addition to the generation of lipid-derived adducts, ROS can also directly damage MT proteins, causing protein modifications such as carbonyls, deamidation, and modification of tyrosine to nitrotyrosine. This damage can impair the ability of MT to generate energy by affecting proteins of the ETC, TCA cycle, beta oxidation, or affect calcium buffering capacity by inactivating the calcium transporters located in the inner membrane. ROS-induced damage to mtDNA includes double- and single-strand breaks and formation of DNA adducts, which can interrupt translation and replication of the MT genome. mtDNA damage (discussed in detail in Sect. 10.2) can ultimately cause mutations or deletions in key MT-encoded proteins or tRNA that are required for synthesis of MT proteins.
10.2 Influence of mtDNA on Retinal Disease
To date, the nuclear DNA, with approximately 20,000 protein-coding genes, has been studied extensively for its relationship to development and diseases. In contrast, significantly less is known about the role that mtDNA plays in human pathologies. For the past 50–60 years, it has been thought that the main cellular contribution of mtDNA was energy production. However, more recent studies have demonstrated that mtDNA plays important roles in retrograde signaling (from mitochondria to nucleus), modulating nuclear genes, and encoding for MT-Derived Peptides (MDPs) that are cyto-protective against aging and diseases [21,22,23,24]. This section will provide an overview of mtDNA structure and discuss how mtDNA influences retinal diseases, and in particular AMD.
10.2.1 mtDNA Structure
MT are unique in that they have their own DNA that is inherited through the maternal lineage. The human mtDNA forms a closed circle of double stranded DNA, with 16,569 nucleotide pairs, comprised of two strands (Heavy and Light Strands) that are differentiated by their nucleotide content (Fig. 10.4). The heavy strand is guanine rich and encodes for 28 genes while the light strand is cytosine rich and encodes for 9 genes. Unlike the nuclear genome, mtDNA lacks introns and has a major Non-Coding Region (NCR) that includes the OH (the Heavy-strand origin) for replication, Heavy-Strand promoter (HSP) and Light-Strand promoter (LSP) for transcription, along with the Displacement-loop (D-loop), which is actually a stable, third short DNA strand (also referred to as 7S DNA) [25]. The hypervariable regions -1 (HVR1) and -2 (HVR2) are also located within the NCR and are frequent sites of mutations and polymorphisms. The coding region of mtDNA codes for 37 genes including 13 protein subunits essential for oxidative phosphorylation, 2 ribosomal RNAs and 22 transfer RNAs [26,27,28].
Map of human mtDNA showing non-coding region (NCR) and coding genome. The displacement loop (D-loop) is found within the NCR and has a short, third strand of DNA (7S DNA). The Mitochondrial Derived Peptides are encoded from the 16s RNA and 12s RNA regions of the mtDNA. The region encompassing the “Common Deletion” includes 4977 base pairs. OH, Heavy strand origin; OL Light strand origin; HSP, Heavy strand promoter; LSP, Light strand promoter; CSB, Conserved sequence block; TAS, Termination associated sequences; HVS, Hypervariable region (Modified from www.mitomap.org and Modified/Adapted with Permission from ScienceDirect, Nicholls and Minczuk 2014. Illustration by S. Atilano)
Within a cell there is a single DNA copy of the nuclear genome but multiple copies of mtDNA because there can be thousands of MT per cell, and within each mitochondrion, 1 to 10 copies of mtDNA. With aging and exposure to oxidative stress, mtDNA molecules can be damaged, which results in a mixture of nonmutated (wildtype) and mutant mtDNA within the same cell. This mixture of damaged and undamaged mtDNA is termed heteroplasmy. When cells with heteroplasmic mitochondria divide, the two types of mtDNA are randomly or in some instances nonrandomly distributed into the daughter cells [29,30,31,32,33]. Cells can function with relatively low levels of heteroplasmy but once this threshold is breached, abnormal functions, cell death and diseases can occur.
10.2.2 Mechanisms That Influence Disease Processes
The contribution of the mtDNA to disease and pathology can be the result of inheritable recent mutations, the ancient adaptive polymorphism changes (haplogroups) or the somatic changes associated with aging. Each mechanism will be discussed below.
10.2.2.1 Maternally Inherited mtDNA Disease-Causing Mutations
There are well described categories of diseases caused by specific mutations within the mtDNA genome, such as MELAS (Mitochondrial encephalopathy lactic acidosis and stroke-like episodes) and MERRF (Myoclonic epilepsy and ragged red fibers) [34, 35]. Individuals born with these mtDNA mutations have deficiencies in the various respiratory complexes of the ETC, resulting in decreased ATP production. They often have numerous systemic abnormalities including deafness, diabetes, myopathies, and neuropathies. The common ocular signs of these mtDNA mutations are retinal pigmentary degenerations and optic nerve atrophy. To date, there are no reports of these types of maternally inherited mtDNA mutations associated with AMD.
10.2.2.2 Maternally Inherited mtDNA Haplogroups and Their Association with AMD
Haplogroups are defined as an accumulation of mtDNA single nucleotide polymorphisms (SNP) that can be traced through maternal lineages and represent populations of different geographic origins (Fig. 10.5). These SNP variations have occurred over more than 150,000 years in response to migration patterns and climate adaptations [3]. The oldest haplogroups are located in Africa (L0, L1, L2, L4-6). The L3 population migrated north and gave rise to founder haplogroups M, N and R. From these founder haplogroups, the European (H, T, U, V, W, X, I, J, K haplogroups), Asian (A, B, C, D, F, G haplogroups) and the most recent Native American haplogroups (A, B, C, D) have evolved. Therefore, no matter the present-day location of an individual, mtDNA analyses can identify their maternal-origin lineage. This is significant because haplogroup-defining SNP variants within the mtDNA coding region can be nonsynonymous (amino acid changing), which can alter efficiencies of energy production, causing increased ROS formation, apoptosis and cell death. If the haplogroup-defining SNP variants are found in the mtDNA Non-Coding Region, then the mtDNA replication and transcription rates can be affected. This means that each haplogroup, with its different set of SNPs, can produce unique bioenergetic properties, which may play a role in developing diseases and responding to medications.
Schematic of the major, high branching haplogroup-defining mitochondrial SNPs. The SNPs on the left side of the position number is the rCRS nucleotide (GeneBank NC_012920) and defines that haplogroup. The diverging haplogroup is represented by the SNP on the right of the number (Modified from www.mitomap.org, Illustration by S. Atilano)
Since AMD is found most commonly in Caucasian populations, the European haplogroups have been analyzed and it has been reported that J, T and U haplogroups are associated with AMD [36,37,38,39,40,41] while the H haplogroup has a protective effect [39]. Large, soft drusen and retinal pigment abnormalities, which are characteristics of AMD retinas, have been associated with J and U haplogroups [37]. The haplogroup T-associated SNP (A4917G), located in the NADH dehydrogenase subunit 2 of complex I (MT-ND2), is an independent predictor of AMD [36]. Two variants of the T2 haplogroup, A11812G of MT-ND4 and A14233G of MT-ND6 located in respiratory complex I, are 2.5 times more likely to be associated with advanced AMD than the age-matched control subjects [40]. Additionally, Udar and coworkers found that patients with late AMD showed strong association with noncoding mtDNA D-loop SNPs in the neural retina [41].
Our knowledge and understanding of the effects of mtDNA upon cellular homeostasis has been advanced through the use of the transmitochondrial cybrid model, which are cell lines with identical nuclei, but contain MT from different subjects. Using human retinal pigment epithelial cell cybrids, it has been demonstrated that mtDNA can greatly affect growth rates of cells, bioenergetics and expression levels of nuclear genes for complement, inflammation and angiogenesis, pathways which are important in human retinal diseases [23, 42,43,44,45]. In addition, cybrids with H (protective against AMD) versus J (high risk for AMD) mtDNA haplogroups have different responses to heat stress, hydrogen peroxide, and ultraviolet radiation [42, 46,47,48]. These studies demonstrate that an individual’s mtDNA background sets up baseline cellular homeostasis, making the cells differentially susceptible to identical stressors.
The mechanism(s) by which mtDNA haplogroups influence AMD risk is unclear. It is known that different SNP variants in MT can alter cellular bioenergetics causing partial uncoupling of oxidative phosphorylation and decreased efficiency of ATP production [49, 50]. If MT-generated energy levels decline below a specific bioenergetic threshold, the MT permeability transition pore (mtPTP) can be opened and the apoptosis pathway activated. Models of conplastic strains of mice (where the nuclear genome from one strain has been crossed onto the cytoplasm of another, designated as Nuclear genome-mtCytoplasmic genome) have also demonstrated that the mtDNA affects pathways related to behavior, immunity and cognition [51].
There are a number of ocular diseases associated with specific haplogroups [52,53,54,55,56,57] but the literature lacks reports examining thousands of patients. The analyses for mtDNA haplogroups and variants are often difficult because the mitochondrial protocols are more limited than those available for nuclear gene analyses. The common approaches to characterize mtDNA patterns are to isolate the total DNA from blood or saliva, then characterize the lineages by identifying the haplogroup defining SNPs (Fig. 10.5) using PCR/RFLP (polymerase chain reaction/restriction fragment length polymorphism), custom designed TaqMan probes and/or whole mitochondrial DNA sequencing. At present, the platforms/chips used for GWAS (genome wide association studies) do not have enough SNPs on them to determine haplogroups accurately, making it very difficult to use that format for large scale mtDNA classification. In addition, the mtDNA SNP numbering on the commercial arrays is variable, making analyses of the data inconsistent and unreliable. Until this technical problem is resolved, it is unlikely that analyses for large numbers of AMD populations will be available.
10.2.2.3 Somatic Age-Related mtDNA Damage Associated with AMD
The third mechanism by which mtDNA can affect disease processes is through the cumulative mtDNA damage associated with aging. Human and animal studies show, the levels of mtDNA fragmentation/degradation/deletion is increased significantly with aging [58, 59]. Environmental factors, such as oxidative stress and ultraviolet radiation, can cause strand breaks, deletions and new SNP variants. In addition, one of the errors of double-strand break repair is the deletion of a 4977 base pair region of the MT genome called the “Common Deletion” (Fig. 10.4). This deletion accumulates with age in postmitotic tissue, such as brain, skeletal muscle, heart and RPE [60, 61]. Several factors make mtDNA more susceptible to damage, including the lack of protective histones and its location within the MT matrix, where it is in close proximity to sites of ROS formation. Importantly, MT have poor DNA repair processes, thereby allowing the damage to accumulate with aging.
There are numerous reports of MT abnormalities, including mtDNA damage, in both the neural retina and RPE with aging and AMD [6, 11, 58, 60, 62,63,64,65]. It was reported that sequence analyses of the mtDNA D-loop from the neural retina found a significantly greater number of SNPs per person in the AMD population compared to either older or younger normal groups [62]. Increased oxidative DNA damage in retinas from donors with AMD was also shown using immune-histochemical staining with antibodies that recognize 8-OH-dG, a specific DNA adduct generated by ROS that serves as a biomarker of oxidative stress [41]. In the RPE from donors with AMD, an accumulation of mtDNA oxidative damage was found compared to RPE from age-matched normal donors [60, 65]. These findings were supported by analysis of primary cultures of RPE cells where increased DNA damage and fivefold increased somatic mutations were observed in RPE from AMD donors compared with age-matched controls [66]. Interestingly, the increased mtDNA damage associated with AMD progression occurs in the RPE but not the neural retina, and is localized to specific MT genome regions, including the regulatory D-loop [65]. Deletions occurring in coding regions for subunits of Complex I (ND3, ND4, ND4L, ND5), Complex IV (Cytochrome Oxidase III), or Complex V (ATPase 6, ATPase 8), could have tremendous impact on cellular bioenergetics and functions. Furthermore, this type of damage can increase heteroplasmy (mixture of damaged and undamaged mtDNA) and decrease copy numbers.
Using a model of AMD cybrid cells (RPE cell lines with identical nuclei but MT from individual AMD patients) the destructive impact that damaged MT have on retinal cells has been demonstrated [24, 67]. When MT from AMD subjects are placed into human retinal cell lines, there was a significant loss of viability, impaired MT functions, upregulation of pro-apoptosis, pro-autophagy, pro-ER stress genes, and enhanced vulnerability to oxidative stress. Importantly, Nashine and coworkers showed that although damaged, these AMD MT can be rescued by treatment with Humanin-G, a cyto-protective peptide that increased the cellular longevity [24]. Humanin is one of several MT-derived peptides that are encoded by the MT genome within the 16S ribosomal RNA gene. The role of these MT-derived peptides in MT metabolic control and cytoprotection is currently being explored [68]. Initial results are encouraging researchers to pursue the MT-targeting therapies to prevent cell death associated with AMD. It is highly likely that future investigations will yield multiprong approaches to further characterize the damaged AMD MT and identify novel therapeutic agents to restore the defective MT to healthier status.
10.3 Introduction to the Metabolic Ecosystem Concept and Its Link to AMD Pathology
Section 10.3 will discuss the concept of a retina/RPE ecosystem by first providing an overview of the pertinent metabolic features and pathways within individual retinal cells. The final section will consider the idea that a bioenergetic crisis in the RPE is the primary event that tips the metabolic balance toward AMD [69] and addresses the critical question of why the macula is preferentially and most severely affected with AMD.
The idea of a metabolic ecosystem within the eye makes sense when one considers the many important and diverse metabolic interactions that occur between organs and tissues in a whole organism. All living cells convert fuels into metabolic energy. Each type of cell adapts its biochemical machinery to make the most effective use of the types of fuels available to it and to the timing and magnitude of its energy needs. Cells within an organism generally have metabolic features that allow them to support each other. For example, when muscles are highly active, they can produce pyruvate at a rate faster than it can be oxidized by the cell’s MT. Glycolysis requires a continuous supply of NAD+ to oxidize glucose. The muscles are able to sustain energy production through glycolysis by regenerating NAD+ via reduction of pyruvate to lactate. Remarkably, the liver can recycle the lactate. Lactate is exported from muscles to the blood and imported into liver cells. Energy from fatty acid oxidation in the liver cells can then be used to reduce the lactate to glucose. The glucose is exported from the liver and then taken up by the muscles to support the continuing energy demands of the active muscle. The metabolic interdependence in this classic biochemistry example highlights how distinct cells with specialized metabolic features can support each other and how they function as a metabolic ecosystem.
10.3.1 Metabolic Features of the Retina
10.3.1.1 Aerobic Glycolysis in the Retina
Cells within the vertebrate eye also have specialized metabolic features. This was discovered in the early 1920s by Krebs and Warburg when they were exploring glycolytic and oxidative metabolism in a variety of tissues [70, 71]. They noted that tumors and retinas are especially glycolytic, meaning that the ratio of lactate production to O2 consumption is substantially higher in these tissues compared to other tissues they had examined. They observed that lactate was produced even when plenty of O2 was available. Since then that finding has been confirmed many times over [72,73,74,75,76,77]. Production of lactate caused by a shortage of O2 is common and is referred to as “anaerobic glycolysis.” Production of lactate by retinas or tumors is referred to as “aerobic glycolysis” because it occurs even when O2 is readily available [78]. In recognition of its discovery by Warburg, this type of metabolism is referred to as the “Warburg Effect.”
The site of aerobic glycolysis in retinas hasn’t been identified directly, but several observations indicate it occurs in photoreceptors. Most of the lactate produced by an eye comes from the outer retina, which is made up primarily of photoreceptors [76]. Hexokinase, which catalyzes the first step in glycolysis is enriched in the outer retina [79,80,81] and specialized isoforms of glycolytic enzymes like hexokinase II [80, 81] and pyruvate kinase M2 [72, 82,83,84] are specifically expressed in photoreceptors. Lactate dehydrogenase A is expressed at high levels in photoreceptors [72, 82, 85]. Inactivating pyruvate kinase, lactate dehydrogenase [72] or hexokinase II [80] expression in rods alters their morphology in ways that appear to be cell-autonomous [72]. A fluorescent glucose derivative gavaged into the stomachs of either mice or zebrafish [86] or injected into the tail veins of mice [87] accumulates in the retinas, primarily in photoreceptors.
10.3.1.2 Mitochondria in Photoreceptors
Retinas can consume other fuels including glutamine, lactate, pyruvate [88], fatty acids and ketone bodies [89, 90]. Those fuels require MT to oxidize them and the retinas are very capable of actively consuming O2. Linsenmeier’s and Cringle’s in vivo measurements [91, 92] of the distributions of O2 in retinas have shown that nearly all of the O2 available from the choroid circulation can be consumed by the outer retina, most likely by the photoreceptors.
MT are abundant in rods and cones [93]. They are in a region of the photoreceptor just below the base of the outer segment known as the ellipsoid. When there is inner retinal vascularization, as is the case for humans, mitochondria also accumulate at photoreceptor synaptic terminals [94]. In animals such as frogs, rabbits, guinea pigs, horses and birds, the inner retinas are mostly avascular [95]. Those retinas do not have a substantial source of O2 from the inner retina. Photoreceptor synapses in avascular retinas can obtain ATP made at the ellipsoid and transferred to the synapse via a phosphocreatine shuttle [96]. An isoform of creatine kinase that localizes to mitochondria can make phosphocreatine from ATP at the ellipsoid, which then can diffuse from the ellipsoid to the synaptic terminal where the phosphocreatine is used by another isoform of creatine kinase to make ATP. Measurements of metabolic flux through mitochondrial intermediates revealed that some aspects of mitochondrial function are more active in darkness than in light [74]. This could be a consequence of the higher cytosolic free Ca2+ levels in darkness. Higher cytosolic Ca2+ can alter the concentration of free Ca2+ in the mitochondrial matrix [97]. Flux measurements suggest oxidation of α-ketoglutarate to succinyl CoA, a step in the TCA cycle, is stimulated in darkness compared to in light [74], consistent with findings that Ca2+ can stimulate α-ketoglutarate dehydrogenase activity in vitro [98]. Mitochondria in retinas appear relatively uncoupled [74] and consumption of O2 does not appear to be limited by ATP demand [74, 99, 100].
MT morphology in photoreceptors is striking and diverse [86, 96, 101]. For example, MT in mammalian rods is highly elongated and line up close to the plasma membrane in parallel with the major axis of the rod cell. In contrast, MT in cones of zebrafish retinas coalesces into a tightly packed cluster just below the outer segment. The tight cluster of MT in the zebrafish cones can maintain distinct pools of Ca2+ in the outer vs. inner segments of a cone cell [97]. There also is evidence that MT can influence the optical properties of photoreceptors [93]. Any advantages that these unique and diverse structures provide to photoreceptors still need to be identified. MT are more abundant and may be more active in cones [101]. In primate retinas the sizes and volumes of MT vary throughout the fovea, perifovea and peripheral regions of the retina [93]. The morphologies of these MT change when photoreceptors become affected by AMD and the changes can be followed by Optical Coherence Tomography (OCT) [102].
10.3.1.3 Müller Cells
Müller glia span the retina radially with their apical processes extending around the inner segments of the photoreceptors and their end feet at the inner surface of the retina. Just below the apical processes the junctions between Müller cells create a visible structure referred to as the outer limiting membrane. Within the retina the Müller cells extend horizontal processes of diverse morphology that infiltrate between neurons.
Immunohistochemical studies suggest that Müller cells may not have some of the enzymes needed for glycolysis. Pyruvate kinase and hexokinase, which are detected readily in photoreceptors, have not yet been detected in Müller cells [83, 85]. Immunocytochemistry indicates that Müller cells also lack a key enzyme required for the malate-aspartate shuttle that shuttles electrons from glycolysis into MT [103]. It is important to know whether the portion of Müller cells in the outer retina has glucose transporters, which are required for direct uptake of glucose. However, the overlap of Müller cell apical processes with the ellipsoid region of rods, which contain the GLUT-1 glucose transporter, makes it difficult to resolve unambiguously the localization in the apical processes of enzymes and transporters. Co-labeling with several rod and Müller cell-specific antibodies suggest that GLUT-1 in Müller cells is abundant in the end-feet but not in the apical processes [86]. However, more precise and higher-resolution studies will be needed to resolve this definitively. Müller cell MT are small and abundant just underneath the apical processes [86]. Glial cells are the only cells in the retina with the enzyme, glutamine synthetase, needed to synthesize glutamine. The ability of retinas to synthesize glutamine from lactate and aspartate indicates that Müller glia are capable of taking up and metabolizing lactate [83].
10.3.1.4 Glycogen in Retinas
Glycogen is present in retinas. It is most abundant in the end-feet of Müller cells. Glycogen also can be detected in cone photoreceptors. The amount of glycogen stored in Müller cell end feet can vary depending on the amount of glucose available to the retina [104]. There is a report that Müller cells express the enzymes needed for gluconeogenesis [105]. This is an important observation, because gluconeogenesis from lactate could produce glucose for glycogen storage to support the activity of inner retinal neurons. However, that line of investigation has not been pursued beyond the initial reports 30 years ago and needs further confirmation. Glycogen and gluconeogenesis in Müller cells could be fundamentally important for retinal survival and function.
10.3.2 Metabolic Features of the RPE
RPE cells form a cellular monolayer between the choriocapillaris and retina [106]. They are sealed together by tight junctions and function as a blood retina barrier for the outer retina. To reach the retina, fuels and metabolites from blood in the choriocapillaris must pass through membrane transporters in both the basolateral and apical plasma surfaces of RPE cells. Glucose from the blood must be able to reach the retina to support the high glycolytic activity of the retina. The glucose transporter, GLUT-1, is exceptionally abundant on both surfaces of the RPE [107, 108]. Monocarboxylate transporters are differentially distributed to the two surfaces of the RPE. MCT1 is on the apical side whereas MCT3 is on the basolateral side [109].
RPE cells perform vital functions that support the retina [106]. Once a day they phagocytose the tips of the rod outer segments and either metabolize their contents or recycle them back to the retina. RPE cells also have the capacity to esterify and isomerize all-trans retinol, a product of photobleaching visual pigments [110]. This is an essential process in the pathway for regeneration of rhodopsin after it has undergone photobleaching in rod outer segments.
10.3.2.1 Glycolysis in the RPE
The exceptional abundance of glucose transporters on both sides of the RPE [107, 108] highlights how RPE cells are adapted to facilitate transport of as much glucose as possible from the choriocapillaris to the retina. Consistent with this important metabolic function, RPE cells consume glucose much more slowly than retina. Like all cells, RPE cells need fuel for energy and anabolic activities. Compared to retinas, RPE cells have a limited ability to use glucose. However, they can oxidize lactic acid, glutamine, and fatty acids as alternative fuels [74, 86, 90, 111]. RPE cells also actively consume proline and they can export a variety of metabolites involved in energy production and anabolic activities [111]. RPE cell metabolism is more enriched than the retina in two other important metabolic pathways, reductive carboxylation of α-ketoglutarate to produce citrate [112], and carboxylation of pyruvate to produce oxaloacetate [86]. All of these metabolic features appear to be adaptations that can sustain metabolic requirements of RPE cells while minimizing their dependence on glucose.
10.3.2.2 Glycogen in the RPE
RPE cells store glycogen. The amount of stored glycogen in cultured human retinal epithelial cells increases when there is abundant glucose in the medium in which they grow and decreases when glucose levels in the medium have been depleted [113]. These findings suggest that glycogen stored in RPE cells can ensure a steady supply of glucose to the retina, but this potentially critically important activity of RPE cells has not yet been investigated directly.
10.3.3 Metabolic Synergism in the Eye: A Retina/RPE Ecosystem
As described in the preceding sections RPE cells and the neurons and glia of the retina have diverse and specialized metabolic features. The distinct metabolic functions of cells in the eye appear generally analogous to the distinct and diverse metabolic roles of various types of plants and animals in the ecosystem of a forest or ocean.
Based on currently available data it is likely that the retina/RPE metabolic ecosystem functions as portrayed in the schematic model in Fig. 10.6 [86]. Glucose from the choriocapillaris enters and leaves RPE cells through abundant GLUT-1 glucose transporters on the basolateral and apical surfaces of the RPE cells. When glucose reaches photoreceptors most of it is oxidized rapidly by glycolysis and then reduced to lactate. The lactate, with metabolic energy still remaining within its chemical bonds, is exported from the photoreceptors to both the RPE and to Müller cells where it can be oxidized by MT to drive synthesis of ATP.
Summary of the metabolic interactions between RPE, photoreceptors and MÜller cells. Glucose from the choroidal blood supply passes through glucose transporters on the basolateral and apical surfaces of the RPE to reach the retina. The glucose that reaches the Interphotoreceptor Matrix (IPM) is taken up by glucose transporters on the plasma membrane of the rod inner segments. Glucose in the rod cell is converted rapidly to lactate by aerobic glycolysis and then released from the photoreceptor. The lactate released can be used either by Müller cells or by the RPE. Studies have shown that lactate can suppress glycolysis in the RPE to minimize consumption of glucose by the RPE so that more glucose can reach the retina
A significant advantage of this type of metabolic strategy is that oxidation of lactate imported into RPE cells can deplete NAD+ in the RPE cells. Since NAD+ is needed for glycolysis, depletion of NAD+ by lactate further suppresses glycolysis in RPE cells so that more glucose can pass through the RPE and reach the retina.
10.3.3.1 Findings from In Vivo Experiments that Support the Concept of an Essential Retina/RPE Ecosystem
This concept of metabolic interdependence is generally supported by the results of several recent experiments. Genetic manipulations that affect a specific type of cell in the retina or RPE affect the viability not only of that specific type of cell, but also of other types of cells that were not affected directly by the mutation. Some of these examples are described briefly in the following paragraphs.
Example 1
Cone viability depends on the presence of rods. Rods are more abundant than cones in human and mouse retinas. Even within the human macula there are 9 times more rods than cones [114]. Although rods and cones carry out similar functions in the eye, many of the phototransduction genes that have specific roles in rods are not needed and are not expressed in cones because other genes fulfill those functions in cones [115]. For example, PDE6β, PDE6γ and rhodopsin are expressed only in rods, not in cones. A mutation in any of these genes can directly disrupt rod viability. However, after the rods degenerate, cones then also begin to degenerate, even though the mutation did not influence cones directly. Recent findings provide clues about the nature of this “bystander effect.” As rods die, cones show evidence of nutrient deprivation [116]. The evidence is a change in the phosphorylation state of mTOR, a protein that senses the availability of nutrients. Recent findings have identified two biochemical mechanisms that may be responsible for depriving cones of essential nutrients. First, rods in a normal retina produce and release a polypeptide, rod-derived cone viability factor (RdCVF) [117, 118]. RdCVF interacts with a protein, basigin, on cones, that stabilizes or stimulates the activity of glucose transporters on the cone plasma membrane. When rods degenerate, RdCVF no longer is released by rods, glucose transporters on cones are not stabilized and cones starve. The absence of rods also can cause cones to starve because glucose becomes trapped within the RPE. Normally, when a fluorescent glucose derivative is injected into the tail veins of mice it passes through the RPE and accumulates in photoreceptors in the retina [87]. However, as rods degenerate the fluorescent glucose becomes trapped within the RPE making it inaccessible to the cones [117, 118]. Part of this glucose trapping may be caused by failure of the ecosystem (Fig. 10.6). Less lactate will be made by the retina in the absence of rods. The RPE will rely on glucose instead of lactate for fuel so less glucose reaches the retina. Other effects of rod loss on RPE function also may contribute to glucose trapping.
Example 2
The viability of rods relies on mitochondrial activity in RPE cells. MT require a specific transcription factor, TFAM, for expression of genes encoded by mitochondrial DNA. When TFAM is inactivated specifically in RPE cells, the metabolism of the RPE cells become less specialized and consumption of glucose by glycolysis increases [119]. This causes rod photoreceptors in the retina to degenerate. This observation is consistent with two ideas: (i) RPE minimizes its consumption of glucose so sufficient glucose is available to fuel the retina and (ii) maintenance of ATP production and other MT functions in the RPE is essential for retinal health.
Example 3
The viability of rods relies on restraint of glycolytic activity in RPE cells. Unrestrained glycolytic activity in RPE cells could cause the RPE to consume so much glucose that not enough glucose passes through the RPE to fuel the retina. Hypoxia-Inducible factor (HIF1α) can stimulate transcription of genes that encode the enzymes that catalyze glycolysis. Von Hippel-Lindau Factor (VHF) is a protein that suppresses glycolysis by helping to degrade HIF. When VHF expression is blocked specifically in the RPE, HIF1α is stabilized. That makes the RPE more glycolytic and it causes photoreceptors to degenerate [120], consistent with the model in Fig. 10.6.
Example 4
Robustness is enhanced when glycolytic activity is increased in rods. Photoreceptor degeneration can be induced by deleterious mutations in genes that encode subunits of a rod cGMP phosphodiesterase. Normal retinas are highly glycolytic. However, expression of glycolysis genes normally is limited by expression of a protein, SIRT6. When SIRT6 expression is blocked in photoreceptors, the rate of glycolysis in retinas increases several folds above its already high level [121]. Degeneration caused by a PDE6 mutation is slowed substantially when glycolysis is enhanced by SIRT6 inactivation. This is consistent with the model in Fig. 10.6 because enhanced lactate production from the mutant rods can suppress RPE glycolysis so more glucose can reach the retina.
Taken together, all of these findings support the idea that diverse metabolic features of cells in the retina and RPE contribute specific metabolic roles that sustain the viability and function of cells in a metabolic ecosystem.
10.3.4 Relationship Between AMD and the Metabolic Ecosystem of the RPE and Retina
The observations that enhanced glycolysis in RPE cells induces rod degeneration and that enhanced glycolysis in photoreceptors makes them more robust suggest that the state of the retina/RPE metabolic ecosystem can influence degenerative diseases, such as AMD. The distribution of rods and cones and the distributions and volumes of ellipsoid MT in human retinas are not homogeneous [93, 122,123,124,125]. It is likely that the efficiency of the RPE/retina metabolic ecosystem depends heavily on RPE MT being able to use lactate, glutamine, fatty acids and proline as fuels to minimize consumption of glucose. Human RPE MT accumulate DNA damage with advancing age [60, 66] and cumulative damage compromises their metabolic activity [8, 15]. Since the RPE MT have a central role in supporting the metabolic ecosystem sketched in Fig. 10.6, the regions of the nonhomogeneous retina that are most dependent on the metabolic ecosystem ought to be the first regions where photoreceptor stress and damage become evident. Since cones in the central fovea are more enriched in MT than rods [93] they may be less dependent on the metabolic ecosystem, that is, they may be more capable of efficiently using limited amounts of glucose than other regions of the retina. Just outside the central fovea, rods become more abundant, but since they are more sparse in this region their net output of lactate would be limited. This could make the rods in the perifoveal region the most reliant on RPE MT and therefore most sensitive to damage of RPE MT. This hypothesis may help explain why AMD correlates with RPE MT damage and why it initially affects perifoveal rods. Further studies in which the metabolic capabilities of the central fovea, perifovea, and peripheral retinas are compared in nonhuman primate retinas and in human donor retinas will be required to evaluate the validity of this hypothesis.
10.4 Enhancing MT Function and Bioenergetics As a Therapeutic Strategy for Treating AMD
Studies suggest that metabolic dysregulation may be a major contributing factor to disease pathogenesis in AMD [15, 65, 69, 86]. Mutations in over 200 genes have been associated with retinal degenerations (RetNet, [126]), and diseases such as AMD are multifactorial, with genes and environment contributing to disease. Due to the large number of mutations and factors that can lead to retinal degeneration, gene independent strategies that focus on targeting the biological pathways that lead to retinal degeneration or retinal neuroprotection are crucial. Based on strong evidence that dysfunction of the metabolic ecosystem leads to the retinal degeneration associated with AMD, pathways related to energy metabolism, MT biogenesis, and oxidative stress may be ideal targets to treat AMD. This section will discuss several studies that have examined the role of these pathways in AMD and how targeting these pathways may be neuroprotective.
10.4.1 Energy Metabolism
10.4.1.1 AMPK
A viable strategy for treating AMD is to target pathways that stimulate or shift the metabolic ecosystem back to its homeostatic state. An ideal target for enhancing metabolic function is a key pathway involved in regulating energy levels in the cell. One such protein involved in regulating cellular energy metabolism in a variety of tissues is 5′ adenosine monophosphate protein kinase (AMPK). AMPK is activated by cellular metabolic stress and is expressed ubiquitously all eukaryotic cells. AMPK directly binds AMP, ADP, or ATP allowing it to detect energy levels in the cell. AMPK is activated by upstream kinases, including liver kinase B1 (LKB1) and Ca/calmodulin-activated protein kinase (CaMMKKβ), as well as pharmacological activators, such as AICAR and metformin [127]. Activation of AMPK promotes downstream energy producing pathways, including glucose metabolism, MT function, and autophagy, and inhibits downstream energy consuming pathways, including protein synthesis and fatty acid metabolism. This makes AMPK an ideal target for diseases such as AMD, where MT and metabolic dysfunction likely plays an important role in disease pathogenesis.
Several studies have examined the role of AMPK and upstream kinase LKB1 in the retina. Interestingly, LKB1 expression in the retina decreases with age in mice [128]. Studies examining conditional deletion of LKB1 in retinal progenitor cells found that deletion of LBK1 results in loss of ONL thickness, shifts in synaptic positioning, a reduction in the total number of synapses, and loss of rod and cone function. Knocking down AMPK, produced a similar phenotype, suggesting that AMPK plays an important role in this process. Interestingly, the authors were able to reverse the phenotype by 20% using metformin to activate AMPK and by 50% using caloric restriction to activate AMPK. Conversely, feeding the animals a high fat diet resulted in a 70% increase in synaptic mislocalization. The authors found a reduction of phosphorylated, or active, AMPK in older mice, suggesting that levels of activated AMPK decrease in the retina with age [128].
Other studies have investigated metformin-mediated activation of AMPK as a neuroprotective therapy. Studies have shown that systemic metformin treatment is able to activate AMPK in the retina [129]. Metformin treatment preserved retinal function and morphology in mouse models of light-induced retinal degeneration, the Rd10 model of inherited retinal degeneration, and was protective to the RPE in a model of RPE injury using sodium iodate [129]. Metformin protection was dose-dependent, and was associated with increased mitochondrial DNA copy number, increased ATP levels, and reduced levels of oxidative stress and DNA damage. Importantly, metformin was no longer protective with deletion of AMPKα1 and α2 subunits in the retina, suggesting that AMPK is necessary for metformin-induced protection, and that metformin protection is due to local activation of AMPK in the retina [129]. These results suggest that promotion of MT function and retinal metabolism with metformin treatment is neuroprotective to the retina. Therefore, caloric restriction, caloric restriction mimetics, or activators of AMPK are potential neuroprotective therapies that could stimulate retinal energy metabolism homeostasis to prevent retinal degeneration in AMD.
10.4.1.2 Insulin/mTOR
Another metabolic pathway that is a potential neuroprotective target is the mTOR (mammalian target of rapamycin) pathway. mTOR is a key regulator of metabolism and cell growth and is found in two complexes, mTORC1 or mTORC2. mTORC1 regulates cell metabolism and protein synthesis while mTORC2 regulates pro-cell survival mechanisms. Deletion of mTORC1 or mTORC2 individually in the retina does not affect function or survival of cones up to 1 year of age but does result in alterations of outer and inner segment morphology [130]. Deletion of both mTORC1 and mTORC2 in the retina leads to a loss of cone function, but not cone death [131]. These findings suggest that due to the high metabolic activity of healthy cones, the role of mTOR in regulation of metabolism in healthy cones is minimal.
Although the insulin/mTOR signaling pathway may play a minor role in healthy cones, insulin/mTOR signaling may play a larger role in cones under conditions of stress. Using four models of retinitis pigmentosa, Punzo et al. examined changes in gene expression at various points of cone degeneration [116]. Interestingly, 36% of the genes that were upregulated at least twofold upon secondary death of cones were related to cellular metabolism. The insulin/mTOR signaling pathway had the highest number of hits, suggesting that metabolism and mTOR signaling play an important role in the cone death process. Increases in levels of heterodimeric transcription factor hypoxia inducible factor 1 (HIF-1a/b), which increases glycolysis under stressful conditions such as those of low oxygen, and GLUT1, a glucose transporter expressed in the photoreceptors, were observed. This suggests that cone death may be due to compromised glucose uptake in the cones, resulting in starvation. Stimulation of the mTOR pathway with insulin treatment resulted in enhanced survival of cones, while inhibition of insulin with injection of streptozotocin, a drug that kills the insulin-producing beta cells of the pancreas, resulted in decreased survival of cones [116].
Other studies have shown that constitutive activation of mTORC1 is able to preserve function and survival of cones in mouse models of retinitis pigmentosa, and that mTORC1 is required for the protective effect of activating insulin/mTOR pathway [130]. Activation of mTORC1 resulted in increased uptake and utilization of glucose and elevated levels of NADPH. Loss of the mTORC1 accessory protein, RAPTOR, resulted in accelerated retinal degeneration in disease models, but had no effect on retinal function or cell survival when it was deleted in a wildtype retina. Activation of mTORC1 was associated with increased expression of genes related to uptake, retention, and utilization of glucose [130]. These findings suggest that mTORC1 specifically plays an important role in maintaining cone function in disease, and this protection may be mediated through enhanced uptake, retention, and utilization of glucose.
Although these studies focus on models of retinitis pigmentosa rather than AMD, the dysregulation of metabolism observed in these models may reflect pathological features of AMD under conditions of stress, particularly, when there is a reduction in glucose levels reaching the photoreceptors due to increased glycolysis in the RPE. These results suggest that metabolic imbalance is a major contributing factor to loss of cones, and stimulation of pathways such as the insulin/mTOR signaling pathway may be protective to cones under conditions of reduced glucose levels. Further studies examining the role of insulin/mTOR signaling in energy metabolism in AMD are necessary to further investigate this possibility.
10.4.1.3 CoQ10
Coenzyme Q10 (CoQ10), also known as ubiquinone, is ubiquitously expressed, and is localized primarily in the MT. CoQ10 is a component of the electron transport chain acting as an electron carrier. It also has antioxidant capabilities and can affect metabolism-related gene expression. Studies have shown that CoQ10 levels decline in a variety of tissues in the human body with age, including the retina [132]. A randomized, double-blind, placebo controlled clinical trial investigated the efficacy of using a combination of acetyl-L-carnitine, n-3 fatty acids, and CoQ10 in subjects with early AMD [133]. The aim of this therapy was to target MT lipid metabolism for a metabolic therapy to prevent AMD progression. Treatment resulted in preservation of visual field function, as compared to placebo-treated controls, and a reduction in drusen area [133]. These results suggest that using CoQ10 to improve retinal metabolism may reduce progression of early AMD and may be a potential therapeutic target for the disease. Targeting pathways that enhance energy metabolism or promote energy homeostasis in the retina may represent a suitable target for therapies for AMD.
10.4.2 Oxidative Stress and Mitochondrial Biogenesis
10.4.2.1 Nrf2
Other neuroprotective avenues for AMD include reducing levels of oxidative stress through targeting antioxidant pathways. A reduction in oxidative stress in the RPE and retina would likely help prevent oxidative DNA damage to mtDNA, nuclear DNA, and prevent oxidation of lipids and proteins. Nrf2 is a master transcription factor that regulates the antioxidant response in virtually all cell types. Several studies have focused on targeting Nrf2 to reduce levels of oxidative stress in AMD. In unstressed conditions, Nrf2 is held in the cytoplasm by Keap1, which facilitates its ubiquitination and subsequent degradation. However, in conditions of oxidative stress, Keap1 structure is disrupted, preventing it from interacting with Nrf2, which allows Nrf2 to translocate to the nucleus and activate the antioxidant response element. Studies have shown that AAV-mediated gene therapy delivering an Nrf2-derived peptide that interacts with Keap1 to prevent endogenous Nrf2 degradation is able to increase antioxidant expression and protect retinal function and morphology in response to sodium iodate-induced RPE damage [134]. Other groups have shown that Nrf2 upregulation in the retinas of mice, via AAV-mediated gene delivery, resulted in protection from damaging light levels [135]. Targeting Nrf2 to reduce levels of oxidative stress may protect MT from oxidative damage and help preserve RPE function in AMD.
10.4.2.2 PGC-1a
Another potentially neuroprotective target to enhance mitochondrial bioenergetics is peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α). PGC-1α is a transcriptional coactivator that is involved in regulation of many genes involved in energy metabolism, including those involved in MT biogenesis. PGC-1α is activated by increased levels of oxidative stress and by direct phosphorylation by AMPK [136].
Studies have shown that overexpression of PGC-1α in the RPE leads to increased expression of genes associated with fatty acid oxidation, MT respiration, oxidative metabolism, and antioxidants [137]. This increase in antioxidant expression correlated with an increased ability for the RPE cells to cope with oxidative insults, including H2O2 and hydroquinone (an oxidant found in cigarette smoke) [137]. As MT dysfunction and elevated levels of oxidative stress in the RPE are associated with AMD, overexpression of PGC-1α may represent an important target for potential therapies for the disease.
Other studies suggest that PGC-1α plays an important role in vivo. Mice heterozygous for PGC-1α that were fed a high fat diet for 4 months exhibited RPE and photoreceptor degeneration. This was associated with an accumulation of lipofuscin, basal laminar deposits, Bruch’s membrane thickening, and deposits containing proteins with oxidative damage [138]. This phenotype may be due to reduced MT function, as these mice had decreased levels of mtDNA copy number, reduced activity of MT oxidative phosphorylation Complex I, and increased levels of reactive oxygen species [138]. These findings suggest that decreased PGC-1α expression in AMD may contribute to disease phenotypes. Targeting PGC-1α, through indirect activation using an AMPK activator, such as metformin, other pharmacological activators, or gene therapies may represent a possible neuroprotective therapy for AMD that could enhance MT function and possibly prevent RPE dysfunction.
10.4.3 Neuroprotection in AMD
Dysregulation of MT and energy metabolism in the RPE is likely a contributing factor to AMD pathogenesis. As predicted by the Metabolic Ecosystems Model, metabolic dysfunction in the RPE can lead to dysregulation of metabolism in photoreceptors and a bioenergetic crisis in the entire retina. Therapies that target pathways involved in regulation of energy metabolism or MT activity in the photoreceptors and especially RPE, such as AMPK, insulin/mTOR, CoQ10 and PGC-1α, have the potential to restore or preserve energy homeostasis, and thus retinal health and function (Fig. 10.7). Targeting other pathways such as those involved in regulation of oxidative stress response, like Nrf2, may protect the RPE from oxidative insults that could disrupt energy homeostasis. Although these pathways are strong candidates for targets for neuroprotective therapies for AMD, a better understanding of the underlying mechanisms of the pathology of AMD is essential to develop targeted therapies to prevent or treat AMD.
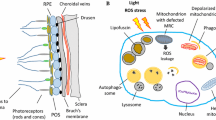
Potential neuroprotective targets for AMD to enhance mitochondrial function and cellular metabolism in the RPE and retina. In the RPE, targeting AMPK, CoQ10, Nrf2, or PGC-1α may promote mitochondrial function and antioxidant responses. The retina, targeting AMPK or mTOR pathways may promote glucose metabolism, enhance mitochondrial function, and NADPH metabolism. Red arrows = inhibitory effect, green arrow = promoting effect
10.5 Summary
MT regulate cellular energy production, oxidative stress, inflammation, signal transduction, and apoptotic pathways.
MT alterations, including mtDNA damage/mutations and dysfunction, likely play an important role in AMD pathogenesis.
Alterations in MT in AMD may disrupt the highly coordinated metabolic ecosystem of the retina, which can lead to further retinal metabolic stress and retinal degeneration.
Targeting MT function and retinal metabolism may be neuroprotective to the retina and as these pathways may represent the optimal neuroprotective therapies for AMD.
References
Burger G, Gray MW, Lang BF (2003 Dec 1) Mitochondrial genomes: anything goes. Trends Genet 19(12):709–716
Gray MW, Burger G, Lang BF (1999 Mar 5) Mitochondrial evolution. Science 283(5407):1476–1481
Wallace DC (2005 Dec 15) A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet 39:359–407
Toft-Kehler AK, Skytt DM, Svare A, Lefevere E, Van Hove I, Moons L, Waagepetersen HS, Kolko M (2017 Sep 30) Mitochondrial function in Müller cells-does it matter? Mitochondrion 36:43–51
Carelli V, La Morgia C, Ross-Cisneros FN, Sadun AA (2017 Jul 26) Optic neuropathies: the tip of the neurodegeneration iceberg. Hum Mol Genet 26(R2):R139–R150
Feher J, Kovacs I, Artico M, Cavallotti C, Papale A, Gabrieli CB (2006 Jul 1) Mitochondrial alterations of retinal pigment epithelium in age-related macular degeneration. Neurobiol Aging 27(7):983–993
Viiri J, Amadio M, Marchesi N, Hyttinen JM, Kivinen N, Sironen R, Rilla K, Akhtar S, Provenzani A, D'Agostino VG, Govoni S (2013 Jul 29) Autophagy activation clears ELAVL1/HuR-mediated accumulation of SQSTM1/p62 during proteasomal inhibition in human retinal pigment epithelial cells. PLoS One 8(7):e69563
Golestaneh N, Chu Y, Xiao YY, Stoleru GL, Theos AC (2017 Jan) Dysfunctional autophagy in RPE, a contributing factor in age-related macular degeneration. Cell Death Dis 8(1):e2537
Yates Iii JR, Gilchrist A, Howell KE, Bergeron JJ (2005 Sep) Proteomics of organelles and large cellular structures. Nat Rev Mol Cell Biol 6(9):702
Schlame M, Ren M (2009 Oct 31) The role of cardiolipin in the structural organization of mitochondrial membranes. Biochim Biophys Acta (BBA) Biomembr 1788(10):2080–2083
Nordgaard CL, Karunadharma PP, Feng X, Olsen TW, Ferrington DA (2008 Jul 1) Mitochondrial proteomics of the retinal pigment epithelium at progressive stages of age-related macular degeneration. Invest Ophthalmol Vis Sci 49(7):2848–2855
Schrier SA, Falk MJ (2011 Sep) Mitochondrial disorders and the eye. Curr Opin Ophthalmol 22(5):325
Nordgaard CL, Berg KM, Kapphahn RJ, Reilly C, Feng X, Olsen TW, Ferrington DA (2006 Mar 1) Proteomics of the retinal pigment epithelium reveals altered protein expression at progressive stages of age-related macular degeneration. Invest Ophthalmol Vis Sci 47(3):815–822
Rhee HW, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, Ting AY (2013 Mar 15) Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339(6125):1328–1331
Ferrington DA, Ebeling MC, Kapphahn RJ et al (2017) Altered bioenergetics and enhanced resistance to oxidative stress in human retinal pigment epithelial cells from donors with age-related macular degeneration. Redox Biol 13:255–265
Ivannikov MV, Macleod GT (2013 Jun 4) Mitochondrial free Ca2+ levels and their effects on energy metabolism in Drosophila motor nerve terminals. Biophys J 104(11):2353–2361
Crabb JW, Miyagi M, Gu X, Shadrach K, West KA, Sakaguchi H, Kamei M, Hasan A, Yan L, Rayborn ME, Salomon RG (2002 Nov 12) Drusen proteome analysis: an approach to the etiology of age-related macular degeneration. Proc Natl Acad Sci 99(23):14682–14687
Gu X, Meer SG, Miyagi M, Rayborn ME, Hollyfield JG, Crabb JW, Salomon RG (2003 Aug 15) Carboxyethylpyrrole protein adducts and autoantibodies, biomarkers for age-related macular degeneration. J Biol Chem 278(43):42027
Hollyfield JG, Bonilha VL, Rayborn ME, Yang X, Shadrach KG, Lu L, Ufret RL, Salomon RG, Perez VL (2008 Feb) Oxidative damage–induced inflammation initiates age-related macular degeneration. Nat Med 14(2):194
Ethen CM, Reilly C, Feng X, Olsen TW, Ferrington DA (2007 Aug 1) Age-related macular degeneration and retinal protein modification by 4-hydroxy-2-nonenal. Invest Ophthalmol Vis Sci 48(8):3469–3479
Cagin U, Duncan OF, Gatt AP, Dionne MS, Sweeney ST, Bateman JM (2015 Nov 3) Mitochondrial retrograde signaling regulates neuronal function. Proc Natl Acad Sci 112(44):E6000–E6009
Hunt RJ, Bateman JM (2018 Mar) Mitochondrial retrograde signaling in the nervous system. FEBS Lett 592(5):663–678
Kenney MC, Chwa M, Atilano SR, Falatoonzadeh P, Ramirez C, Malik D, Tarek M, Cáceres-del-Carpio J, Nesburn AB, Boyer DS, Kuppermann BD (2014a Feb 28) Inherited mitochondrial DNA variants can affect complement, inflammation and apoptosis pathways: insights into mitochondrial–nuclear interactions. Hum Mol Genet 23(13):3537–3551
Nashine S, Cohen P, Chwa M, Lu S, Nesburn AB, Kuppermann BD, Kenney MC (2017 Jul) Humanin G (HNG) protects age-related macular degeneration (AMD) transmitochondrial ARPE-19 cybrids from mitochondrial and cellular damage. Cell Death Dis 8(7):e2951
Nicholls TJ, Minczuk M (2014 Aug 1) In D-loop: 40 years of mitochondrial 7S DNA. Exp Gerontol 56:175–181
McFarland R, Turnbull DM (2009 Feb) Batteries not included: diagnosis and management of mitochondrial disease. J Intern Med 265(2):210–228
Wallace DC (1992 Jul) Diseases of the mitochondrial DNA. Annu Rev Biochem 61(1):1175–1212
Wallace DC (1994 Jun 1) Mitochondrial DNA mutations in diseases of energy metabolism. J Bioenerg Biomembr 26(3):241–250
Bua E, Johnson J, Herbst A, Delong B, McKenzie D, Salamat S, Aiken JM (2006 Sep 1) Mitochondrial DNA–deletion mutations accumulate intracellularly to detrimental levels in aged human skeletal muscle fibers. Am J Hum Genet 79(3):469–480
Chinnery PF, Zwijnenburg PJ, Walker M, Howell N, Taylor RW, Lightowlers RN, Bindoff L, Turnbull DM (1999 Aug 27) Nonrandom tissue distribution of mutant mtDNA. Am J Med Genet 85(5):498–501
Durham SE, Samuels DC, Cree LM, Chinnery PF (2007 Jul 1) Normal levels of wild-type mitochondrial DNA maintain cytochrome c oxidase activity for two pathogenic mitochondrial DNA mutations but not for m.3243A→G. Am J Hum Genet 81(1):189–195
Fan W, Waymire KG, Narula N, Li P, Rocher C, Coskun PE, Vannan MA, Narula J, MacGregor GR, Wallace DC (2008 Feb 15) A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science 319(5865):958–962
Shoubridge EA, Karpati G, Hastings KE (1990 Jul 13) Deletion mutants are functionally dominant over wild-type mitochondrial genomes in skeletal muscle fiber segments in mitochondrial disease. Cell 62(1):43–49
Chinnery PF (2010) Mitochondrial disorders overview. In: RA BTP, Dolan CR (eds) GeneReviews (Internet). National Library of Medicine, National Institutes of Health, Seattle, WA
Yu-Wai-Man P, Griffiths PG, Chinnery PF (2011 Mar 1) Mitochondrial optic neuropathies–disease mechanisms and therapeutic strategies. Prog Retin Eye Res 30(2):81–114
Canter JA, Olson LM, Spencer K, Schnetz-Boutaud N, Anderson B, Hauser MA, Schmidt S, Postel EA, Agarwal A, Pericak-Vance MA, Sternberg P Jr (2008 May 7) Mitochondrial DNA polymorphism A4917G is independently associated with age-related macular degeneration. PLoS One 3(5):e2091
Jones MM, Manwaring N, Wang JJ, Rochtchina E, Mitchell P, Sue CM (2007 Sep 1) Mitochondrial DNA haplogroups and age-related maculopathy. Arch Ophthalmol 125(9):1235–1240
Kenney MC, Hertzog D, Chak G, Atilano SR, Khatibi N, Soe K, Nobe A, Yang E, Chwa M, Zhu F, Memarzadeh M (2013 Dec) Mitochondrial DNA haplogroups confer differences in risk for age-related macular degeneration: a case control study. BMC Med Genet 14(1):4
Mueller EE, Schaier E, Brunner SM, Eder W, Mayr JA, Egger SF, Nischler C, Oberkofler H, Reitsamer HA, Patsch W, Sperl W (2012a Feb 13) Mitochondrial haplogroups and control region polymorphisms in age-related macular degeneration: a case-control study. PLoS One 7(2):e30874
SanGiovanni JP, Arking DE, Iyengar SK, Elashoff M, Clemons TE, Reed GF, Henning AK, Sivakumaran TA, Xu X, DeWan A, Agrón E (2009 May 12) Mitochondrial DNA variants of respiratory complex I that uniquely characterize haplogroup T2 are associated with increased risk of age-related macular degeneration. PLoS One 4(5):e5508
Udar N, Atilano SR, Memarzadeh M, Boyer DS, Chwa M, Lu S, Maguen B, Langberg J, Coskun P, Wallace DC, Nesburn AB (2009 Jun 1) Mitochondrial DNA haplogroups associated with age-related macular degeneration. Invest Ophthalmol Vis Sci 50(6):2966–2974
Bellizzi D, Taverna D, D’Aquila P, De Blasi S, De Benedictis G (2009 May 1) Mitochondrial DNA variability modulates mRNA and intra-mitochondrial protein levels of HSP60 and HSP75: experimental evidence from cybrid lines. Cell Stress Chaperones 14(3):265–271
Chen A, Raule N, Chomyn A, Attardi G (2012 Oct 29) Decreased reactive oxygen species production in cells with mitochondrial haplogroups associated with longevity. PLoS One 7(10):e46473
Kenney MC, Chwa M, Atilano SR, Falatoonzadeh P, Ramirez C, Malik D, Tarek M, del Carpio JC, Nesburn AB, Boyer DS, Kuppermann BD (2014b Feb 1) Molecular and bioenergetic differences between cells with African versus European inherited mitochondrial DNA haplogroups: implications for population susceptibility to diseases. Biochim Biophys Acta (BBA) Mol Basis Dis 1842(2):208–219
Pacheu-Grau D, Gómez-Durán A, Iglesias E, Lopez-Gallardo E, Montoya J, Ruiz-Pesini E (2012 Dec 7) Mitochondrial antibiograms in personalized medicine. Hum Mol Genet 22(6):1132–1139
Lin TK, Lin HY, Chen SD, Chuang YC, Chuang JH, Wang PW, Huang ST, Tiao MM, Chen JB, Liou CW (2012 Dec) The creation of cybrids harboring mitochondrial haplogroups in the Taiwanese population of ethnic Chinese background: an extensive in vitro tool for the study of mitochondrial genomic variations. Oxidative Med Cell Longev 6. https://doi.org/10.1155/2012/824275
Malik D, Hsu T, Falatoonzadeh P, Cáceres-del-Carpio J, Tarek M, Chwa M, Atilano SR, Ramirez C, Nesburn AB, Boyer DS, Kuppermann BD (2014 Jun 11) Human retinal transmitochondrial cybrids with J or H mtDNA haplogroups respond differently to ultraviolet radiation: implications for retinal diseases. PLoS One 9(6):e99003
Mueller EE, Brunner SM, Mayr JA, Stanger O, Sperl W, Kofler B (2012b Dec 26) Functional differences between mitochondrial haplogroup T and haplogroup H in HEK293 cybrid cells. PLoS One 7(12):e52367
Mishmar D, Ruiz-Pesini E, Golik P, Macaulay V, Clark AG, Hosseini S, Brandon M, Easley K, Chen E, Brown MD, Sukernik RI (2003 Jan 7) Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci 100(1):171–176
Ruiz-Pesini E, Mishmar D, Brandon M, Procaccio V, Wallace DC (2004 Jan 9) Effects of purifying and adaptive selection on regional variation in human mtDNA. Science 303(5655):223–226
Ballinger SW Beyond retrograde and anterograde signalling: mitochondrial–nuclear interactions as a means for evolutionary adaptation and contemporary disease susceptibility. Biochem Soc Trans. https://doi.org/10.1042/BST20120227
Abu-Amero KK, Cabrera VM, Larruga JM, Osman EA, González AM, Al-Obeidan SA (2011a) Eurasian and sub-Saharan African mitochondrial DNA haplogroup influences pseudoexfoliation glaucoma development in Saudi patients. Mol Vis 17:543
Abu-Amero KK, González AM, Osman EA, Larruga JM, Cabrera VM, Al-Obeidan SA (2011b) Mitochondrial DNA lineages of African origin confer susceptibility to primary open-angle glaucoma in Saudi patients. Mol Vis 17:1468
Achilli A, Olivieri A, Pala M, Kashani BH, Carossa V, Perego UA, Gandini F, Santoro A, Battaglia V, Grugni V, Lancioni H (2011 Jun 9) Mitochondrial DNA backgrounds might modulate diabetes complications rather than T2DM as a whole. PLoS One 6(6):e21029
Estopinal CB, Chocron IM, Parks MB, Wade EA, Roberson RM, Burgess LG, Brantley MA, Samuels DC (2014 Sep 1) Mitochondrial haplogroups are associated with severity of diabetic retinopathy. Invest Ophthalmol Vis Sci 55(9):5589–5595
Kofler B, Mueller EE, Eder W, Stanger O, Maier R, Weger M, Haas A, Winker R, Schmut O, Paulweber B, Iglseder B (2009 Dec) Mitochondrial DNA haplogroup T is associated with coronary artery disease and diabetic retinopathy: a case control study. BMC Med Genet 10(1):35
Wolf C, Gramer E, Müller-Myhsok B, Pasutto F, Wissinger B, Weisschuh N (2010 Dec) Mitochondrial haplogroup U is associated with a reduced risk to develop exfoliation glaucoma in the German population. BMC Genet 11(1):8
Bravo-Nuevo A, Williams N, Geller S, Stone J (2003) Mitochondrial deletions in normal and degenerating rat retina. Adv Exp Med Biol 533:241–248
Wang AL, Lukas TJ, Yuan M, Neufeld AH (2010 Nov 1) Age-related increase in mitochondrial DNA damage and loss of DNA repair capacity in the neural retina. Neurobiol Aging 31(11):2002–2010
Karunadharma PP, Nordgaard CL, Olsen TW, Ferrington DA (2010 Nov 1) Mitochondrial DNA damage as a potential mechanism for age-related macular degeneration. Invest Ophthalmol Vis Sci 51(11):5470–5479
Meissner C, Bruse P, Mohamed SA, Schulz A, Warnk H, Storm T, Oehmichen M (2008 Jul 1) The 4977 bp deletion of mitochondrial DNA in human skeletal muscle, heart and different areas of the brain: a useful biomarker or more? Exp Gerontol 43(7):645–652
Kenney MC, Atilano SR, Boyer D, Chwa M, Chak G, Chinichian S, Coskun P, Wallace DC, Nesburn AB, Udar NS (2010 Aug 1) Characterization of retinal and blood mitochondrial DNA from age-related macular degeneration patients. Invest Ophthalmol Vis Sci 51(8):4289–4297
Liang FQ, Godley BF (2003 Apr 1) Oxidative stress-induced mitochondrial DNA damage in human retinal pigment epithelial cells: a possible mechanism for RPE aging and age-related macular degeneration. Exp Eye Res 76(4):397–403
Nag TC, Wadhwa S, Chaudhury S (2006 Dec 11) The occurrence of cone inclusions in the ageing human retina and their possible effect upon vision: an electron microscope study. Brain Res Bull 71(1-3):224–232
Terluk MR, Kapphahn RJ, Soukup LM, Gong H, Gallardo C, Montezuma SR, Ferrington DA (2015 May 6) Investigating mitochondria as a target for treating age-related macular degeneration. J Neurosci 35(18):7304–7311
Lin H, Xu H, Liang FQ, Liang H, Gupta P, Havey AN, Boulton ME, Godley BF (2011 May 1) Mitochondrial DNA damage and repair in RPE associated with aging and age-related macular degeneration. Invest Ophthalmol Vis Sci 52(6):3521–3529
Nashine S, Chwa M, Kazemian M, Thaker K, Lu S, Nesburn A, Kuppermann BD, Kenney MC (2016 Aug 3) Differential expression of complement markers in normal and AMD transmitochondrial cybrids. PLoS One 11(8):e0159828
Cobb LJ, Lee C, Xiao J, Yen K, Wong RG, Nakamura HK, Mehta HH, Gao Q, Ashur C, Huffman DM, Wan J (2016 Apr) Naturally occurring mitochondrial-derived peptides are age-dependent regulators of apoptosis, insulin sensitivity, and inflammatory markers. Aging (Albany NY) 8(4):796
Fisher CR, Ferrington DA (2018 Mar 20) Perspective on AMD pathobiology: a bioenergetic crisis in the RPE. Invest Ophthalmol Vis Sci 59(4):AMD41–AMD47
Krebs HA (1927) On the metabolism of the retina. Biochem Z 189:57–59
Warburg O (1924) Über den Stoffwechsel der Carzinomzelle. Naturwissenschaften 12:1131. https://doi.org/10.1007/BF01504608
Chinchore Y, Begaj T, Wu D, Drokhlyansky E, Cepko CL (2017 Jun 9) Glycolytic reliance promotes anabolism in photoreceptors. Elife 6:e25946
Cohen LH, Noell WK (1960 May) Glucose catabolism of rabbit retina before and after development of visual function. J Neurochem 5(3):253–276
Du J, Rountree A, Cleghorn WM, Contreras L, Lindsay KJ, Sadilek M, Gu H, Djukovic D, Raftery D, Satrústegui J, Kanow M (2016a Feb 26) Phototransduction influences metabolic flux and nucleotide metabolism in mouse retina. J Biol Chem 291(9):4698–4710
Narayan DS, Chidlow G, Wood JP, Casson RJ (2017 Sep) Glucose metabolism in mammalian photoreceptor inner and outer segments. Clin Exp Ophthalmol 45(7):730–741
Wang L, Törnquist P, Bill A (1997 Apr) Glucose metabolism in pig outer retina in light and darkness. Acta Physiol Scand 160(1):75–81
Winkler BS (1981 Jun 1) Glycolytic and oxidative metabolism in relation to retinal function. J Gen Physiol 77(6):667–692
Schuster S, Boley D, Möller P, Stark H, Kaleta C (2015 Dec 1) Mathematical models for explaining the Warburg effect: a review focussed on ATP and biomass production. Biochem Soc Trans 43(6):1187–1194
Lowry OH, Roberts NR, Schulz DW, Clow JE, Clark JR (1961 Oct 1) Quantitative histochemistry of retina II. Enzymes of glucose metabolism. J Biol Chem 236(10):2813–2820
Petit L, Ma S, Cipi J, Cheng SY, Zieger M, Hay N, Punzo C (2018 May 29) Aerobic glycolysis is essential for normal rod function and controls secondary cone death in retinitis pigmentosa. Cell Rep 23(9):2629–2642
Rajala A, Gupta VK, Anderson RE, Rajala RV (2013 Nov 30) Light activation of the insulin receptor regulates mitochondrial hexokinase. A possible mechanism of retinal neuroprotection. Mitochondrion 13(6):566–576
Casson RJ, Wood JP, Han G, Kittipassorn T, Peet DJ, Chidlow G (2016 Jan 1) M-type pyruvate kinase isoforms and lactate dehydrogenase A in the mammalian retina: metabolic implications. Invest Ophthalmol Vis Sci 57(1):66–80
Lindsay KJ, Du J, Sloat SR, Contreras L, Linton JD, Turner SJ, Sadilek M, Satrústegui J, Hurley JB (2014 Oct 28) Pyruvate kinase and aspartate-glutamate carrier distributions reveal key metabolic links between neurons and glia in retina. Proc Natl Acad Sci 111(43):15579–15584
Rajala RV, Rajala A, Kooker C, Wang Y, Anderson RE (2016 Nov 24) The Warburg effect mediator pyruvate kinase M2 expression and regulation in the retina. Sci Rep 6:37727
Rueda EM, Johnson JE Jr, Giddabasappa A, Swaroop A, Brooks MJ, Sigel I, Chaney SY, Fox DA (2016) The cellular and compartmental profile of mouse retinal glycolysis, tricarboxylic acid cycle, oxidative phosphorylation, and ~P transferring kinases. Mol Vis 22:847
Kanow MA, Giarmarco MM, Jankowski CS, Tsantilas K, Engel AL, Du J, Linton JD, Farnsworth CC, Sloat SR, Rountree A, Sweet IR (2017 Sep 13) Biochemical adaptations of the retina and retinal pigment epithelium support a metabolic ecosystem in the vertebrate eye. Elife 6:e28899
Wang W, Lee SJ, Scott PA, Lu X, Emery D, Liu Y, Ezashi T, Roberts MR, Ross JW, Kaplan HJ, Dean DC (2016 Apr 12) Two-step reactivation of dormant cones in retinitis pigmentosa. Cell Rep 15(2):372–385
Du J, Cleghorn W, Contreras L, Linton JD, Chan GC, Chertov AO, Saheki T, Govindaraju V, Sadilek M, Satrústegui J, Hurley JB (2013 Oct) Cytosolic reducing power preserves glutamate in retina. Proc Natl Acad Sci 9:201311193
Adijanto J, Du J, Moffat C, Seifert EL, Hurley JB, Philp NJ (2014 Jul 25) The retinal pigment epithelium utilizes fatty acids for ketogenesis implications for metabolic coupling with the outer retina. J Biol Chem 289(30):20570–20582
Reyes-Reveles J, Dhingra A, Alexander D, Bragin A, Philp NJ, Boesze-Battaglia K (2017 Mar 16) Phagocytosis dependent ketogenesis in retinal pigment epithelium. J Biol Chem 292:8038
Wangsa-Wirawan ND, Linsenmeier RA (2003 Apr 1) Retinal oxygen: fundamental and clinical aspects. Arch Ophthalmol 121(4):547–557
Yu DY, Cringle SJ (2001 Mar 1) Oxygen distribution and consumption within the retina in vascularised and avascular retinas and in animal models of retinal disease. Prog Retin Eye Res 20(2):175–208
Hoang QV, Linsenmeier RA, Chung CK, Curcio CA (2002 Jul) Photoreceptor inner segments in monkey and human retina: mitochondrial density, optics, and regional variation. Vis Neurosci 19(4):395–407
Stone J, van Driel D, Valter K, Rees S, Provis J (2008 Jan 16) The locations of mitochondria in mammalian photoreceptors: relation to retinal vasculature. Brain Res 1189:58–69
De Schaepdrijver L, Simoens P, Lauwers H, De Geest JP (1989 Jul 1) Retinal vascular patterns in domestic animals. Res Vet Sci 47(1):34–42
Linton JD, Holzhausen LC, Babai N, Song H, Miyagishima KJ, Stearns GW, Lindsay K, Wei J, Chertov AO, Peters TA, Caffe R (2010 May 11) Flow of energy in the outer retina in darkness and in light. Proc Natl Acad Sci 107(19):8599–8604
Giarmarco MM, Cleghorn WM, Sloat SR, Hurley JB, Brockerhoff SE (2017 Jan) Mitochondria maintain distinct Ca2+ pools in cone photoreceptors. J Neurosci 23:2689–2616
Denton RM, McCormack JG (1990 Mar) Ca2+ as a second messenger within mitochondria of the heart and other tissues. Annu Rev Physiol 52(1):451–466
Kooragayala K, Gotoh N, Cogliati T, Nellissery J, Kaden TR, French S, Balaban R, Li W, Covian R, Swaroop A (2015 Dec 1) Quantification of oxygen consumption in retina ex vivo demonstrates limited reserve capacity of photoreceptor mitochondria. Invest Ophthalmol Vis Sci 56(13):8428–8436
Pearsall EA, Cheng R, Zhou K, Takahashi Y, Matlock HG, Vadvalkar SS, Shin Y, Fredrick TW, Gantner ML, Meng S, Fu Z (2017 Dec) PPARα is essential for retinal lipid metabolism and neuronal survival. BMC Biol 15(1):113
Perkins GA, Ellisman MH, Fox DA (2004 Sep 30) The structure–function correlates of mammalian rod and cone photoreceptor mitochondria: observations and unanswered questions. Mitochondrion 4(5):695–703
Litts KM, Zhang Y, Freund KB, Curcio CA (2018 Mar 1) Optical coherence tomography and histology of age-related macular degeneration support mitochondria as reflectivity sources. Retina 38(3):445–461
Xu Y, Ola MS, Berkich DA, Gardner TW, Barber AJ, Palmieri F, Hutson SM, LaNoue KF (2007 Apr) Energy sources for glutamate neurotransmission in the retina: absence of the aspartate/glutamate carrier produces reliance on glycolysis in glia. J Neurochem 101(1):120–131
Kuwabara T, Cogan DG (1961) Retinal glycogen. Trans Am Ophthalmol Soc 59:106
Goldman SS (1990 Feb 1) Evidence that the gluconeogenic pathway is confined to an enriched Müller cell fraction derived from the amphibian retina. Exp Eye Res 50(2):213–218
Strauss O (2005 Jul) The retinal pigment epithelium in visual function. Physiol Rev 85(3):845–881
Gospe SM, Baker SA, Arshavsky VY (2010 Jan 1) Facilitative glucose transporter Glut1 is actively excluded from rod outer segments. J Cell Sci 123:3639
Rajala A, Wang Y, Brush RS, Tsantilas K, Jankowski CS, Lindsay KJ, Linton JD, Hurley JB, Anderson RE, Rajala RV (2018 Feb 14) Pyruvate kinase M2 regulates photoreceptor structure, function, and viability. Cell Death Dis 9(2):240
Deora AA, Philp N, Hu J, Bok D, Rodriguez-Boulan E (2005 Nov 8) Mechanisms regulating tissue-specific polarity of monocarboxylate transporters and their chaperone CD147 in kidney and retinal epithelia. Proc Natl Acad Sci 102(45):16245–16250
Wright CB, Redmond TM, Nickerson JM (2015 Jan 1) A history of the classical visual cycle. In: Progress in molecular biology and translational science, vol 134. Academic Press, New York, pp 433–448
Chao JR, Knight K, Engel AL, Jankowski C, Wang Y, Manson MA, Gu H, Djukovic D, Raftery D, Hurley JB, Du J (2017 Aug 4) Human retinal pigment epithelial cells prefer proline as a nutrient and transport metabolic intermediates to the retinal side. J Biol Chem 292(31):12895–12905
Du J, Yanagida A, Knight K, Engel AL, Vo AH, Jankowski C, Sadilek M, Manson MA, Ramakrishnan A, Hurley JB, Chao JR (2016b Dec 20) Reductive carboxylation is a major metabolic pathway in the retinal pigment epithelium. Proc Natl Acad Sci 113(51):14710–14715
deS Senanayake P, Calabro A, Hu JG, Bonilha VL, Darr A, Bok D, Hollyfield JG (2006 Aug 1) Glucose utilization by the retinal pigment epithelium: evidence for rapid uptake and storage in glycogen, followed by glycogen utilization. Exp Eye Res 83(2):235–246
Curcio CA, Owsley C, Jackson GR (2000 Jul 1) Spare the rods, save the cones in aging and age-related maculopathy. Invest Ophthalmol Vis Sci 41(8):2015–2018
Ingram NT, Sampath AP, Fain GL (2016 Oct 1) Why are rods more sensitive than cones? J Physiol 594(19):5415–5426
Punzo C, Kornacker K, Cepko CL (2009 Jan) Stimulation of the insulin/mTOR pathway delays cone death in a mouse model of retinitis pigmentosa. Nat Neurosci 12(1):44
Aït-Ali N, Fridlich R, Millet-Puel G, Clérin E, Delalande F, Jaillard C, Blond F, Perrocheau L, Reichman S, Byrne LC, Olivier-Bandini A (2015 May 7) Rod-derived cone viability factor promotes cone survival by stimulating aerobic glycolysis. Cell 161(4):817–832
Léveillard T, Sahel JA (2017 Oct 1) Metabolic and redox signaling in the retina. Cell Mol Life Sci 74(20):3649–3665
Zhao C, Yasumura D, Li X, Matthes M, Lloyd M, Nielsen G, Ahern K, Snyder M, Bok D, Dunaief JL, LaVail MM (2011 Jan 4) mTOR-mediated dedifferentiation of the retinal pigment epithelium initiates photoreceptor degeneration in mice. J Clin Invest 121(1):369–383
Kurihara T, Westenskow PD, Gantner ML, Usui Y, Schultz A, Bravo S, Aguilar E, Wittgrove C, Friedlander MS, Paris LP, Chew E (2016 Mar 15) Hypoxia-induced metabolic stress in retinal pigment epithelial cells is sufficient to induce photoreceptor degeneration. Elife 5:e14319
Zhang L, Du J, Justus S, Hsu CW, Bonet-Ponce L, Wu WH, Tsai YT, Wu WP, Jia Y, Duong JK, Mahajan VB (2016 Dec 1) Reprogramming metabolism by targeting sirtuin 6 attenuates retinal degeneration. J Clin Invest 126(12):4659–4673
Curcio CA, Medeiros NE, Millican CL (1996 Jun 1) Photoreceptor loss in age-related macular degeneration. Invest Ophthalmol Vis Sci 37(7):1236–1249
Curcio CA, Millican CL, Allen KA, Kalina RE (1993 Nov 1) Aging of the human photoreceptor mosaic: evidence for selective vulnerability of rods in central retina. Invest Ophthalmol Vis Sci 34(12):3278–3296
Curcio CA, Sloan KR, Kalina RE, Hendrickson AE (1990 Feb 22) Human photoreceptor topography. J Comp Neurol 292(4):497–523
Packer O, Hendrickson AE, Curcio CA (1989 Oct 1) Photoreceptor topography of the retina in the adult pigtail macaque (Macaca nemestrina). J Comp Neurol 288(1):165–183
Daiger SP, Sullivan LS, Bowne SJ, Rossiter BJ RetNet: Retinal Information Network, 1996. Data services and software for identifying genes and mutations causing retinal degeneration. http://www.sph.uth.tmc.edu/RetNet/ [updated Dec 08 2014]
Calabrese MF, Rajamohan F, Harris MS, Caspers NL, Magyar R, Withka JM, Wang H, Borzilleri KA, Sahasrabudhe PV, Hoth LR, Geoghegan KF (2014 Aug 5) Structural basis for AMPK activation: natural and synthetic ligands regulate kinase activity from opposite poles by different molecular mechanisms. Structure 22(8):1161–1172
Samuel MA, Voinescu PE, Lilley BN, De Cabo R, Foretz M, Viollet B, Pawlyk B, Sandberg MA, Vavvas DG, Sanes JR (2014 Sep) LKB1 and AMPK regulate synaptic remodeling in old age. Nat Neurosci 17(9):1190
Xu L, Kong L, Wang J, Ash JD (2018 Oct 9) Stimulation of AMPK prevents degeneration of photoreceptors and the retinal pigment epithelium. PNAS 115:10475–10480
Venkatesh A, Ma S, Le YZ, Hall MN, Rüegg MA, Punzo C (2015 Apr 1) Activated mTORC1 promotes long-term cone survival in retinitis pigmentosa mice. J Clin Invest 125(4):1446–1458. https://doi.org/10.1172/JCI79766
Ma S, Venkatesh A, Langellotto F, Le YZ, Hall MN, Rüegg MA, Punzo C (2015 Jun 1) Loss of mTOR signaling affects cone function, cone structure and expression of cone specific proteins without affecting cone survival. Exp Eye Res 135:1–3. https://doi.org/10.1016/j.exer.2015.04.006
Qu J, Kaufman Y, Washington I (2009 Apr 1) Coenzyme Q10 in the human retina. Invest Ophthalmol Vis Sci 50(4):1814–1818. https://doi.org/10.1167/iovs.08-2656
Feher J, Kovacs B, Kovacs I, Schveoller M, Papale A, Gabrieli CB (2005) Improvement of visual functions and fundus alterations in early age-related macular degeneration treated with a combination of acetyl-L-carnitine, n-3 fatty acids, and coenzyme Q10. 219(3):154–166
Ildefonso CJ, Jaime H, Brown EE, Iwata RL, Ahmed CM, Massengill MT, Biswal MR, Boye SE, Hauswirth WW, Ash JD, Li Q (2016 Feb 1) Targeting the Nrf2 signaling pathway in the retina with a gene-delivered secretable and cell-penetrating peptide. Invest Ophthalmol Vis Sci 57(2):372–386
Liang KJ, Woodard KT, Weaver MA, Gaylor JP, Weiss ER, Samulski RJ (2017 Mar 1) AAV-Nrf2 promotes protection and recovery in animal models of oxidative stress. Mol Ther 25(3):765–779
Jäger S, Handschin C, Pierre JS, Spiegelman BM (2007 Jul 17) AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1α. Proc Natl Acad Sci 104(29):12017–12022
Iacovelli J, Rowe GC, Khadka A, Diaz-Aguilar D, Spencer C, Arany Z, Saint-Geniez M (2016 Mar 1) PGC-1α induces human RPE oxidative metabolism and antioxidant capacity. Invest Ophthalmol Vis Sci 57(3):1038–1051. https://doi.org/10.1167/iovs.15-17758
Zhang M, Chu Y, Mowery J, Konkel B, Galli S, Theos AC, Golestaneh N (2018 Jan 1) PGC-1α repression and high fat diet induce age-related macular degeneration-like phenotypes in mice. Dis Models Mech. https://doi.org/10.1242/dmm.032698
Acknowledgements
This work was supported by grants from the National Institutes of Health / National Eye Institute (R01 EY026012 and R01 EY028554 to DAF; R01EY0127363 to MCK, R01 EY06641 and R01 EY017863 to JBH, R01EY016459-11 and R01EY031720 to JDA), the Elaine and Robert Larson Endowed Vision Research Chair, an anonymous benefactor for AMD research, the Lindsay Family Foundation (to DAF); the Discovery Eye Foundation, Polly and Michael Smith, Iris and B. Gerald Cantor Foundation (to MCK), and an unrestricted grant from Research to Prevent Blindness to the Department of Ophthalmology, University of Florida and to the Gavin Herbert Eye Institution, University of California.
The authors wish to acknowledge the insightful discussions at the meetings of the “Ryan’s Initiative for Macular Research” as the basis for the collaborative writing of this chapter.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply
About this chapter
Cite this chapter
Ferrington, D.A., Kenney, M.C., Atilano, S.R., Hurley, J.B., Brown, E.E., Ash, J.D. (2021). Mitochondria: The Retina’s Achilles’ Heel in AMD. In: Chew, E.Y., Swaroop, A. (eds) Age-related Macular Degeneration. Advances in Experimental Medicine and Biology, vol 1256. Springer, Cham. https://doi.org/10.1007/978-3-030-66014-7_10
Download citation
DOI: https://doi.org/10.1007/978-3-030-66014-7_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-66013-0
Online ISBN: 978-3-030-66014-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)