Abstract
Fatty Acid Binding-Protein 5 (FABP5) is a cytoplasmic protein, which binds long-chain fatty acids and other hydrophobic ligands. This protein is implicated in several physiological processes including mitochondrial β-oxidation and transport of fatty acids, membrane phospholipid synthesis, lipid metabolism, inflammation and pain. In the present study, we used molecular docking tools to determine the possible interaction of FABP5 with six selected compounds retrieved form Drugbank. Our results showed that FABP5 binding pocket included 31 polar and non-polar amino acids, and these residues may be related to phosphorylation, acetylation, ubiquitylation, and mono-methylation. Docking results showed that the most energetically favorable compounds are NADH (−9.12 kcal/mol), 5′-O-({[(Phosphonatooxy)phosphinato]oxy}phosphinato)adenosine (−8.62 kcal/mol), lutein (−8.25 kcal/mol), (2S)-2-[(4-{[(2-Amino-4-oxo-1,4,5,6,7,8-hexahydro-6-pteridinyl)methyl]amino}benzoyl)amino]pentanedioate (−7.17 kcal/mol), Pteroyl-L-glutamate (−6.86 kcal/mol) and (1S,3R,5E,7Z)-9,10-Secocholesta-5,7,10-triene-1,3,25-triol (−6.79 kcal/mol). Common interacting residues of FABP5 with nutraceuticals included SER16, LYS24, LYS34, LYS40 and LYS17. Further, we used the SwissADME server to determine the physicochemical and pharmacokinetic characteristics and to predict the ADME parameters of the selected nutraceuticals after molecular analysis by docking with the FABP5 protein. Amongst all compounds, pteroyl-L-glutamate is the only one meeting the Lipinski’s rule of five criteria, demonstrating its potential pharmacological use. Finally, our results also suggest the importance of FABP5 in mediating the anti-inflammatory activity of the nutraceutical compounds.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
29.1 Introduction
Fatty Acid-Binding Protein 5 is a cytoplasmic protein belonging to the family Fatty acid-binding proteins (FABPs). These proteins are structurally conserved cytosolic proteins with a broad specificity for ligands including eicosanoids, peroxisome proliferators, bile salts, long-chain fatty acids and cannabinoids [11, 26]. To date, at least 12 of FABP different genes have been identified [33]. FABPs have differential expression in different tissues and organs, including liver (FABP1), intestine (FABP2), heart (e.g FABP3), adipocytes and macrophages (FABP4), epidermis (FABP5), Ileum (FABP6) and brain (FABP7) among others [14, 22, 33].
FABP5 is also known as psoriasis-associated fatty acid-binding protein, epidermal, or cutaneous fatty acid-binding protein (PA-, E-, or C-FABP), has been shown to be present or expressed in several tissues including epidermis, nociceptive dorsal root ganglia, spinal cord and liver [22]. It is a small protein of 135 amino acids, with a structural conformation of ten β-strands (β1-β10), two α-helices (α1-α2) and ten loops (L1-L10, [6]). FABP5 binds to long-chain fatty acids and other hydrophobic ligands such as saturated FAs, MUFAs (monounsaturated FAs), n-3, and n-6 PUFAs (polyunsaturated FAs) through its binding pocket, which includes ARG109, ARG129 and TYR131 [3]. Its main functions include fatty acid uptake, transport, and metabolism. Moreover, this protein may modulate the actions of the PPAR β/δ (nuclear receptor peroxisome proliferator-activated receptor), and promote cell proliferation, survival and migration by exhibiting pro-oncogenic activities in colorectal, ovarian, breast and prostate cancers [1, 12, 22, 29]. Finally, FABP5 has been explored as a potential pharmacological target for inflammation, pain and amelioration of drug withdrawal symptoms [4, 17, 34].
Previous computational studies have been developed to understand the regulation of FABP5 and other FABPs by different ligands [24, 31]. For example, Yan et al. [31] used a bioinformatic approach based on molecular dynamics in combination with molecular mechanics generalized Born surface area (MM-GBSA) to determine the binding selectivity of three FABP4/FABP5 inhibitors with therapeutic potential for arteriosclerosis and inflammation [31]. Similarly, Shinoda et al. [24] reported the affinity of ten polyphenolic ligands for FABP3, FABP4 and FABP5. Using computational docking simulations and experimental methods, the potential anti-inflammatory and protective effects of FABP ligands in neurodegenerative disorders and peripheral ischemic injury were explored [24]. Briefly, nutraceuticals are naturally occurring compounds present in food with possible medical benefits, which include amino acids like N-acetylcysteine, carotenoids, polyphenols, vitamins, minerals and fatty acids [27]. Nutraceuticals have been used to improve health and prevent chronic diseases including diabetes, cardio and cerebrovascular diseases, cancer , and the neuroinflammatory diseases Alzheimer and Parkinson [2, 5, 15, 16, 18, 20, 21, 25, 28, 32]. At this point, targeting FABP5 using pharmacological approaches could be a potential way to mitigate inflammation and lipid metabolism abnormalities in different diseases. Therefore, in the present study, we used molecular docking tools to explore the possible interaction of FABP5 with six selected nutraceutical compounds. Identification of the residues important for this interaction could pave the way for drug design against lipid dysfunction and neuroinflammatory diseases.
29.2 Materials and Methods
29.2.1 Ligand Preparation
A list with a total of 60 naturally occurring ligands compounds database (version 5.1.7) was retrieved from DrugBank (www.drugbank.ca) in SDF format and then converted into.mol using Molecular Operating Environment (MOE 2015.10, Chemical Computing Group). Polar hydrogens and charges were added before docking. The molecular structure of all compounds was built using the ligand builder plugin in MOE.
29.2.2 Structure Preparation
The 3D structure of the human protein Fatty acid-binding protein 5 (FABP5, entry: 5HZ5) were downloaded from Protein Data Bank (https://www.rcsb.org/). The protein structure was prepared using the “Structure preparation” plugin in MOE for protonation and energy minimization, the protein-associated ligands were removed, and the missing hydrogen atoms were added. To determine the protein’s binding pocket we used the CASTp server (http://sts.bioe.uic.edu/castp).
29.2.3 Molecular Docking of FABP5 with Nutraceuticals
For docking, chain A of the FABP5 protein was used. Initially we performed a blinded docking to determine the possible interaction site of the ligands to the FABP5 protein using MOE. Based on the interaction energy and best conformations, we selected 6 compounds (Fig. 29.1) for a more exhaustive study. A second docking analysis was carried out, now with exhaustiveness of 100 different poses using the GBVI/WSA dG and London dG scores as parameters. The analyzed docking parameters were: Root-mean-square deviation (RMSD), water accessible surface area (ASA), potential energy (E), electrostatic potential energy (E_ele), electrostatic interaction energy (E_rele), van der Waals interaction energy (E_rvdw), van der Waals potential energy (E_vdw), total SCF energy (kcal/mol) calculated using the MNDO Hamiltonian (MNDO_E), energy of the highest occupied molecular orbital (HOMO), energy of the lowest unoccupied molecular orbital (LUMO) and radius of gyration (rgyr). Validation of docking results were performed by running the same experiment using Autock Vina on PyRx.
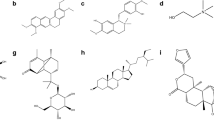
Molecular structure of naturally occurring compounds. (a) NADH(2-); (b) 5′-O-({[(Phosphonatooxy)phosphinato]oxy}phosphinato)adenosine; (c) lutein; (d) (2S)-2-[(4-{[(2-Amino-4-oxo-1,4,5,6,7,8-hexahydro-6-pteridinyl)methyl]amino}benzoyl)amino]pentanedioate; (e) Pteroyl-L-glutamate; and (f) (1S,3R,5E,7Z)-9,10-Secocholesta-5,7,10-triene-1,3,25-triol
29.2.4 Studies of Toxicity/ADMET of Nutraceutical Compounds
The SwissADME server (http://www.swissadme.ch/) was used to determine the physicochemical and pharmacokinetic characteristics and to predict the ADME parameters of the selected nutraceuticals after molecular analysis by docking with the FABP5 protein.
29.3 Results and Discussion
29.3.1 Binding Pocket of FABP5
The use of molecular docking for screening possible compounds for drug repurposing has been rapidly expanding. In a straightforward manner, it generates a great amount of data, making it feasible to identify possible sites and/or domains of interaction between ligands and target proteins. This makes this approach very useful as to selection of drug candidates and druggable targets. Initially, in the present study, using the CASTp server we determined that the FABP5 binding pocket included the amino acid residues PHE19, TYR22, MET23, LEU26, VAL28, LEU32, MET35, GLY36, ALA39, PRO41, CYS43, ILE54, THR56, SER5, LYS61, THR62, GLN64, PHE65, GLU75, THR76, THR77, ALA78, ASP79, ARG81, GLN98, ILE107, ARG109, VAL118, CYS120, ARG129, and TYR131. Previous structural studies have shown that FABP5 binding pocket for linoleic acid consists of ARG129, TYR131 (hydroxyl moiety) and ARG109 [3], which is consistent with our results. Moreover, our results are also in line with previous studies reporting that the binding site of both FABP4 and 5 is within the β-barrel, containing the loops β3–β4 and β5–β6, in combination with α1-loop-α2 domain forming a sort of like controlling gate to allow the entrance and exit of ligands involved in the interaction [13, 31]. This study also described that the residues ARG126, ARG106 and TYR128 interact in the barrel’s cavity through electrostatic interactions [31]. Finally, a recent study using molecular dynamics approaches showed the energy contributions of key residues such as F19, Y22, M23, P41, T56 and L60 from K61, R109 and R129 to be important in the interaction with studied ligands [6].
29.3.2 Molecular Docking of Nutraceuticals and FABP5
We started from an initial list of 60 different ligands of natural origin obtained from the DrugBank database, from which by blinded docking we selected a few for a more detailed analysis. Interestingly, some of the residues that make part of the FABP5 binding pocket are directly involved in the interaction with some ligands, which suggests that they can interact with the protein through their active site. In this first approximation, the 10 best binding poses from each ligand were analyzed, from which we selected a total of 6 ligands (Fig. 29.1) that presented the best interaction energy. After this, we performed a more exhaustive docking, now with 100 different poses and found that NADH (2-) (−9.12 kcal/mol) presents the greatest interaction energy, followed by 5′-O - ({[(Phosphonatooxy) phosphinato] oxy} phosphinato) adenosine (−8.62 kcal/mol), lutein (−8.25 kcal/mol), (2S) -2 - [(4 - {[(2-Amino-4-oxo-1,4,5, 6,7,8-hexahydro-6-pteridinyl) methyl] amino} benzoyl) amino] pentanedioate (−7.17 kcal/mol), Pteroyl-L-glutamate (−6.86 kcal/mol) and (1S, 3R, 5E, 7Z) -9,10-Secocholesta-5,7,10-triene-1,3,25-triol (−6.79 kcal/mol) (Table 29.1). Interestingly, lutein, which is a xanthophyll-type carotenoid found in leafy vegetables and yellow fruits, has been shown to exert neuroprotective and anti-inflammatory effects in animal models of ocular diseases [23] through the suppression of reactive oxygen species (ROS) and inflammatory signaling. Moreover, pteroyl-L-glutamate (folic acid) is important in the metabolism of amino acids and nucleic acids, and has been used as an adjuvant to cytotoxic agents in cancer treatment [10], and in the modulation of inflammatory response in microglia [8]. Similarly, 9,10-Secocholesta-5,7,10-triene-1,3,25-triol (the active metabolite of vitamin D-3) has potential benefits against carcinogenic cells proliferation, and anti-inflammatory effects through the inhibition of NF-κB signaling, and the suppression of prostaglandin metabolism [30]. Finally, both NADH and ATP (5′-O - ({[(Phosphonatooxy) phosphinato] oxy} phosphinato) adenosine) have shown to be important in the regulation of pro-inflammatory cytokines, the inflammatory related kinases IKKβ, JNK and ERK and enzymes such as Sirt1, Sirt6, PARP-1, ART-1 [7, 19]. These newly presented results further support the importance of FABP5 in the regulation of inflammatory processes. However, further research is needed in order to establish the physiological and molecular mechanisms of this regulatory process.
The FABP5 residues that interact through H-bonds with NADH (2-) are GLU21, VAL14, ASP15, ASP20, SER16, LYS24, LYS34, ARG33, and ionically with LYS34 Y LYS24; through H-bonds, 5′-O - ({[((Phosphonatooxy) phosphinato] oxy} phosphinato) adenosine interacts with GLY18, SER16, PH19, LYS17, LYS40, SER16, ALA39, ARG129, and by ionic bonds with LYS17, LYS40, ARG129. For the ligand (2S) -2 - [(4 - {[(2-Amino-4-oxo-1,4,5,6,7,8-hexahydro-6-pteridinyl) methyl] amino} benzoyl) amino] pentanedioate the interaction by H-bonds includes the residues ASP15, MET38, SER16, PHE19, ASP20, LYS17, GLY18, LYS34, LYS40, and by ionic with LYS34 and pi-H with SER16. The Pteroyl-L-glutamate forms H-bonds with ASP15, SER16, PHE19, GLY18, LYS34, LYS17) and ionic with LYS34. Finally, the ligand (1S, 3R, 5E, 7Z) -9,10-Secocholesta-5,7,10-triene-1,3,25-triol forms H-bonds with GLU21 and LYS40 (Figs. 29.2, and 29.3 and Table 29.1). Binding sites of a ligand onto a protein can induce post-translational modifications, potentially changing the functionality and conformation of protein. Interestingly, some of the residues through which the ligands interact with FABP5 are sites of these modifications, suggesting that in addition to binding to them, the selected nutraceuticals can modulate their response depending on the type of modification. We found that the residue SER16 may be related to phosphorylation, while LYS17 (site for acetylation, ubiquitylation and succinylation), LYS24 (site for ubiquitylation and mono-methylation), LYS34 (ubiquitylation) and finally LYS40 might be implicated in acetylation and ubiquitylation (Table 29.1).
2D representation of the binding interaction of FABP5 with selected nutraceuticals. (a) NADH(2-); (b) 5′-O-({[(Phosphonatooxy)phosphinato]oxy}phosphinato)adenosine; (c) lutein; (d) (2S)-2-[(4-{[(2-Amino-4-oxo-1,4,5,6,7,8-hexahydro-6-pteridinyl)methyl]amino}benzoyl)amino]pentanedioate; (e) Pteroyl-L-glutamate; and (f) (1S,3R,5E,7Z)-9,10-Secocholesta-5,7,10-triene-1,3,25-triol
3D representation of the binding interacting site of FBP5 with nutraceuticals. (a) NADH(2-); (b) 5′-O-({[(Phosphonatooxy)phosphinato]oxy}phosphinato)adenosine; (c) lutein; (d) (2S)-2-[(4-{[(2-Amino-4-oxo-1,4,5,6,7,8-hexahydro-6-pteridinyl)methyl]amino}benzoyl)amino]pentanedioate; (e) Pteroyl-L-glutamate; and (f) (1S,3R,5E,7Z)-9,10-Secocholesta-5,7,10-triene-1,3,25-triol
Based on a more detailed analysis of the interaction of six nutraceuticals with FABP5, we calculated different docking scores (Table 29.2), noting that 5′-O - ({[((Phosphonatooxy) phosphinato] oxy} phosphinato) adenosine has a lower RMSD with a major electrostatic potential energy, van der Waals potential energy and overall potential energy (E). For the radius of gyration, a measure that determines the conformation of a protein in terms of its stability and folding, we observed that 5′-O - ({[(Phosphonatooxy) phosphinato] oxy} phosphinato) adenosine is the one with the lowest value, while lutein shows the highest value compared with other ligands. ASA of a protein, the parameter that denotes the water accessible surface, suggests that lutein followed by NADH (2-) are the ones that present the highest values when compared to the others.
29.3.3 ADME Properties of All Nutraceuticals
ADME serves as a score to determine the ligand’s physicochemical and pharmacokinetic characteristics, being useful to determine whether a drug could be considered as a potential therapeutic agent. The properties that ADME determines include absorption, bioavailability, hepatic metabolism, and excretion. According to the Lipinski’s rule of five, a potential drug must meet the following criteria: no more than 5 h-bond donors, no more than 10 h-bond acceptors, a molecular weight of less than 500 daltons, a logP not exceeding 5 (high lipophilicity) and molar refractivity between 40 and 130. Within the selected ligands, only pteroyl-L-glutamate fulfilled the criteria, which suggests its potential for pharmacological therapy (Table 29.3). Importantly, ADME parameters (Absorption, Distribution, Metabolism and Excretion) are essential in the discovery phase of potential drugs as they increase the success rate in the search of novel compounds that can pass to clinical phases [9].
29.4 Conclusions
In conclusion, the present study evaluated in silico interactions of FABP5 with 6 putative ligands with nutraceutical properties. Our results suggest that NADH (2-) presents the greatest interaction energy, followed by 5′-O - ({[(Phosphonatooxy) phosphinato] oxy} phosphinato) adenosine, lutein, (2S) -2 - [(4 - {[(2-Amino-4-oxo-1,4,5, 6,7,8-hexahydro-6-pteridinyl) methyl] amino} benzoyl) amino] pentanedioate, pteroyl-L-glutamate and (1S, 3R, 5E, 7Z) -9,10-Secocholesta-5,7,10-triene-1,3,25-triol. We further evaluated the physicochemical and pharmacokinetic characteristics of the ligands and found that only pteroyl-L-glutamate fulfilled the Lipinski’s criteria, suggesting its potential application for pharmacological uses. However, additional computational and experimental validation studies are needed to establish the in vivo and in vitro interactions of FABP5 and its regulatory mechanisms against anti-inflammatory processes.
References
Adhikary T, Brandt DT, Kaddatz K, Stockert J, Naruhn S, Meissner W et al (2013) Inverse PPARbeta/delta agonists suppress oncogenic signaling to the ANGPTL4 gene and inhibit cancer cell invasion. Oncogene 32(44):5241–5252. https://doi.org/10.1038/onc.2012.549
Areiza-Mazo N, Robles J, Zamudio-Rodriguez JA, Giraldez L, Echeverria V, Barrera-Bailon B et al (2018) Extracts of Physalis peruviana protect astrocytic cells under oxidative stress with rotenone. Front Chem 6:276. https://doi.org/10.3389/fchem.2018.00276
Armstrong EH, Goswami D, Griffin PR, Noy N, Ortlund EA (2014) Structural basis for ligand regulation of the fatty acid-binding protein 5, peroxisome proliferator-activated receptor beta/delta (FABP5-PPARbeta/delta) signaling pathway. J Biol Chem 289(21):14941–14954. https://doi.org/10.1074/jbc.M113.514646
Berger WT, Ralph BP, Kaczocha M, Sun J, Balius TE, Rizzo RC et al (2012) Targeting fatty acid binding protein (FABP) anandamide transporters – a novel strategy for development of anti-inflammatory and anti-nociceptive drugs. PLoS One 7(12):e50968. https://doi.org/10.1371/journal.pone.0050968
Bibak B, Shakeri F, Barreto GE, Keshavarzi Z, Sathyapalan T, Sahebkar A (2019) A review of the pharmacological and therapeutic effects of auraptene. Biofactors 45(6):867–879. https://doi.org/10.1002/biof.1550
Chen J, Liu X, Zhang S, Chen J, Sun H, Zhang L, Zhang Q (2020) Molecular mechanism with regard to the binding selectivity of inhibitors toward FABP5 and FABP7 explored by multiple short molecular dynamics simulations and free energy analyses. Phys Chem Chem Phys 22(4):2262–2275. https://doi.org/10.1039/c9cp05704h
Chen W, Yi C, Jin L (2018) The role of nicotinamide adenine dinucleotide in the pathogenesis of rheumatoid arthritis: potential implications for treatment. Eur Med J 3(3):90–97
Cianciulli A, Salvatore R, Porro C, Trotta T, Panaro MA (2016) Folic acid is able to polarize the inflammatory response in LPS activated microglia by regulating multiple signaling pathways. Mediat Inflamm 2016:5240127. https://doi.org/10.1155/2016/5240127
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7:42717. https://doi.org/10.1038/srep42717
Danenberg PV, Gustavsson B, Johnston P, Lindberg P, Moser R, Odin E et al (2016) Folates as adjuvants to anticancer agents: chemical rationale and mechanism of action. Crit Rev Oncol Hematol 106:118–131. https://doi.org/10.1016/j.critrevonc.2016.08.001
Deutsch DG (2016) A personal retrospective: elevating anandamide (AEA) by targeting fatty acid amide hydrolase (FAAH) and the fatty acid binding proteins (FABPs). Front Pharmacol 7:370. https://doi.org/10.3389/fphar.2016.00370
Di-Poi N, Michalik L, Tan NS, Desvergne B, Wahli W (2003) The anti-apoptotic role of PPARbeta contributes to efficient skin wound healing. J Steroid Biochem Mol Biol 85(2–5):257–265. https://doi.org/10.1016/s0960-0760(03)00215-2
Floresta G, Pistara V, Amata E, Dichiara M, Marrazzo A, Prezzavento O, Rescifina A (2017) Adipocyte fatty acid binding protein 4 (FABP4) inhibitors. A comprehensive systematic review. Eur J Med Chem 138:854–873. https://doi.org/10.1016/j.ejmech.2017.07.022
Furuhashi M, Saitoh S, Shimamoto K, Miura T (2014) Fatty acid-binding protein 4 (FABP4): pathophysiological insights and potent clinical biomarker of metabolic and cardiovascular diseases. Clin Med Insights Cardiol 8(Suppl 3):23–33. https://doi.org/10.4137/CMC.S17067
Ghanaatian N, Lashgari NA, Abdolghaffari AH, Rajaee SM, Panahi Y, Barreto GE et al (2019) Curcumin as a therapeutic candidate for multiple sclerosis: molecular mechanisms and targets. J Cell Physiol 234(8):12237–12248. https://doi.org/10.1002/jcp.27965
Jurado-Coronel JC, Avila-Rodriguez M, Echeverria V, Hidalgo OA, Gonzalez J, Aliev G, Barreto GE (2016) Implication of green tea as a possible therapeutic approach for Parkinson disease. CNS Neurol Disord Drug Targets 15(3):292–300. https://doi.org/10.2174/1871527315666160202125519
Kaczocha M, Rebecchi MJ, Ralph BP, Teng YH, Berger WT, Galbavy W et al (2014) Inhibition of fatty acid binding proteins elevates brain anandamide levels and produces analgesia. PLoS One 9(4):e94200. https://doi.org/10.1371/journal.pone.0094200
Keshavarzi Z, Shakeri F, Barreto GE, Bibak B, Sathyapalan T, Sahebkar A (2019) Medicinal plants in traumatic brain injury: neuroprotective mechanisms revisited. Biofactors 45(4):517–535. https://doi.org/10.1002/biof.1516
Lee JH, Zhang Y, Zhao Z, Ye X, Zhang X, Wang H, Ye J (2017) Intracellular ATP in balance of pro- and anti-inflammatory cytokines in adipose tissue with and without tissue expansion. Int J Obes 41(4):645–651. https://doi.org/10.1038/ijo.2017.3
Mazo NA, Echeverria V, Cabezas R, Avila-Rodriguez M, Tarasov VV, Yarla NS et al (2017) Medicinal plants as protective strategies against Parkinson’s disease. Curr Pharm Des 23(28):4180–4188. https://doi.org/10.2174/1381612823666170316142803
Nasri H, Baradaran A, Shirzad H, Rafieian-Kopaei M (2014) New concepts in nutraceuticals as alternative for pharmaceuticals. Int J Prev Med 5(12):1487–1499. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/25709784
Ohata T, Yokoo H, Kamiyama T, Fukai M, Aiyama T, Hatanaka Y et al (2017) Fatty acid-binding protein 5 function in hepatocellular carcinoma through induction of epithelial-mesenchymal transition. Cancer Med 6(5):1049–1061. https://doi.org/10.1002/cam4.1020
Ozawa Y, Sasaki M, Takahashi N, Kamoshita M, Miyake S, Tsubota K (2012) Neuroprotective effects of lutein in the retina. Curr Pharm Des 18(1):51–56. https://doi.org/10.2174/138161212798919101
Shinoda Y, Wang Y, Yamamoto T, Miyachi H, Fukunaga K (2020) Analysis of binding affinity and docking of novel fatty acid-binding protein (FABP) ligands. J Pharmacol Sci 143(4):264–271. https://doi.org/10.1016/j.jphs.2020.05.005
Singh SK, Barreto GE, Aliev G, Echeverria V (2017) Ginkgo biloba as an alternative medicine in the treatment of anxiety in dementia and other psychiatric disorders. Curr Drug Metab 18(2):112–119. https://doi.org/10.2174/1389200217666161201112206
Smathers RL, Petersen DR (2011) The human fatty acid-binding protein family: evolutionary divergences and functions. Hum Genomics 5(3):170–191. https://doi.org/10.1186/1479-7364-5-3-170
Souyoul SA, Saussy KP, Lupo MP (2018) Nutraceuticals: a review. Dermatol Ther (Heidelb) 8(1):5–16. https://doi.org/10.1007/s13555-018-0221-x
Uddin MS, Al Mamun A, Kabir MT, Jakaria M, Mathew B, Barreto GE, Ashraf GM (2019) Nootropic and anti-Alzheimer’s actions of medicinal plants: molecular insight into therapeutic potential to alleviate Alzheimer’s neuropathology. Mol Neurobiol 56(7):4925–4944. https://doi.org/10.1007/s12035-018-1420-2
Wang D, Wang H, Guo Y, Ning W, Katkuri S, Wahli W et al (2006) Crosstalk between peroxisome proliferator-activated receptor delta and VEGF stimulates cancer progression. Proc Natl Acad Sci U S A 103(50):19069–19074. https://doi.org/10.1073/pnas.0607948103
Xu J, Li W, Ma J, Liu J, Sha H, Zhou S et al (2013) Vitamin D – pivotal nutraceutical in the regulation of cancer metastasis and angiogenesis. Curr Med Chem 20(33):4109–4120. https://doi.org/10.2174/09298673113209990194
Yan F, Liu X, Zhang S, Su J, Zhang Q, Chen J (2018) Molecular dynamics exploration of selectivity of dual inhibitors 5M7, 65X, and 65Z toward fatty acid binding proteins 4 and 5. Int J Mol Sci 19(9):2496. https://doi.org/10.3390/ijms19092496
Yaribeygi H, Zare V, Butler AE, Barreto GE, Sahebkar A (2019) Antidiabetic potential of saffron and its active constituents. J Cell Physiol 234(6):8610–8617. https://doi.org/10.1002/jcp.27843
Zhang Y, Zhang J, Ren Y, Lu R, Yang L, Nie G (2020) Tracing the evolution of fatty acid-binding proteins (FABPs) in organisms with a heterogeneous fat distribution. FEBS Open Bio 10(5):861–872. https://doi.org/10.1002/2211-5463.12840
Zhou Y, Elmes MW, Sweeney JM, Joseph OM, Che J, Hsu HC et al (2019) Identification of fatty acid binding protein 5 inhibitors through similarity-based screening. Biochemistry 58(42):4304–4316. https://doi.org/10.1021/acs.biochem.9b00625
Acknowledgments
None.
Conflict of Interests
None of the authors has a competing interest directly related to the content of this study.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Cabezas, R., Sahebkar, A., Echeverria, V., Santos, J.G., Ashraf, G.M., Barreto, G.E. (2021). In Silico Identification of Novel Interactions for FABP5 (Fatty Acid-Binding Protein 5) with Nutraceuticals: Possible Repurposing Approach. In: Barreto, G.E., Sahebkar, A. (eds) Pharmacological Properties of Plant-Derived Natural Products and Implications for Human Health. Advances in Experimental Medicine and Biology, vol 1308. Springer, Cham. https://doi.org/10.1007/978-3-030-64872-5_29
Download citation
DOI: https://doi.org/10.1007/978-3-030-64872-5_29
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-64871-8
Online ISBN: 978-3-030-64872-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)