Abstract
Automated extraction of cerebrovascular is of great importance in understanding the mechanism, diagnosis, and treatment of many cerebrovascular pathologies. However, segmentation of cerebrovascular networks from magnetic resonance angiography (MRA) imagery continues to be challenging because of relatively poor contrast and inhomogeneous backgrounds, and the anatomical variations, complex geometry and topology of the networks themselves. In this paper, we present a novel cerebrovascular segmentation framework that consists of image enhancement and segmentation phases. We aim to remove redundant features, while retaining edge information in shallow features when combining these with deep features. We first employ a Retinex model, which is able to model noise explicitly to aid removal of imaging noise, as well as reducing redundancy within an image and emphasizing the vessel regions, thereby simplifying the subsequent segmentation problem. Subsequently, a reverse edge attention module is employed to discover edge information by paying particular attention to the regions that are not salient in high-level semantic features. The experimental results show that the proposed framework enables the reverse edge attention network to deliver a reliable cerebrovascular segmentation.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Cerebrovascular diseases, such as stroke, aneurysm, and arteriovenous malformation, are some of the most common fatal diseases threatening human health worldwide [1]. MRA is a common imaging technique for observing the cerebrovascular system, and an accurate detection of the cerebrovascular from MRA imagery is essential for many clinical applications to support early diagnosis, optimal treatment, and neurosurgery planning for vascular-related diseases. However, manual annotation of cerebrovascular networks is an exhausting task even for experts, and existing computer-aided systems cannot reliably extract and segment these networks, due to the high degree of anatomical variation.
In the last two decades, we have witnessed the rapid development of vessel segmentation methods for different medical imaging modalities, as evidenced by extensive reviews [2,3,4,5]. However, the extraction of cerebrovascular networks is a less explored topic. Most techniques tend to over-segment or mis-segment, primarily due to the complex geometry involved, as well as problems posed by varying scales of noise, or imbalanced illumination within an image, as well as low image contrast and spatial resolution. As a result, it is desirable to design a fully automated cerebrovascular segmentation method.
Conventional cerebrovascular segmentation approaches have been based on handcrafted features, including vessel intensity distribution, gradient features, morphological features and many others [6]. For example, the cerebrovascular were segmented by means of active contour models [7] and geometric models [8]. In addition, a variety of filtering methods have been proposed, including Hessian matrix-based filters [9], a symmetry filter [4], and a tensor-based filter [10]. These approaches aim to remove undesired intensity variations in the image, and suppress background structures and image noise. However, these methods require elaborate design, which depends heavily on the user’s domain knowledge and expertise, and errors can be propagated and accumulated, especially in the case of small vessels, due to poor imaging quality.
In recent years, several deep learning-based methods have been proposed for cerebrovascular segmentation. Phellan et al. [11] explored a relatively shallow Convolutional Neural Network (CNN) on MRA scans to segment the vessels at 2D slice level. Livne et al. [12] utilized the U-Net deep learning framework [13] to segment the vessel region on each slice of an MRA volume. Nevertheless, 2D CNN in taken from an MRA slice necessarily discards valuable three-dimensional(3D) context information that is crucial for tracking curvilinear structures. Sanches et al. [14] presented a Uception model for segmentation of an arterial cerebrovascular network. Zhang et al. [15] presented an efficacious framework that applies deep 3D CNN to automatic cerebrovascular segmentation with sparsely labeled data: however, they neglected to prioritize the extraction of blood vessel edges.
The aforementioned methods have not yet completely addressed the issues posed by the high degree of anatomical variation across the population, and poor contrast and varying scales of noise within an image. In this paper, we propose a novel cerebrovascular segmentation framework that hybridizes image enhancement and segmentation steps. First, we employ a Retinex model to improve image contrast and also model noise explicitly to aid the removal of imaging noise. Subsequently, we make use of a reverse attention mechanism, and introduce a Reverse Edge attention Network (RE-Net) capable of discovering the missing edge features and residual details effectively. This leads in turn to a significant improvement in the cerebrovascular segmentation.
2 Methodology
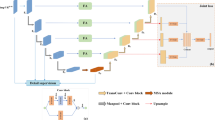
In this section, we detail the proposed cerebrovascular segmentation method. Figure 1 illustrates the pipeline of the proposed framework.
2.1 Image Enhancement via Retinex Model
In general, it is observed that laminar flow within cerebrovascular vessels causes velocity variations in blood flow, which leads to highly varying contrast distribution in MRA imagery. In addition, degrading noise is usually inherited from the image acquisition process. To this end, the enhancement of MRA images is essential so as to obtain a more precise segmentation result. In this section, we introduce a novel noise-suppression Retinex model to enhance MRA images.
The classic Retinex model [16] assumes that an image \(\mathbf{S }\) can be decomposed into two components, the reflectance \(\mathbf{R }\) and the illumination \(\mathbf{L }\), with the range \((0, \infty )\): \(\mathbf{S =\mathbf{L} \cdot \mathbf{R} }\), and it follows that \(\mathbf{S} \le \mathbf{L} \). By removing the influence of illumination \(\mathbf{L }\), the resulting \(\mathbf{R }\) is able to reveal the reflectance of the object of interest more objectively, and it can thus be regarded as the enhanced image. In this work, we utilized the Retinex model proposed by Elad et al. [17] for MRA volume enhancement.
2.2 Cerebrovascular Segmentation via RE-Net
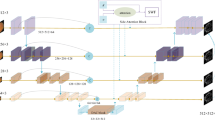
In this section, we introduce the proposed Reverse Edge Attention network for cerebrovascular segmentation. The proposed RE-Net consists of three phases: the encoder module, the Reverse Edge Attention Module (REAM), and the decoder module, as shown in Fig. 2.
The encoder module contains four encoder stages based on the ResNet Block [18]. For each stage, the inputs are first fed into a stack of \(3\,\times \, 3\,\times \, 3\,-\,3 \,\times \, 3\,\times \, 3\) convolutional layers, and are then summed with the shortcut of inputs to generate the final outputs, followed by a max-pooling of \( 2\times 2\times 2 \) to increase the receptive field for better extraction of global features. With the residual connection, the model can avoid the gradient vanishing and accelerate the network convergence. In the decoder module, each of the three stages consists of a deconvolution of \(2\times 2\times 2\) with a stride of 2, followed by two \(3\times 3\times 3\) convolutions activated with ReLU. To assist the decoding process, skip connections copy feature maps generated by the encoder module to the decoder module. Afterwards, a REAM is embedded in a skip connection to extract edge information from encoder layers.
Reverse Edge Attention Module: Consecutive pooling and striding convolutional operations may enlarge the receptive field, and obtain more global information. However, they lead to the loss of edge information. To maintain such information, the features generated by the deep layers in the decoder module are usually concatenated with features in the encoder module. Unfortunately, these features encoded by the shallow layers not only contain edges, but also keep textural features, which might then constitute interference factors compromising the robustness of high-level features [19].
Inspired by the reverse attention model in [20], we propose to make use of a reverse edge attention module, as shown in Fig. 2 (b), to extract the edge information from the feature maps generated by the encoding layers. This edge information is then fused with the features maps generated by the decoder for the corresponding positions, so that the vessel edge features will be enhanced and the original image edge details will be more accurately restored.
\(X_{\text{ i }} \in R^{h \times w \times d \times c_{x}}\), \(2 \le \mathrm {\textit{i}} \le 4\) denotes the features generated in the ith encoder stage. A \(1\, \times \, 1 \,\times \, 1\) convolution is first used to fuse the features into a single channel. The features are then upsampled to the same resolution as the outputs of the (\( i-1 \))th encoder stage, thus generating \({F} \in R^{h \times w \times d \times c}\). The corresponding weight \(A_{\mathrm{i}-1}\) in the (\( i-1 \))th encoder stage is simply generated by subtracting the upsampled prediction of the ith stage from 1, as below:
In REAM, the edge information was discovered by paying attention to the regions that are not salient in high-level semantic features. Let \(X_{\mathrm{i}-1} \in R^{h \times w \times d \times c_{x}}\) denote features generated in the (\( i-1 \))th encoder stage. Then edge feature \(E_{\mathrm{i}-1}\) can be captured by element-wise multiplication, expressed as follows:
The edge feature is fused into the features of the decoder at the corresponding position by skip connection after summing with \(X_{i-1}\). Comparably, the feature map after concatenation at the same position has more edge information with the aid of REAM, thus improving the segmentation accuracy.
3 Experimental Results
Our RE-Net was implemented on a PyTorch framework with a single GPU (TITAN RTX). Adaptive moment estimation (Adam) was employed for network optimization. The initial learning rate was set to 0.0001, with a weight decay of 0.0005. A poly learning rate policy [21] with power 0.9 was used, and the maximum epoch was 4000. Dice loss was adopted as the loss function. The batch size was set to 4 during training, because of the limitations of GPU memory.
Data and metrics: a publicly available datasetFootnote 1 with 42 time-of-flight MRA volumes was used in our work. These images were captured by a 3 T unit under standardized protocols, with a voxel size of \(0.5\,\times \,0.5\,\times \,0.8\) \(\mathrm{mm}^3\), and reconstructed with a \(448\times 448\times 128\) matrix. Manual annotations were available online, and the cerebrovascular labels were obtained via an open source toolkit - TubeTK [22]. In the experiment, patches of size \(96\,\times \,96\,\times \,96\) were randomly cropped from the MRA volume, followed by a 90-\(^\circ \) rotation for data augmentation. To facilitate better observation and objective evaluation of the cerebrovascular segmentation method, the following metrics were calculated: sensitivity (Sen) = TP/ (TP + FN); specificity (Spe) = TN/ (TN + FP); Precision (Pre) = TP/(TP + FP); and Dice similarity coefficient (DSC) = 2*TP/ (FP + FN + 2*TP). In addition, the Average Hausdorffs Distance (AHD) was adopted, because it is sensitive to the edge of segmentation results.
3.1 Evaluation of Image Enhancement Performance
Firstly, we analyze the effect of the Retinex (NSR) pre-processing step in the proposed method.
Figure 3 illustrates the sagittal, axial, and coronal views of maximum intensity projection (MIP) of a sample MRA, before and after the NSR method was applied. Overall, our method provides similar performance with raw data on the large vessels. However, when scrutinizing the smaller vessels, we can see that the proposed method provides relatively stronger responses; i.e., it has successfully enhanced some small vessels, as indicated by the green arrows. This is because our NSR enhancement is able to reduce noise, and normalize the entire background to a similar level, so as to increase the contrast between the vessel regions and their background. In addition to visual inspection of the enhancement results, objective evaluation was also undertaken by comparing the segmentation results with a state-of-the-art 3D segmentation model, 3D U-Net [24]. Table 1 shows the segmentation results in comparison with two classic image enhancement methods, i.e. gamma correction, and guided filter [23]. The proposed method improves the segmentation of the original images (i.e. raw data) by 2.80% and 3.24% in terms of Sen and DSC. By contrast, relatively more significant margins of segmentation results have been shown, when compared with two alternative enhancement methods, which indicates that our enhancement method demonstrates larger improvement than its competitors. It can be seen that our NSR method achieves the best performance in terms of all metrics.
3.2 Evaluation of Cerebrovascular Segmentation Performance
In this section, we compare performance in segmenting cerebrovascular networks using different 3D segmentation models. Three state-of-the-art 3D segmentation networks - including 3D U-Net [24], V-Net [25] and Uception [14] - as well as the proposed RE-Net, and RE-Net without REAM (backbone) were used for comparison. Figure 4 illustrates the segmentation results on two MRA volumes. It should be noted that, for purposes of fair comparison, 3D U-Net, V-Net, and Uception were applied to the same enhanced data. Overall, all methods demonstrate a similar performance on large vessels. However, the proposed RE-Net produces better segmentation results than other models in preserving small vessels, and maintaining vascular continuity, as highlighted by the green arrows with close-ups shown in the green rectangles in Fig. 4. RE-Net is capable of discovering lost edge features and residual details effectively, which leads to a significant improvement on detecting small vessels.
The above findings are reinforced by the evaluation scores in Table 2. The RE-Net outperforms 3D U-Net, V-Net, and Uception by 3.51%, 4.73%, and 8.45% in terms of Sen respectively. In addition, the DSC score of the Backbone method (RE-Net without REAM) is approximately 1.87% lower than that of RE-Net, which constitutes additional proof that the reverse attention mechanism benefits the performance of cerebrovascular segmentation. Overall, the proposed RE-Net yields the best segmentation results in terms of all metrics.
4 Conclusion
Cerebrovascular segmentation is an important step for the diagnosis of many cerebrovascular diseases. In this paper, we have proposed a novel cerebrovascular segmentation framework, which consists of Retinex-based method for MRA image enhancement and a newly designed 3D segmentation network. The former takes into account a noise term in conventional Retinex model, so as to suppress noise in MRA imagery and further ease the segmentation task. The RE-Net implements a self-attention mechanism, and pays more attention to regions that are not salient in high-level global features. The experimental results based on a publicly available dataset demonstrate that the proposed method significantly improves cerebrovascular segmentation.
References
Yasugi, M., Hossain, B., Nii, M., Kobashi, S.: Relationship between cerebral aneurysm development and cerebral artery shape. J. Adv. Comput. Intell. Intell. Inform. 22(2), 249–255 (2018)
Fraz, M., et al.: Blood vessel segmentation methodologies in retinal images - a survey. Comput. Methods Programs Biomed. 108(1), 407–433 (2012)
Zhao, Y., Rada, L., Chen, K., Harding, S.P., Zheng, Y.: Automated vessel segmentation using infinite perimeter active contour model with hybrid region information with application to retinal images. IEEE Trans. Med. Imaging 34(9), 1797–1807 (2015)
Zhao, Y., et al.: Automatic 2-D/3-D vessel enhancement in multiple modality images using a weighted symmetry filter. IEEE Trans. Med. Imaging 37(2), 438–450 (2017)
Zhao, Y., et al.: Retinal artery and vein classification via dominant sets clustering-based vascular topology estimation. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11071, pp. 56–64. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00934-2_7
Zhao, Y., et al.: Automated tortuosity analysis of nerve fibers in corneal confocalmicroscopy. In: IEEE Transactions on Medical Imaging (2020)
Yang, X., Cheng, K.T., Chien, A.: Geodesic active contours with adaptive configuration for cerebral vessel and aneurysm segmentation. In: 2014 22nd International Conference on Pattern Recognition, pp. 3209–3214. IEEE (2014)
Forkert, N.D., et al.: 3D cerebrovascular segmentation combining fuzzy vessel enhancement and level-sets with anisotropic energy weights. Magn. Reson. Imaging 31(2), 262–271 (2013)
Frangi, A.F., Niessen, W.J., Vincken, K.L., Viergever, M.A.: Multiscale vessel enhancement filtering. In: Wells, W.M., Colchester, A., Delp, S. (eds.) MICCAI 1998. LNCS, vol. 1496, pp. 130–137. Springer, Heidelberg (1998). https://doi.org/10.1007/BFb0056195
Cetin, S., Unal, G.: A higher-order tensor vessel tractography for segmentation of vascular structures. IEEE Trans. Med. Imaging 34(10), 2172–2185 (2015)
Phellan, R., Peixinho, A., Falcão, A., Forkert, N.D.: Vascular segmentation in TOF MRA images of the brain using a deep convolutional neural network. In: Cardoso, M.J., et al. (eds.) LABELS/CVII/STENT -2017. LNCS, vol. 10552, pp. 39–46. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-67534-3_5
Livne, M., et al.: A u-net deep learning framework for high performance vesselsegmentation in patients with cerebrovascular disease. Frontiers Neurosci. 13, 97 (2019)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Sanchesa, P., Meyer, C., Vigon, V., Naegel, B.: Cerebrovascular network segmentation of mra images with deep learning. In: IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), pp. 768–771. IEEE (2019)
Zhang, B., et al.: Cerebrovascular segmentation from TOF-MRA using model-and data-driven method via sparse labels. Neurocomputing, (2019)
Land, E.H., McCann, J.J.: Lightness and retinex theory. J. Opt. Soc. Am. 61(1), 1–11 (1971)
Elad, M.: Retinex by two bilateral filters. In: Kimmel, R., Sochen, N.A., Weickert, J. (eds.) Scale-Space 2005. LNCS, vol. 3459, pp. 217–229. Springer, Heidelberg (2005). https://doi.org/10.1007/11408031_19
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 770–778. IEEE (2016)
Pang, Y., Li, Y., Shen, J., Shao, L.: Towards bridging semantic gap to improve semantic segmentation. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 4230–4239. IEEE (2019)
Chen, S., Tan, X., Wang, B., Hu, X.: Reverse attention for salient object detection. In: Proceedings of the European Conference on Computer Vision (ECCV), pp. 234–250 (2018)
Zhao, H., Shi, J., Qi, X., Wang, X., Jia, J.: Pyramid scene parsing network. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 2881–2890. IEEE (2017)
Aylward, S.R., Bullitt, E.: Initialization, noise, singularities, and scale in height ridge traversal for tubular object centerline extraction. IEEE Trans. Med. Imaging 21(2), 61–75 (2002)
He, K., Sun, J., Tang, X.: Guided image filtering. IEEE Trans. Pattern Anal. Mach. Intell. 35, 1397–1409 (2013)
Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T., Ronneberger, O.: 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 424–432. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_49
Milletari, F., Navab, N., Ahmadi, S.A.: V-net: fully convolutional neural networks for volumetric medical image segmentation. In: 2016 Fourth International Conference on 3D Vision (3DV), pp. 565–571. IEEE (2016)
Acknowledgment
This work was supported by Beijing Natural Science Foundation (4202011), National Natural Science Foundation of China (61572076), Key Research Grant of Academy for Multidisciplinary Studies of CNU (JCKXYJY2019018), Zhejiang Provincial Natural Science Foundation of China (LZ19F010001), and Ningbo “2025 S&T Megaprojects” (2019B10033, 2019B10061).
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Zhang, H. et al. (2020). Cerebrovascular Segmentation in MRA via Reverse Edge Attention Network. In: Martel, A.L., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2020. MICCAI 2020. Lecture Notes in Computer Science(), vol 12266. Springer, Cham. https://doi.org/10.1007/978-3-030-59725-2_7
Download citation
DOI: https://doi.org/10.1007/978-3-030-59725-2_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-59724-5
Online ISBN: 978-3-030-59725-2
eBook Packages: Computer ScienceComputer Science (R0)