Abstract
DNA binding proteins under starvation (Dps) are proteins belonging to the ferritin family with the capacity for DNA binding, in addition to iron storage and ferroxidation. Present only in the prokaryotes, these multifaceted proteins have been assigned with a number of roles, from pathogenesis to nucleoid condensation and protection. They have a significant role in protecting the cells from free radical assaults, indirectly by sequestration of iron and by directly binding to the DNA. Due to their symmetry, stability and biomineralization capacity, these proteins have ever increasing potential applications in biotechnology and drug delivery. This chapter tries to bring together all these aspects of Dps in the view of current understanding and older perspectives by studies of our group as well as other experts in the field.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
Introduction
Dps (DNA-binding protein from starved cells) are homo-dodecamers belonging to the ferritin family which can bind DNA, carry out ferroxidation and store iron. These proteins are present only in members of eubacteria and archaea. In nature, iron starvation and oxidative stress are two hurdles that bacteria must circumvent to establish growth. To overcome the paradox of iron and oxidative stress, as free iron contributes to further oxidative stress by its participation in the Fenton reaction (Fenton 1894), iron is stored in a non-toxic form by Dps proteins.
Dps also binds DNA in a sequence independent manner and brings about its condensation. Thus, it is classed as a nucleoid associated protein (NAP) similar to HU, IHF, H-NS etc. Dps seems to play a multi-faceted role in bacterial cells, as it performs a number of diverse roles in various organisms. This section attempts to give a general introduction from the discovery of Dps to a description of its role in protecting cells under various stresses and its rapid degradation in the cells during logarithmic growth phase through the N-end rule degradation pathway.
Discovery of DNA Binding Proteins Under Starvation (Dps)
The Kolter group (Almiron et al. 1992) while studying gene expression of starved Escherichia coli cells discovered the over expression of a certain DNA binding protein whose expression depended on a stationary phase specific transcription factor σS. In negative staining electron microscopy, it had a double layered hexameric ring like appearance and formed two dimensional arrays with DNA (Fig. 3.1a, b). Later a homolog discovered in nitrogen starved Synechococcus sp. PCC7942 named DpsA showed the presence of a carboxyl domain homologous to the C termini of bacterioferritins (Pena and Bullerjahn 1995), giving first indications of its iron binding property. Subsequently, a dodecameric ferritin-like protein isolated from the gram-positive Listeria innocua able to sequester around 500 iron atoms and oxidise iron, was shown to be a Dps-like protein (Bozzi et al. 1997).
More comprehensive information about the Dps structure which gave an experimental basis for its functions, was obtained after the crystal structure of Dps from E. coli was solved by Hogle, Kolter and colleagues. The X-ray crystal structure showed Dps monomers to have a ferritin-like fold assembling into 12-mers with tetrahedral (23) symmetry, enclosing a hollow core with pores at the three-folds (Grant et al. 1998) (Fig. 3.2a, b). The structure of another Dps homolog from L. innocua gave the first direct evidence of a ferroxidase center indicating a structural basis of ferroxidase activity (Ilari et al. 2000). X-ray structures of more Dps homologs revealed these features to be consistent within the family.
Iron Oxidation and Storage
Iron, one of the most abundant metals on earth, plays a vital role in the biology of living organisms. It is a cofactor in many redox reactions and can be found in the active sites of a number of enzymes. Iron can act as an electron source and a sink, and has a central role in various cellular processes like metabolism, respiration, nucleotide synthesis, gene regulation etc. Iron starvation or a lack of this valuable metal in the environment poses an adverse threat to existence.
Free iron can exist in two states, ferrous (Fe2+) and ferric (Fe3+) in organisms. The ferrous state is soluble and readily assimilated in cells, but the ferric state is highly insoluble and unavailable for physiological functions. But the more bioavailable ferrous form has a tendency to oxidise to ferric thereby limiting its bioavailability and also generating hydroxyl radicals in this process. H2O2, a product of oxidative respiration reacts with ferrous iron to generate hydroxyl free radicals which are toxic to cells, by a process called the Fenton reaction (Eq. 3.1) (Martinez and Kolter 1997).
Thus, it is essential that iron is stored in a readily accessible but non-toxic form in cells. Ferritin, a family of iron storage proteins are the primary iron storage compartments in vertebrates, invertebrates, plants and microorganisms. They are structurally adept at storing iron in their hollow protein shells in a state that is available and non-deleterious. The ferritin family of proteins includes ferritins and other ferritin-like proteins such as the haem containing bacterioferritins and Dps which can also bind DNA (Andrews 2010).
Ferritins and other ferritin-like proteins have a hollow central cavity where iron is oxidised and stored as ferric oxide. Based on their capacity to store iron, the ferritin-like proteins can be also be widely categorised as the 24-meric maxiferritins which include ferritins and bacterioferritins and 12-meric miniferritins which are the Dps. The miniferritins or Dps have been recognised only in eubacteria and archaea, unlike the maxiferritins which are present in bacteria, archaea and the eukaryotes. The maxiferritins due to their bigger protein shells which consequently enclose a larger cavity, can store >4000 iron atoms per protein. Miniferritins on the other hand can store only around 500 iron atoms per dodecamer. Hydrogen peroxide is the preferred reagent for iron oxidation in miniferritins, unlike maxi ferritins where dioxygen is the major electron acceptor (Zhao et al. 2002).
In Dps, iron incorporation is a multi-step process of Fe (II) binding, Fe(II) oxidation, nucleation and growth of the mineral core, in a reaction pathway similar to classical ferritins (Zhao et al. 2002).
Fe(II) Binding (Phase 1)
24 Fe(II) binds at the 12 di-iron binding ferroxidation sites in the protein
where [Fe(II)2–P]Z+2FS represents a di-Fe(II)-protein complex at each of the 12 putative ferroxidase sites. For more information about the ferroxidase centre and di-iron binding sites see section Conserved ferroxidase active site, below.
Fe(II) Oxidation (Phase 2)
Two di-nuclear ferroxidase sites bring about the rapid pairwise oxidisation of two Fe (II) by one molecule of H2O2. Thus, two Fe (II) are oxidised per H2O2 reduced, preventing the generation of hydroxyl radicals through Fenton chemistry (Eq. 3.1).
Nucleation and Growth of Mineral Core (Phase 3)
A ferric core of approximately 500 Fe(III) is formed inside the Dps protein shell, according to the mineralisation equation with a 2 Fe(II)/H2O2 stoichiometry:
A visual schematic of the ferroxidation pathway is represented in Fig. 3.14. In Dps, ferroxidation can also take place with O2 as an oxidant, but the reaction happens at a much slower rate.
DNA Protection by Direct and Indirect Modes
DNA is often called the blueprint of life. It is in the interest of cell survival to devise highly efficient DNA protection mechanisms to offset the lethal effects caused by DNA lesions. But during conditions of nutrient depletion the cell cannot afford to repair DNA through energy extravagant DNA defence pathways. In starved E. coli cells for example, Dps accumulates to very large amounts and forms the major component of chromatin in late stationary phase bacteria (Martinez and Kolter 1997). Dps binds DNA and compacts it during stress, thereby exposing fewer unprotected regions to reactive agents. Oxidation of Fe(II) to Fe(III) prevents the ferrous form from participating in Fenton reaction (Eq. 3.1) quenching further production of free radicals deleterious to DNA. The ferroxidation activity also results in the reduction of toxic peroxides by utilization of H2O2 as an oxidant. Dps also has a role in the regulation of the expression of certain proteins under conditions of stress.
A bimodal protection of DNA by Dps was seen in homolog from Mycobacterium smegmatis, MsDps1 (Gupta and Chatterji 2003). This homolog has two oligomeric forms, dimer and dodecamer. The dimeric form has properties of ferroxidation only, whereas the dodecamer could store iron, carry out ferroxidation and bind DNA. The dimer could still protect DNA against the assault of free radicals, as it has intact ferroxidase sites and thus can carry out ferroxidation. In the earlier publication (Gupta and Chatterji 2003) the lower oligomer was identified as a trimer, whereas recently through mass spectrometry it has been correctly assigned as a dimer (Williams et al. 2017).
The DNA binding property and protection can be assessed by simple biochemical assays, which monitor DNA retardation or degradation in the presence and absence of Dps proteins. DNA retardation of pUC19 plasmid by Dps1 from M. smegmatis can be visualised on an agarose gel (Fig. 3.3a). Protection of DNA can be observed by treating DNA with H2O2, where the DNA with bound Dps is shielded (Fig. 3.3b). DNA protection from DNaseI digestion is also accorded by Dps (Fig. 3.3c), which prevents access to DNA by these degrading enzymes (Gupta and Chatterji 2003).
Gel based assays for DNA protection by M.smegmatis Dps a Gel retardation of DNA by Dps. Free pUC19 DNA (lane 1) and Dps-DNA complex shows retardation of DNA in the well (lane 2). b DNA protection from peroxide damage. pUC19 DNA (lane 1) pUC19 DNA treated initially with 50 μM FeSO4 followed by 5 mM H2O2 for 5 min degrades the DNA completely (lane 2) and Dps-DNA complex treated with 50 μM FeSO4 followed by 5 mM H2O2 for 5 min where the DNA is protected (lane3). c DNA protection from DNaseI digestion. pUC19 DNA (lane 1) DNaseI treated pUC19 DNA (lane 2) Dps-DNA complex protected from DNaseI digestion (lane3). Images reproduced from J. Biol. Chem. 2003, 278: 5235–5241
Thus, Dps renders protection by directly binding to DNA and also indirectly by preventing the generation of toxic free radicals through its ferroxidation property. A more detailed description of the mechanisms involved in DNA binding and protection are discussed below.
Dps Across the Prokaryota
The role of Dps in bacterial cells can differ widely, while retaining some of its classic properties, like DNA binding and iron storage. E. coli Dps is one of the best studied Dps homolog and exhibits most of the typical properties of Dps family proteins. E. coli Dps is additionally known to protect the cells from UV and gamma radiations, iron/copper toxicity, thermal stress, acid and base shock in addition to oxidative stress (Nair and Finkel 2004). To better appreciate the numerous homologs of Dps in bacteria and their varied roles, we have summarised some of the better studied Dps proteins in prokaryotes.
Dps homologs like the L. innocua Dps do not bind DNA but can protect it against the deleterious combination of Fe2+ and H2O2 as evidenced by DNA cleavage assays (Su et al. 2005). The plant pathogen, Agrobacterium tumifaciens has a Dps with a truncated N-terminus which is fixed on the protein surface and not available for DNA binding, unlike the long and flexible N-terminus of E. coli Dps which has DNA binding property. In A. tumifaciens, Dps acts in concert with catalase A to counteract toxic peroxides that are major components of plant defense systems against invading bacteria (Ceci et al. 2003).
The Streptococcus mutans Dps homolog named Dpr (Dps-like peroxide resistance) confers peroxide resistance, helping the bacteria to survive in aerobic conditions. This organism is a principal causative agent of dental caries and lacks a respiratory chain and the enzyme catalase responsible for H2O2 elimination (Yamamoto et al. 2002). Similarly, S. pyogenes does not produce catalase, but can grow in aerobic conditions due to the presence of Dpr. Both these streptococcal Dpr proteins do not bind DNA, but provide protection from H2O2 stress through nullifying the Fenton reaction and sequestration and oxidation of iron (Tsou et al. 2008). The swine pathogen S. suis Dpr also protects DNA from hydrogen peroxide stress by removing free Fe2+ from the cytosol, without any apparent DNA binding ability (Kauko et al. 2006). Bacillus anthracis BaDps1 and BaDps2 acts as a pair to carry out iron sequestration and H2O2 depletion respectively, without binding to DNA (Schwartz et al. 2010). B. cereus has three Dps homologs Dps1, Dps2 and Dps3 of which only the latter shows DNA binding property (Shu et al. 2013).
The Dps-1 from Deinococcus radiodurans an organism best known for its unusual resistance to ionising radiations, fails to protect DNA from hydroxyl radical mediated cleavage, due to continuous release of stored iron from the protein, although it can bind DNA. This release of iron from the protein core was attributed to the presence of a distinct iron-exit channel. The other Dps homolog in the same organism, Dps-2, has a signal peptide which directs it to the perimeter of D. radiodurans cells. Dps-2 shows weak affinity to DNA but displays robust iron storage and ferroxidation. These distinct functional properties and localisation suggests that Dps-1 may participate as a NAP, whereas Dps-2 protects against exogenous reactive oxygen species (ROS) due to its location in the perimeter (Reon et al. 2012).
Lactococcus lactis DpsA and DpsB form Dps:DNA complexes which appear as non-crystalline aggregates in electron micrographs, instead of the ordered crystalline arrays seen in Dps: DNA complexes of E. coli and other homologs. The disordered N-terminus in E. coli and M. smegmatis is required for formation of crystalline array with Dps. The N-terminal region of L. lactis on the other hand has an ordered helical conformation. Also, L. lactis Dps was unable to sequester iron due to a preponderance of positively charged residues near the iron entry pores, which occludes iron due to charge repulsion (Stillman et al. 2005). The non-pathogenic mycobacterial species M. smegmatis has two Dps homologs, MsDps1 and MsDps2. These are differently regulated in the cells, where MsDps1 is expressed in the stationary phase and is controlled by the extra cellular sigma factors sigF and sigH, whereas MsDps2 is constitutively expressed by RNA polymerase under the control of sigma factors sigA and sigB (Chowdhury et al. 2007). Streptomyces coelicolor, a soil dwelling gram positive bacterium, has three Dps homologs, DpsA, DpsB and DpsC out of which DpsA is over-expressed during osmotics shock and DpsC is a tailless protein that does not oligomerise (Facey et al. 2009).
The Dps from Helicobacter pylori is a neutrophil activating protein HP-NAP, and drives inflammation in allergic bronchial asthma in humans and mice (Kottakis et al. 2008). HP-NAP of H. pylori does not bind DNA, but addition of Fe2+ was shown to promote DNA binding. Borrelia burgdorferi Dps NapA, is also a neutrophil activating protein and elicits host immune response (Li et al. 2007). Microbacterium arborescens Dps termed AAH (acyl amino acid hydrolase) displays additional catalytic activities such as amide hydrolysis and synthesis (Pesek et al. 2011). Porphyromonas gingivalis Dps, PgDps binds heme through its conserved cysteine residue and confers resistance to heme toxicity (Gao et al. 2012). Campylobacter jejuni Dps has a role in biofilm formation and cecal colonization in poultry, in addition to protecting DNA in the acidic conditions of the host digestive tract and inside phagolysosomes (Theoret et al. 2012).
The role of Dps in extremophiles like D. radiodurans, which survive high ionising radiation has been mentioned above. In thermophiles like Streptococcus thermophilus, Dps functions as a cold shock protein (Nicodeme et al. 2004). An archaeal Dps protein DpsA from the halophilic marine archaea Halobacterium salinarum, renders protection for aerobic survival (Zeth et al. 2004). The Dps from Sulfolobus solfataricus a thermophilic archaeon, has an active site similar to manganese catalases, which can bind Mn2+ in addition to Fe2+ (Hayden and Hendrich 2010).
In pathogenic bacteria, Dps helps the cells to evade host defense pathways. In Candidatus Legionella jeonii, DpsX protects the cells from oxidative stress generated by the phagocytic activities of the host (Park et al. 2006). In Legionella pneumophilia, DpsL is upregulated during iron starvation and helps the pathogen to circumvent iron limiting conditions (Yu et al. 2009). Bacteriodes fragilis, an opportunistic pathogen of the GI tract causes infection when the organism escapes the anaerobic colon to aerobic sites like the peritoneum with high O2 concentrations. Here, resistance to oxidative stress is important and is involved in the initiation and persistence of infection. One of the proteins involved in detoxification of oxygen radicals is Dps and signifies its role in oxidative stress response of aerobic and facultative organisms (Rocha et al. 2003). Dps in Helicobacter hepaticus, a bacterium associated with chronic hepatitis and hepatocellular carcinoma in mice, plays an important role in protecting DNA from oxidative damage (Hong et al. 2006). In Salmonella enterica, Dps helps to resist iron dependent killing by hydrogen peroxide and promotes virulence and survival of the organism in macrophages (Pacello et al. 2008). MrgA, a Dps homolog in Staphylococcus aureus, also resists phagocytic killing by macrophages (Gaupp et al. 2012).
The cyanobacterium Nostoc punctiforme features five Dps proteins, NpDps 1–5, each having distinct physiological differences and cell-specific expression (Howe et al. 2018). In photosynthetic cyanobacteria like Synechococcus sp strain 7942, Dps localizes preferentially to the cytosolic side of thylakoids to provide iron to iron-rich photosystem I (Durham and Bullerjahn 2002). Anabena sp. PCC 7120 has two Dps homologs, which plays a role in alleviating oxidative stress due to environmental changes like temperature and light in addition to its usual function of iron storage and oxidation (Narayan et al. 2010). Thermosynechococcus elongatus Dps homologs, Dps-Te and DpsA-Te, inhibits Fenton mediated damage and this is of special significance in the protection of DNA and photosystems I and II from hydrogen peroxide mediated oxidative damage, as it lacks the catalase gene (Franceschini et al. 2006). In the N2 fixing cyanobacterium Trichodesmium erythraeum, Dpstery protects the cells from oxidative damage (Castruita et al. 2006).
N-end Rule Degradation
We have seen that Dps is an important multifunctional protein in bacteria which accumulates to a variety of stresses in the cells, such as nutrient and oxidative stress. Dps is typically over expressed during the stationary phase of the cells and rapidly degraded during logarithmic phase by ClpX/ClpP (Stephani et al. 2003) and ClpS/ClpA/ClpP proteases (Schmidt et al. 2009). The N-terminus of Dps has ClpX and ClpS recognition motifs which promotes degradation by ClpP and ClpA/ClpP proteases, respectively. In E. coli Dps, ClpX recognition motif is the N terminus without the first methionine residue i.e. Dps2-167, is degraded by ClpXP proteolysis. ClpS targets the N-terminally truncated Dps variant with the first five residues missing, Dps6-167 harbouring the destabilizing Leu6 at the N-terminus.
The N-terminus of Dps in E.coli not only serves as a protease recognition motif, but is also important for DNA binding (Ceci et al. 2004). It is proposed that the N-terminal tail of Dps is sequestered during stationary phase when it binds to DNA and this could protect it from degradation. Generation of Dps2-167 or Dps6-167 through degradation of unprotected N- termini could initiate the disassembly of Dps-DNA complexes and proteolysis of Dps when the cells enter exponential phase (Schmidt et al. 2009). Thus, Dps is a substrate for the N-end rule degradation pathway, where the half-life of a protein is determined by the nature of its N-terminal residue.
Non-specific DNA Binding Property
DNA binding proteins found in cells have various interacting modes with DNA and can be broadly classified as sequence specific and non-sequence specific interactions. Sequence specific DNA protein interactions involve special structural motifs on a protein which is recognised by a consensus sequence on the DNA, e.g. restriction endonucleases recognising palindromic sequences on DNA. On the other hand, non-specific DNA protein interactions predominantly involve ionic interactions with the sugar phosphate backbone of DNA. Some of these non-specific DNA binding proteins gives rise to large supramolecular structures with their association with DNA, e.g. as seen in the case of genome organization by histones in eukaryotes. Also, binding of multiple proteins to long DNA strands allows condensation and makes DNA inaccessible to other environmental factors, giving physical protection (Ganguly et al. 2012).
Studies have shown that when Dps interacts with DNA, highly stable crystals are formed within which DNA is sequestered and protected against various conditions of stress. This has been shown under in vitro and in vivo conditions (Wolf et al. 1999). DNA-Dps co-crystallization presents a binding mode that provides wide range protection by non-specific DNA sequestration. Although the crystalline state is incompatible with life, sequestration of important biological macromolecules in crystalline assemblies may provide adequate protection under conditions of stress. Such ordered Dps-DNA co-crystals have a condensed organisation, which also dramatically enhances the stability of DNA and can endure prolonged starvation and other adverse conditions (Frenkiel-Krispin et al. 2001).
This section explores the mechanism of non-sequence specific DNA binding property of Dps, which has been studied by Atomic force microscopy, X-ray crystallography and more recently Cryo electron microscopy.
Studies on Dps Deletion Mutants
Dps does not have any known DNA binding motifs. In this scenario, initial studies on Dps-DNA binding were carried out by looking at how deletions affect binding to DNA. In E. coli, to establish the effect of the electrostatic interactions between the positively charged residues on the surface of the N-terminus and negatively charged DNA, systematic deletions of the lysines at the N-terminus were carried out. Dps with a deletion of two lysines at the N-terminus was able to bind DNA but could not carry out DNA condensation, whereas a deletion of all the three lysines rendered it not able to bind DNA. The property of Dps to self-aggregate was implicated in its ability to condense DNA by protein–protein aggregation (Ceci et al. 2004) (Fig. 3.4). A long flexible N-terminus (Fig. 3.5a) with charged residues like lysines and arginines are seen in a number of Dps homologs, pointing to similar mechanisms of DNA binding. This was demonstrated by Dps homologs in D. radiodurans, where deletion of the N-terminus abrogated DNA binding (Bhattacharyya and Grove 2007).
DNA condensation through E. coli Dps self-aggregation a Model proposed by Ceci et al. for DNA condensation. Dps is represented as blue circles. The binding of Dps to DNA is promoted by lysines at the N terminus of Dps. b AFM images of Dps-DNA complexes. Upper panel shows top view and lower panel shows the corresponding lateral 3D views. Scale bar is 100 nm. Figure adapted with permission from Nucleic Acids Res. 2004. 32(19): 5935–5944
DNA binding regions of Dps homologs from different species. a E. coli Dps dodecamer (PDB ID: 1DPS) with DNA binding flexible N-terminal residues 9–21 represented in green, residues 1–8 are missing in the crystal structure due to flexibility. b M. smegmatis Dps1 dodecamer (PDB ID: 1VEI) with the interlocking N-termini (magenta) and C-termini (blue). c L. lactis Dps (PDB ID: 1ZUJ) dodecamer has a helical ordered N-terminus (cyan). d A. tumifaciens Dps dodecamer (PDB ID: 1O9R) N-terminal region in red is immobilized on the protein surface and unable to bind DNA
A variation is the M. smegmatis Dps protein MsDps1 which seemed to bind DNA through a mobile and positively charged C-terminus with 3 lysines and 2 arginines, and a deletion of the 16 C-terminal residues resulted in a loss of DNA binding activity (Roy et al. 2007). Deletion of the entire C-terminal tail resulted in an open decamer assembly, indicating that the C-terminal tail has a role in DNA binding and oligomerisation (Fig. 3.5b). An ordered alpha helical N-teminus in L. lactis Dps can condense only long stretches of DNA > 4000 bp. The exact mechanism for this is unclear, but the authors proposed that the decreased flexibility of the N-terminus due to its helical conformation could have a role in the size limitation of DNA binding (Stillman et al. 2005) (Fig. 3.5c). The H. plylori Dps, HP-NAP has a positively charged protein surface at neutral pH and binds DNA through the protonated amino acids on the protein surface (Chiancone and Ceci 2010; Ceci et al. 2007). Dps from T. erythraeum lacking a positively charged N-terminus and a truncated C-terminus, could use a mechanism similar to HP-NAP to bind DNA (Castruita et al. 2006).
Some homologs of Dps like the L. innocua Dps, B. anthracis Dlp-1 and Dlp-2, C. jejuni etc are characterised by a short N-terminus and are unable to interact with DNA (Bozzi et al. 1997; Papinutto et al. 2002; Ishikawa et al. 2003). A. tumifaciens Dps N-terminal region is immobilized on the protein surface and is not accessible for DNA binding (Fig. 3.5d) (Ceci et al. 2003). Also, pH and salt compositions are shown to have an influence in the ability of Dps to bind and/or condense DNA in vitro, so these factors have to be taken into consideration for comparison of studies across different Dps homologs.
Higher Order Dps-DNA Assemblies Visualised Through Atomic Force Microscopy
Dps condenses DNA by self-association giving rise to large Dps-DNA aggregates which do not enter agarose gels. This was first demonstrated in E. coli, where Dps binds DNA through its charged lysine residues. Further through self-aggregation it brings about the looping and condensation of DNA (Fig. 3.4). The interaction to DNA is directed towards condensation only at pH values where the lysine residues are fully protonated. But H. pylori Dps HP-NAP uses a different mechanism, as it is not able to self-aggregate, but can bind and condense DNA at slightly acidic pH values where the histidines which mediates DNA binding are protonated (Ceci et al. 2007).
The Dps from M. smegmatis MsDps1 when overexpressed in cells, promoted a toroidal structure of Dps-DNA. Whereas, with the overexpression of the second M. smegmatis Dps MsDps2 a coral reef structure was formed (Fig. 3.6a) (Ghatak et al. 2011). D. radiodurans Dps2-DNA interaction was shown to form toroidal structures with DNA, similar to the arrangement in MsDps1 (Fig. 3.6b) (Santos et al. 2015). The higher-order assemblies of Dps-DNA shown by AFM and EM are a result of DNA condensation or co-crystallization. Thus, looping and condensation of DNA is seen when Dps is added to linear or circular stretches of DNA. Toroids, coral reef structures and co-crystals are formed within bacterial nucleoids under very late stationary phase or when the protein is overexpressed in cells. Additionally, co-crystals with DNA have been shown to form under in vitro conditions.

reproduced from PLOS ONE. 2011. 6(1): e16019) b D. radiodurans Dps1-DNA complex at pH 6.5 shows DNA condensation (left panel) and DrDps2-DNA toroidal assemblies at pH 6.5 (right panel). Color scale represents the height of D. radiodurans Dps-DNA complex. Images reproduced with permission from FEBS J, 282: 4307–4327
Higher order Dps-DNA assemblies visualised by Atomic Force Microscopy. a The lysate of M. smegmatis mc2155 overexpressing Dps1 at late stationary phase, shows toroidal assemblies of Dps-nucleoid with a diameter of 70–100 nm (left panel). Late stationary phase cell lysate of mc2155 overexpressing Dps2 shows coral reef structures with nucleoid (right panel). (Images
It was shown that the effect of Dps on the nucleoid state can vary depending on different stages of growth or on the species of bacteria. The overexpression of Dps in E. coli in the log phase induces neither nucleoid condensation nor DNA-Dps co-crystallisation, whereas in the stationary phase it results in nucleoid condensation. On the other hand, the S. aureus Dps homolog mrgA (Morikawa et al. 2006) causes nucleoid condensation in both log and stationary phases on over-expression in cells. This was due to the action of Fis, a bacterial nucleoid associated protein abundant in the log phase which inhibits nucleoid condensation by Dps during this stage of growth. Also, Fis is present in gamma Proteobacteria which includes E. coli but not S. aureus, which explains why there is no regulation of DNA condensation in the log phase when Dps is overexpressed in the latter organism. Thus, the organisation of Dps into higher order assemblies with DNA is species-specific and also is an interplay of Dps and other nucleoid binding proteins in cells during different stages of growth.
DNA-Dps Biocrystals
Living systems when exposed to severe environmental stresses, may sequester vital macromolecules in intracellular crystalline assemblies for efficient protection. Under these conditions often defense strategies based on enzyme catalysis can no longer work efficiently due to energy restrictions, so DNA protection is often achieved through physical transactions like phase separation and phase transitions (Wolf et al. 1999). A defense strategy involving co-crystallization of DNA and Dps was observed under in vitro and in vivo conditions in E. coli by Wolf and colleagues. They showed that purified Dps and DNA interact and almost instantaneously form stable crystals (Fig. 3.7a) within which DNA is sequestered and protected from possible environmental assaults. Similar crystalline structures are also formed in vivo in cells where Dps is overexpressed or in wild-type bacteria under starvation conditions (Fig. 3.7b).
Electron microscopy images of Dps-DNA co-crystals a Dps incubated for 15 s with closed circular DNA in vitro (left panel), scale bar 100 nm and its higher magnification, scale bar 40 nm (right panel). b Crystalline assemblies of Dps-DNA in a starved WT E. coli cell indicated by black arrows (left panel) and dps knockout cell (right panel), scale bars are 100 nm. Images
Recently, two different types of Dps-DNA co-crystals were observed by cryo-electron tomography (Kamyshinsky et al. 2019). Subtomogram averaging of first type of co-crystals revealed a triclinic crystal lattice (Fig. 3.8a). The second type of co-crystals adopted a cubic crystal lattice (Fig. 3.8b), where each Dps is surrounded by six DNA strands, unlike in triclinic co-crystals where each Dps is surrounded by four DNA molecules. But the exact nature of Dps-DNA interactions could not be revealed in the low-resolution reconstructions and future single particle cryo-EM studies on individual DNA-Dps complexes or 2D crystals may achieve this.
Dps-DNA co-crystals a CryoTEM images of Dps-DNA co-crystals, red arrows indicate ones with a triclinic crystal lattice and the red inset is a high magnification of the same. Blue arrows indicate co-crystals with a cubic crystal lattice, a higher magnification is shown in the inset with blue outline. Scale bars are 200 nm and inset 20 nm. b 3-D reconstructions based on subtomogram averaging of cubic Dps-DNA co-crystals, in two different views. DNA is in orange and Dps are blue spheres with unit cell shown in green (left). Corresponding models of Dps and DNA fitted in the EM densities are shown in the right panel. Images
Modes of DNA Protection and Regulation
The multifarious protection of DNA by Dps is by virtue of three intrinsic properties of the protein: DNA binding, iron sequestration and ferroxidation. Moreover, Dps is proposed to have roles in gene regulation during starvation, by controlling expression at the transcriptional level. This section explores these claims in the light of recent findings, as well as summarising already existing knowledge of DNA protection mechanisms.
Protecting DNA From Damage
Dps prevents DNA damage by direct physical sequestration and indirectly through its ferroxidation activity. These biochemically separate activities function jointly to preserve DNA integrity (Fig. 3.9) (Calhoun and Kwon 2011). Through its DNA binding ability, Dps was shown to confer protection from UV and gamma radiation damage, which induces potentially harmful mutations in DNA (Nair and Finkel 2004). Another instance of Dps endowing resistance to cells enduring acidic conditions is through acid tolerance in E. coli which helps the bacterial passage through gastric barrier of humans. It was shown that acid stress led to damage of chromosomal DNA, accentuated in dps mutants (Jeong et al. 2008), showing a role for Dps in shielding DNA against acid stress. One of the earliest mechanisms which explains Dps protection of DNA against H2O2 mediated oxidative stress was found to be by direct binding of Dps to DNA, reproduced in in vitro systems (Martinez and Kolter 1997). Dps null mutants were also more sensitive to some types of metal toxicity, such as toxicity from copper and iron during stationary phase. Temperature dependent adaptation was also seen to a less extent in these mutants as compared to WT cells, during thermal stress (Nair and Finkel 2004). Dps expression is also upregulated by the OxyR transcriptional regulator, in response to oxidative stress (Altuvia et al. 1994).
Oxidation of Fe(II) occurs via Fenton reaction releasing hydroxyl free radicals, which causes cell damage (top panel). Oxidation of Fe(II) happening inside the Dps shell at ferroxidation sites proceeds without production of free radicals thereby protecting cells from oxidative damage (bottom panel). Figure adapted with permission from Journal of Applied Microbiology. 2010. 110: 375–386
Dps can also protect the genome from a distance using DNA charge transport through the DNA base pair π-stack. This was demonstrated in E. coli Dps, where an intercalating ruthenium photooxidant was used to generate localised DNA damage to guanine repeats, which are the sites of lowest oxidation potential in DNA. When iron loaded Dps was titrated against this modified DNA as opposed to apo-Dps, it significantly reduced oxidative damage of DNA. The authors propose that the iron loaded Dps is oxidised by DNA mediated oxidation through charge transport, as there was no direct contact between the ruthenium photooxidant and Dps (Fig. 3.10) (Arnold and Barton 2013). Thus through DNA charge transport, Dps could respond to an oxidative affront happening around a 100 base-pairs away.
Schematic of DNA protection through charge transport. Steps: 1. A DNA intercalated ruthenium(II) photooxidant is excited by visible light. 2. Ru(II) is oxidatively quenched to Ru(III) by a diffusing quencher (Q). 3. Ru(III) abstracts an electron from DNA and the electron hole equilibrates along the π-stack and localizes to the most easily oxidised base like guanine (G•). 4. DNA charge transport from Dps to guanine radical to fill the electron hole is a possible long distance protection mechanism. Lower panel shows the structure of Ru(II) covalently tethered to DNA via diaminononane linkage. Figure
Conserved aromatic residues like tyrosine and tryptophan near the ferroxidase site, can act as electron transfer intermediaries, allowing for rapid transfer of electrons between the ferroxidase centre and outside of the protein shell. In L. innocua Dps, a double mutant lacking both the aromatic residues near the ferroxidase site showed attenuation of DNA protection and the DNA was degraded by hydroxyl radicals generated through ferrous oxidation by H2O2. DNA protection assays indicated that the presence of aromatic residues limited the release of hydroxyl radicals into the solution and subsequent oxidative damage of DNA. Thus the aromatic amino acids near the ferroxidase centre could act as a trap for electron holes generated by oxidation of ferrous iron by H2O2 (Bellapadrona et al. 2010). Also, these residues could play a role in DNA mediated oxidation of Dps through charge transport. These findings further highlight the interplay of ferroxidation and DNA binding property for DNA protection.
Nucleoid Condensation in Response to Stress
The genome in bacteria is folded into a compact structure called the nucleoid. In order to reduce the volume to fit inside a cell, nucleoid associated proteins or NAPs bind and condense DNA by bridging, bending or wrapping (Fig. 3.11a–f). In addition, other factors like molecular crowding and DNA supercoiling contribute to further compaction of DNA. DNA bending NAPs like IHF, HU and Fis act together with DNA bridging proteins like H-NS or Lrp, with expression varying according to the growth phases of the cells. Fis has optimal expression during exponential phase (1 Fis/460 bp), after which HU is expressed (1 HU/550 bp). In early stationary phase IHF reaches its expression peak (1 IHF/335 bp). Dps is the most abundant protein in late stationary phase (1 Dps dodecamer/300 bp) (Luijsterburg et al. 2006).
During fast exponential phase of growth with high transcriptional activity, the nucleoid is associated with clustered transcriptional foci. In stringent response where inhibition of transcription happens, the nucleoid decondenses to form an elongated structure. In early and stationary phase ring-shaped toroids containing DNA and Dps and eventually the Dps-DNA crystalline lattice appears (Fig. 3.12), where the DNA is transcriptionally inactive (Travers and Muskhelishvili 2005). To summarise the changes in the expression of NAPs, in the exponential growth phase Fis is the most abundant protein with the following order of abundance: Fis > Hfq > HU > StpA > H-NS > CbpB > Dps. In early stationary phase, Fis disappears and the order of abundance changes to: Dps > IHF > Hfq > HU > CbpA > StpA (Ali Azam et al. 1999).
Role in Transcription Regulation
The above sections have described the mechanisms by which multifaceted Dps proteins protect cells against stresses through DNA binding, iron binding and sequestration and ferroxidation. Another possible mode of protection is by the regulation of gene expression by modulating the structure of DNA thereby controlling access to the transcription machinery. Two dimensional gel electrophoresis of cell lysates from WT and dps null mutant shows differential expression of proteins (Almiron et al. 1992). The compacted Dps-DNA structures, along with differences in protein expression patterns suggested a role for Dps as a pleiotropic regulator of transcription when cells enter the stationary phase.
But, a recent study has shown that dps deletion causes no significant change in the gobal transcriptional patterns and only mild alterations in the proteome, in vivo during the stationary phase. Dps had no effect on RNA polymerase (RNAP) initiation at physiologically relevant concentrations. Single molecule transcription assays used in vitro to probe effects of Dps-mediated DNA compaction revealed that Dps binding did not induce RNAP pausing or arrest transcriptional elongation of DNA (Janissen et al. 2018). These findings suggest that Dps is a DNA binding protein that completely decouples DNA condensation from transcriptional regulation, in contrast to histones and other bacterial NAPs like HU and H-NS. This could provide bacteria greater freedom to tailor transcriptional responses to different stresses at the same time as protecting the genome from damage.
Dps shows co-operativity in binding to DNA (Ganguly et al. 2012) and this requires multiple nearest neighbour interactions between Dps dodecamers to stabilize the relatively weak interactions with DNA. Thus, Dps has a cumulative affinity for DNA, despite having low affinity for individual contacts on DNA (Vtyurina et al. 2016). Therefore, proteins such as RNAP which establishes stable interactions with DNA could displace Dps. But it does not explain how some others like the restriction enzymes with high affinity for DNA could be excluded from acting on Dps-DNA complexes. The dynamic behaviour of Dps-DNA binding could indicate that it forms a phase-separated organelle with DNA, permeable to RNAP. These evidences suggest that the ability of Dps to protect cells during stress is a direct effect of DNA binding rather than activation or repression of specific genes. This dynamic nature of Dps assembly on DNA also ensures that transcription can continue even under extreme conditions of stress (Fig. 3.13) (Janissen et al. 2018).
Model of DNA Protection by Dps a Under normal conditions in cells, E. coli Dps binds DNA but is unable to condense the nucleoid. b Under conditions of stress, Dps condenses most sections of the nucleoid, creating phase-separated organelles, which is accessible to RNA polymerase but inaccessible to other nucleoid binding proteins. Figure
Ferroxidation Properties
Iron plays a vital role in the processes of living organisms and the acquisition and storage of metal in a non-toxic and bioavailable form is a primary requirement for cells. Iron starvation is one of the major obstacles for successfully establishing growth. Proteins of the ferritin family, like ferritins, bacterioferritins and Dps are the major iron storage entities in cells, with Dps found only in prokaryotes. Dps are dodecameric nano-compartments having roughly spherical structures with a hollow core for storing iron. Twelve identical monomers assemble through a 23 symmetry to form 12-mers, and are frequently referred to as miniferritins, smaller in size than the 24-meric ferritins. They can store ~500 iron atoms per dodecamer and can oxidise iron to its inert ferric form (Bozzi et al. 1997). Studies that examine the mechanism of iron uptake, ferroxidation and release in response to the needs of cells, has been carried out through mutational and structural analyses. This section summarises the molecular mechanisms of how Dps processes iron. Being iron storehouses in cells, Dps proteins share considerable structural similarity to ferritins. An evolutionary link to ferritins in the light of recent studies showing a structural basis of a switch between Dps and ferritins is also discussed.
Iron Uptake and Release
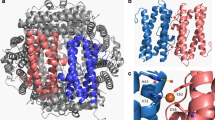
To better understand how Dps sequesters iron, it is necessary to examine the structure of these dodecamers. Each monomer is an alpha helical bundle, with four helices named A–D, and a short BC helix in between B and C helices (Fig. 3.2a). These monomers have a tendency to form dimers, which are thought to be the first intermediate of oligomerisation. The ferroxidase center is placed in the dimeric interface in between two subunits. Due to its symmetrical arrangement, Dps exhibits two type of three-fold interfaces, namely the ferritin-like three-fold and the dps-like three-fold interfaces. The ferritin-like three-fold interface is important in the channelling of iron into the protein, whereas the dps-like interface is responsible for the protein folding into a dodecamer. A model for iron incorporation can be described in three steps: (1) Fe(II) atoms enter the protein cavity through the four ferritin-like three fold interfaces of the dodecamer. (2) Fe(II) atoms are oxidised at the ferroxidase centre. (3) Oxidised Fe(III) atoms move towards nucleation sites in the interior of the cavity. (4) Fe(III) mineral core is formed, with the possibility of further incoming Fe(II) atoms getting oxidised on the surface of this growing mineral core (Fig. 3.14) (Pulliainen et al. 2005).
Schematic of iron oxidation and storage by Dps proteins, with S. suis Dpr as a representative example. Steps: (i) Fe(II) enters the protein cavity through the four hydrophilic pores each within the ferritin-like three-fold interfaces, possibly attracted by negatively charged residues like aspartates which lines the pore. (ii) Fe(II) gets oxidised at the twelve inter-subunit ferroxidation sites, each located between two subunits related by a two-fold interface (iii) and (iv) Fe(III) ions move to nucleation sites where Fe(III) mineral formation is initiated, and the mineral deposition continues to grow with more Fe(II) getting oxidised. “?” represents a path for Fe(II) ions to be oxidised directly on the growing mineral core, which could happen at a faster rate and explains the sigmoidal nature of the ferroxidation curve in Dps proteins. Figure
Like ferritins, Dps has a roughly spherical structure with a hollow core and pores at the ferritin-like interface, making them highly suited for iron sequestration and storage. The ferritin-like interface in Dps forms a funnel-shaped channel with a wide mouth facing the solvent side and which narrows towards the interior of the protein cavity (Fig. 3.15a, b). This channel is lined by hydrophilic residues, mainly negatively charged aspartate and glutamate residues, which trap and internalize iron into the cavity (Williams et al. 2014). A number of studies in ferritins point to the ferritin-like pore as the channel of entry and exit. Mutational analyses in S. suis and L. innocua Dps indicated the importance of the conserved aspartate residues in this channel, whereby any substitutions at these loci led to changes in the rate of iron uptake and release (Pulliainen et al. 2005; Bellapadrona et al. 2009).
Funnel shaped channel for iron entry in Dps a Side view and b Surface view with a continuous blue circle revealing the diameter of the mouth of the funnel-shaped channel and dashed circle shows the narrowest part of the channel. Residues important for iron channelling are shown as sticks (PDB ID: 2Z90)
A mechanism of iron uptake and release was demonstrated on the basis structural evidence in M. smegmatis Dps, MsDps2. An assembly of oppositely charged histidine-aspartate cluster at the ferritin-like interface was identified, which guards the narrowest point of the channel. Site specific variants which disrupted this ionic knot, showed a defective mechanism of iron uptake and release, highlighting a gating mechanism that happens by alterations in the side chains of the residues lining the channel and does not change the over-all stability of the protein. Iron release from iron loaded Dps can be carried out by a combination of reductants and chelators. Reducing agents like sodium dithionite converts Fe(III) to Fe(II) which is chelated by agents such as 2,2′-bipyridyl to form Fe(2,2′-bipyridyl)32+ complex and forms the basis of iron release assays in vitro. The kinetics of iron release exhibits an early fast rate which corresponds to the release of iron bound near the pores and easily accessible to chelators, followed by a slower rate which coincides with the dissolution of iron oxide in the protein core. Replacing the aspartates at the narrow opening with bulkier histidine residues reduce the rate of iron release by several fold (Williams et al. 2014).
However, the molecular mechanisms of iron release in vivo to meet iron requirement in cells during conditions of iron demand, is less understood. In vivo iron release studies in ferritins have demonstrated the cytosolic release of iron by ferritin degradation within lysosomes (Linder 2013). It is thought that once the ferritins are degraded in the lysosomes, the exposed ferrihydrite core maybe reduced/chelated and transported to the cytosol. This mechanism may not translate to Dps, as Dps is present only in prokaryotes. The N-end rule degradation pathway of Dps via ClpX/ClpP could release the iron deposited within, making it accessible to chelators and other iron transporters in cells.
Conserved Ferroxidase Active Site
The first structural evidence of a ferroxidase site was revealed in the X-ray crystal structure of L. innocua Dps (Ilari et al. 2000). This first ferroxidase centre identified contained only one iron atom, contrary to the conventional di-iron centres found in ferritins. Since then di-iron FOCs have been identified in other Dps homologs, where the second iron displayed a partial occupancy. Unlike the FOCs in ferritins which are contained within the four-helix subunit, the FOC in Dps is between two subunits related by a two-fold. The iron in LiDps FOC is coordinated by carboxylate residues and one or two histidine residues that play a role in the redox process. The two FOCs in a two-fold interface are separated by a distance of 21.5 Å.
A di-iron FOC was indicated in the crystal structure of B. brevis Dps, where two neighbouring di-nuclear ferroxidase centers responsible for the oxidation of Fe(II) ions was identified. The iron at site 1 is tightly coordinated to NE2 of His 31 in subunit A, carboxylate group of Asp58 and OE2 of Glu62 from subunit B, where A and B are related by a two-fold symmetry. Iron at site 2 is loosely bound to NE2 of His43 (subunit A), OE1 of Glu62 (subunit B) and OE1 of Glu47 in subunit A via a water molecule (Fig. 3.16). A µ-oxo bridge links the iron atoms of site 1 and 2, representing a reaction intermediate Fe(III)-O-Fe(III) during the oxidation of Fe(II) (Ren et al. 2003). Like most active site residues, the ferroxidation site residues are well conserved among Dps homologs from various species.
Dinuclear ferroxidase centre in B. brevis Dps. Two FOCs are in black discontinuous circles, related by a two-fold interface (PDB ID: 1N1Q). One of the FOCs is magnified on the right panel, showing cordinating residues from subunits A and B. Fe ions at site 1 and site 2 are labelled Fe1 and F2, respectively. Figure adapted with permission from J. Mol. Biol. 2003. 329: 467–477
Iron Gating Mechanisms
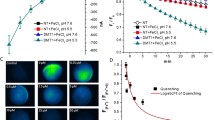
Dps achieves iron channelling by very little changes to its overall conformation, being very stable protein cages. A recent study by our group through co-crystallisation of Dps with iron, showed that flexible aspartates could propel iron from the entry site to the ferroxidation centre. A conserved arginine residue forms a network of interactions which stabilizes the interface between the ferroxidation and iron entry sites (Fig. 3.17) (Williams and Chatterji 2017). The iron can bind to three sites at the entry channel as shown in Fig. 3.18a. In M. smegmatis MsDps2, Asp138 which lines the narrowest region of the pore, shows flexibility which could help in propelling iron to the interior and any substitutions at this site caused a marked reduction in the rate of iron entry. Another, Asp68 at the ferroxidation site was shown to exhibit alternate conformations. Asp68 in its “outward-facing” conformation where the side chain faces away from the iron binding centre, is a possible state when no iron is bound at the site. In an “inward-facing” conformation, the ferroxidation site satisfies the coordination chemistry for iron binding (Fig. 3.18b). A three-tier arrangement of flexible asparates, each tier having three aspartates related by a three-fold interface, lines the path of iron from its entry to the ferroxidation site (Fig. 3.19) (Williams and Chatterji 2017). Further biophysical studies looking at how the movement of aspartate side chains are linked to the passage of iron might throw valuable insights on iron channelling in Dps.
View from the inside of a Dps dodecamer showing conserved arginine stabilizing interactions between the ferroxidation centre (FOC) shown by a red circle and iron entry channel indicated by a blue dashed circle. Each monomer and its residues forming the interface is colored differently. Arg73 is the conserved arginine in the dps homolog MsDps2 (PDB ID: 2Z90)
Three tiers of flexible aspartates help to propel iron to the ferroxidase site (FOC). Iron entry site residues are in magenta sticks, FOC residues are in green. The aspartate residues in blue are in a site between the active site and entry site, could help the movement of iron between these sites. Fe ions are shown as orange spheres. The three aspartates of each tier are related by a trimeric interface (PBB ID: 5WW5)
Structural analysis of Dps loaded with iron to saturation levels did not show any appreciable changes in X-ray crystallographic structure. Possibly because an amorphous core of iron cannot be detected by X-rays, no density accounting for iron could be found in any of the co-crystal structures. Co-crystals of varying ratios of iron: dps did not show any appreciable structural variations from low iron-bound forms to higher iron loaded forms. This indicates the extraordinary stability of the Dps cages which can take iron to saturation levels without major conformational adjustments. These proteins could allow iron entry by fluctuations in side chains of the hydrophilic residues lining the pore gates (Williams and Chatterji 2017).
Interestingly, small iron oxide clusters have been reported in H. salinarum DpsA iron loaded protein crystals (Zeth et al. 2004). A greater mobilization of iron could be required in a hypersaline environment faced by this organism, where iron reduction and release could occur faster in small clusters having higher surface to volume ratios, compared to large iron deposits. As such clusters have not been observed in other Dps homologs, this could be an adaptation strategy unique to this organism in surviving extreme conditions.
Binding to Other Metals
Metal ions like terbium and zinc were shown to inhibit iron incorporation into mammalian ferritins. Listeria innocua Dps has also been found to be weakly inhibited by Zn(II) and strongly be Tb(III) (Stefanini et al. 1999). S. mutans Dpr could incorporate 11.2 zinc atoms per dodecamer in addition to its capacity to incorporate up to 480 iron atoms (Yamamoto et al. 2002). This led researchers to propose that other metal ions could compete with iron binding at the ferroxidation sites. The structural basis of Zn and Tb mediated inhibition of ferroxidation was shown in S. suis where crystal structures with Zn and Tb incorporated Dpr showed the presence of Zn and Tb at the ferroxidase centre and in the same location where iron normally binds (Havukainen et al. 2008). In vitro studies also confirmed that both ions abolish iron oxidation in Dps at concentrations >0.2 mM. A di-zinc binding in the ferroxidase site was also reported in S. pyogenes Dpr (Haikarainen et al. 2010). Divalent metal cations Cu2+, Ni2+, Co2+, Mn2+, Zn2+ and Mg2+ bound to ferroxidase sites of S. suis Dpr was characterised by isothermal titration calorimetry and X-ray crystallography. These ions were found coordinated to the ferroxidase site. The exact physiological function of the ferroxidase centre binding other metal ions is not known, but it could have certain indications on how Dps confers resistance against metal stresses of zinc or copper (Haikarainen et al. 2011).
In S. solfataricus, a cellular environment where availability of Mn is higher than Fe, SsDps (S. solfataricus Dps) could load Mn and inhibit mineralisation of Fe (Hayden and Hendrich 2010). In competition experiments where Mn2+ was already bound to the metal sites, Fe2+ was unable to displace the bound manganese, thereby inhibiting ferroxidation. Kineococcus radiotolerans, a radiation resistant bacterium having a high ratio of Mn:Fe in the cytosol, can survive ionizing radiations. It was discovered that the ferroxidation centre of this organism can have an Mn-Fe composition as opposed to the usual Fe–Fe dinuclear iron bound at the ferroxidase centre. The authors propose that the hetero-binuclear ferroxidase centre with Mn-Fe could have a role in redox cycling and help in combating oxidative stress (Ardini et al. 2013).
The Dps from the cyanobacterium T. elongatus was shown to bind two Zn at the ferroxidase site. The FOC in this dps homolog is unique in having two histidines and only one of the bound zinc can be displaced by incoming Fe ions, giving rise to Zn(II)–Fe(III) complexes as indicated by atomic emission spectra (Alaleona et al. 2010). Another cyanobacterial Dps from N. punctiforme, NpDps4 has an atypical ferroxidase site similar to TeDpsA with two additional histidines, which also can bind Zinc. This led the authors to classify a new type of FOC called the His-type FOC (Howe et al. 2018). Zinc bound at the ferritin-like three-fold interfaces seen in the case of H. pylori HP-NAP, which are iron entry channels in Dps, indicates that other metals could also enter the dodecamer through the usual iron entry sites (Yokoyama and Fujii 2014).
Evolutionary Link with Ferritins
Ferritins are 24-meric proteins with an internal cavity which can store and oxidise iron, like their smaller 12-meric counterparts, Dps. They have a 432 octahedral symmetry with subunits forming four-fold, ferritin-like three-fold and two-fold symmetries. Dps has a 23 tetrahedral symmetry, with subunits related by a ferritin-like three-fold, Dps-like three-fold unique to Dps and two-fold symmetries (Fig. 3.20). The ferritin-like three-fold interface is important for iron channelling, whereas the Dps-like three-fold of Dps and the four-fold interface in ferritins are important for protein folding into higher oligomers. In Dps, the AB loop between helices A and B forms stable interactions at the dps-like trimeric interface, and a mutation of a highly conserved phenyl alanine to a glutamate (F47E) in the AB loop of M. smegmatis Dps1 was shown to alter its assembly from the canonical 12-mer to a ferritin-like 24-mer. The assembly of mutated Dps monomers to ferritin-like cages seems to be stabilised only under crystallization conditions, as in solution these subunits exist as dimers (Williams et al. 2017).
Symmetry related interfaces in MsDps1 (PDB ID: 1VEI), F47E-MsDps1 (PDB ID: 5H46) and frog M ferritin (PDB ID: 3RBC). F47E-MsDps1 has an arrangement similar to ferritins. Dps proteins have a dps-like trimeric interface, distinct from ferritins and F47E-MsDps1which has a tetrameric interface. In Ferritins, E helix is at the centre of the tetrameric interface. F47E-MsDps1 has a large gap in the assembly of the tetramer having no E helices
The crystal structure of F47E-MsDps1 exhibited a 24-meric ferritin-like assembly, with an outer diameter of 120 Å and an inner diameter of 80 Å similar to ferritins. WT Dps has an outer diameter of 90 Å and a central cavity of 45 Å. The F47E-MsDps1 had a 432 octahedral symmetry, with four-fold, ferritin-like three-fold and two-fold symmetries (Fig. 3.20). The point mutation F47E caused the AB loop to lose its rigid conformation due to disruption of the hydrophobic interactions at the dps-like three-fold, pushing its conversion to a four-fold interface similar to ferritins (Fig. 3.21). In ferritins, a C-terminal E helix is poised at the centre of the four-fold and stabilises the tetrameric interface. It is interesting to note that some ferritin homologs have a short AB loop and others have an AB loop which faces away from the four-fold interface, so as to avoid clashes with the E-helix.
Model for assembly of ferritin family proteins. Top panel shows the proposed assembly of a Dps 12-mer, where a four-helix monomer with a “rigid” AB loop associates via the dps-like trimeric interface. Bottom panel shows the monomers associating through a tetrameric interface with the C-terminal E helix at the centre, forming 24-meric ferritins. Figure reproduced with permission from Structure. 2017. 25: 1449–1454
Ferritin family proteins are thought to have originated from rubyerythrin-like ancestors which evolved into 12-meric bacterioferritins (Bfr) that later diverged into 12-meric Dps and 24-meric ferritins/Bfrs (Andrews 2010). Thus, early ferritin-like proteins could have had a dodecameric assembly, and as indicated by the mutational switch F47E, minor changes in secondary structure could have led to the evolution of 24-mers.
Applications in Biotechnology and Nanomedicine
Biomolecules like Dps proteins with cage-like assemblies can be engineered via its inner shell, external surface or subunit interfaces. The hollow interior can be manipulated to load therapeutic drugs and other diagnostic molecules. External surface modifications can help to enhance biocompatibility and cell-specific targeting. Self-assembly can be modulated through altering the subunit interfaces. The iron release pores can be altered to fine tune release of molecular cargos (He and Marles-Wright 2015). This section further explores the applications of Dps in biotechnology, drug delivery and nonomedicine.
Engineering of Nanoscale Devices
Multiprotein complexes like Dps with their well-defined interior cavities are attractive templates for bionanotechnology. They can be produced in large quantities, are amenable to chemical modifications, are structurally and functionally well characterised (He and Marles-Wright 2015). The synthetic flexibility of these miniferritins have been demonstrated by computational design introducing wide-scale mutations for altering polarity of the interior from a hydrophilic to hydrophobic cavity. These mutated Dps could assemble similar to WT proteins and form dodecamers, paving way for engineering proteins with novel functions (Swift et al. 2006). Dps can bind other metal ions in addition to iron and this property has been utilized to generate mineral cores of cobalt and oxygen in L. innocua Dps (Allen et al. 2003).
Ferritin-like proteins are used to mineralise non-physiological metals through self-assembly around a solution of metal ions, chemically mediated redox reactions or photochemistry (Yoshimura 2006). Ferritins can also mineralise iron sulphide, Mn(III) and magnetite (Fe3O4) cores. Ferritin-like proteins with magnetite core called magnetoferritins have uses in cell imaging, providing magnetic contrast and magnetic separation of cells and particles (Deans et al. 2006).
Dps has been used to fabricate inorganic nanodevices like quantum dots and nanowires. Here, Dps cores loaded with a mineral core are deposited on silicon substrates which are silanized and made functional with small peptides or molecules. The protein cage can be removed with heat to leave the core in place, and such semiconductor cores deposited on silicon wafers can be used as memory gates (Okuda et al. 2005). Metals cores isolated from Dps can be used to seed the growth of carbon non-tubes and nanowires (Kim et al. 2011). Dps loaded with mineral nanoparticles can crystallise in 2-dimensional arrays to give a regular array of uniformly sized nanoparticles on solid surfaces.
Due to the capacity for mineralisation of heavy atoms in the core, Dps can be used as contrast agents in electron microscopy. The stability of Dps and its ordered symmetric structure makes it an ideal sample for cryoEM imaging techniques. Apoferritins are routinely used as a standard for estimation of the magnification of micrographs in cryoEM (Wasilewski et al. 2012).
Nanoparticles in Biomedicine
Ferritin family proteins are stable and biocompatible entities that can be manipulated with respect to their assembly, surface modifications and reconstitution. Thus, they have the potential to be developed as drug delivery vehicles. Due to their hydrophilic cavities, incorporation of non-metal containing drugs poses challenges. Addition of charged molecules or drugs complexed with metals such as copper, could overcome this limitation (Maham et al. 2009, 2011). Surface modifications with epitopes and labels to target these loaded nanovehicles to specific cell types or tumours, could be a powerful tool for drug delivery.
Ferritins have been used to develop self-assembling nanoparticles that elicit broader and more potent immunity in comparison to traditional vaccines. In this instance, haemagglutinin from the influenza virus was fused to ferritins, which assembled via the trimeric ferritin-like interfaces to give eight trimeric viral spikes on the ferritin surface. Immunization with the HA-ferritin fusion protein generated an increased HA inhibition antibody titre compared to the traditional inactivated vaccines (Kanekiyo et al. 2013). Also, there was no autoimmune reaction here, due to the use of H. pylori ferritin with high sequence variation from the human ferritin. Similar fusion proteins have been engineered with Dps, with a roles in diagnosis, treatment and prevention of diseases. Dps fusion proteins are soluble, in spite of the poor solubility of the attached peptide/epitope. They also have enhanced thermostability and enhanced immune response due to multimeric assembly, similar to ferritins (Verma et al. 2018).
Biomoleules can often rival synthetic molecules due to their flexibility, biocompatibility, diversity and lower toxicity. Naturally occurring nanocompartments like Dps can thus be exploited to encapsulate nanoparticles, and surface modifications can help to target tissue systems and make them suitable as drug carriers. Understanding the loading and unloading mechanisms, as exemplified in the study of gating mechanisms in Dps can help us understand how to fine tune the release of drugs in a tissue-specific manner (Williams et al. 2014). Also, having a better knowledge of multi-subunit assembly of the ferritin family proteins can help in building higher order assemblies (Williams et al. 2017).
References
Alaleona F, Franceschini S, Ceci P, Ilari A, Chiancone E (2010) Thermosynechococcus elongatus DpsA binds Zn(II) at a unique three histidine-containing ferroxidase center and utilizes O2 as iron oxidant with very high efficiency, unlike the typical Dps proteins. FEBS J 277(4):903–917. https://doi.org/10.1111/j.1742-4658.2009.07532.x
Ali Azam T, Iwata A, Nishimura A, Ueda S, Ishihama A (1999) Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J Bacteriol 181(20):6361–6370
Allen M, Willits D, Young M, Douglas T (2003) Constrained synthesis of cobalt oxide nanomaterials in the 12-subunit protein cage from Listeria innocua. Inorg Chem 42(20):6300–6305. https://doi.org/10.1021/ic0343657
Almiron M, Link AJ, Furlong D, Kolter R (1992) A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev 6(12B):2646–2654. https://doi.org/10.1101/gad.6.12b.2646
Altuvia S, Almiron M, Huisman G, Kolter R, Storz G (1994) The dps promoter is activated by OxyR during growth and by IHF and sigma S in stationary phase. Mol Microbiol 13(2):265–272. https://doi.org/10.1111/j.1365-2958.1994.tb00421.x
Andrews SC (2010) The Ferritin-like superfamily: Evolution of the biological iron storeman from a rubrerythrin-like ancestor. Biochim Biophys Acta 1800(8):691–705. https://doi.org/10.1016/j.bbagen.2010.05.010
Ardini M, Fiorillo A, Fittipaldi M, Stefanini S, Gatteschi D, Ilari A, Chiancone E (2013) Kineococcus radiotolerans Dps forms a heteronuclear Mn-Fe ferroxidase center that may explain the Mn-dependent protection against oxidative stress. Biochim Biophys Acta 1830(6):3745–3755. https://doi.org/10.1016/j.bbagen.2013.02.003
Arnold AR, Barton JK (2013) DNA protection by the bacterial ferritin Dps via DNA charge transport. J Am Chem Soc 135(42):15726–15729. https://doi.org/10.1021/ja408760w
Bellapadrona G, Ardini M, Ceci P, Stefanini S, Chiancone E (2010) Dps proteins prevent Fenton-mediated oxidative damage by trapping hydroxyl radicals within the protein shell. Free Radic Biol Med 48(2):292–297. https://doi.org/10.1016/j.freeradbiomed.2009.10.053
Bellapadrona G, Stefanini S, Zamparelli C, Theil EC, Chiancone E (2009) Iron translocation into and out of Listeria innocua Dps and size distribution of the protein-enclosed nanomineral are modulated by the electrostatic gradient at the 3-fold “ferritin-like” pores. J Biol Chem 284(28):19101–19109. https://doi.org/10.1074/jbc.M109.014670
Bhattacharyya G, Grove A (2007) The N-terminal extensions of Deinococcus radiodurans Dps-1 mediate DNA major groove interactions as well as assembly of the dodecamer. J Biol Chem 282(16):11921–11930. https://doi.org/10.1074/jbc.M611255200
Bozzi M, Mignogna G, Stefanini S, Barra D, Longhi C, Valenti P, Chiancone E (1997) A novel non-heme iron-binding ferritin related to the DNA-binding proteins of the Dps family in Listeria innocua. J Biol Chem 272(6):3259–3265. https://doi.org/10.1074/jbc.272.6.3259
Calhoun LN, Kwon YM (2011) Structure, function and regulation of the DNA-binding protein Dps and its role in acid and oxidative stress resistance in Escherichia coli: a review. J Appl Microbiol 110(2):375–386. https://doi.org/10.1111/j.1365-2672.2010.04890.x
Castruita M, Saito M, Schottel PC, Elmegreen LA, Myneni S, Stiefel EI, Morel FM (2006) Overexpression and characterization of an iron storage and DNA-binding Dps protein from Trichodesmium erythraeum. Appl Environ Microbiol 72(4):2918–2924. https://doi.org/10.1128/AEM.72.4.2918-2924.2006
Ceci P, Cellai S, Falvo E, Rivetti C, Rossi GL, Chiancone E (2004) DNA condensation and self-aggregation of Escherichia coli Dps are coupled phenomena related to the properties of the N-terminus. Nucleic Acids Res 32(19):5935–5944. https://doi.org/10.1093/nar/gkh915
Ceci P, Ilari A, Falvo E, Chiancone E (2003) The Dps protein of Agrobacterium tumefaciens does not bind to DNA but protects it toward oxidative cleavage: x-ray crystal structure, iron binding, and hydroxyl-radical scavenging properties. J Biol Chem 278(22):20319–20326. https://doi.org/10.1074/jbc.M302114200
Ceci P, Mangiarotti L, Rivetti C, Chiancone E (2007) The neutrophil-activating Dps protein of Helicobacter pylori, HP-NAP, adopts a mechanism different from Escherichia coli Dps to bind and condense DNA. Nucleic Acids Res 35(7):2247–2256. https://doi.org/10.1093/nar/gkm077
Chiancone E, Ceci P (2010) Role of Dps (DNA-binding proteins from starved cells) aggregation on DNA. Front Biosci (Landmark Ed) 15:122–131. https://doi.org/10.2741/3610
Chowdhury RP, Gupta S, Chatterji D (2007) Identification and characterization of the dps promoter of Mycobacterium smegmatis: promoter recognition by stress-specific extracytoplasmic function sigma factors sigmaH and sigmaF. J Bacteriol 189(24):8973–8981. https://doi.org/10.1128/JB.01222-07
Deans AE, Wadghiri YZ, Bernas LM, Yu X, Rutt BK, Turnbull DH (2006) Cellular MRI contrast via coexpression of transferrin receptor and ferritin. Magn Reson Med 56(1):51–59. https://doi.org/10.1002/mrm.20914
Durham KA, Bullerjahn GS (2002) Immunocytochemical localization of the stress-induced DpsA protein in the cyanobacterium Synechococcus sp. strain PCC 7942. J Basic Microbiol 42 (6):367–372. doi:https://doi.org/10.1002/1521-4028(200212)42:6<367::AID-JOBM367>3.0.CO;2-T
Facey PD, Hitchings MD, Saavedra-Garcia P, Fernandez-Martinez L, Dyson PJ, Del Sol R (2009) Streptomyces coelicolor Dps-like proteins: differential dual roles in response to stress during vegetative growth and in nucleoid condensation during reproductive cell division. Mol Microbiol 73(6):1186–1202. https://doi.org/10.1111/j.1365-2958.2009.06848.x
Fenton HJH (1894) LXXIII-Oxidation of tartaric acid in presence of iron. J Chem Soc Trans 65:899–910. https://doi.org/10.1039/ct8946500899
Franceschini S, Ceci P, Alaleona F, Chiancone E, Ilari A (2006) Antioxidant Dps protein from the thermophilic cyanobacterium Thermosynechococcus elongatus. FEBS J 273(21):4913–4928. https://doi.org/10.1111/j.1742-4658.2006.05490.x
Frenkiel-Krispin D, Levin-Zaidman S, Shimoni E, Wolf SG, Wachtel EJ, Arad T, Finkel SE, Kolter R, Minsky A (2001) Regulated phase transitions of bacterial chromatin: a non-enzymatic pathway for generic DNA protection. EMBO J 20(5):1184–1191. https://doi.org/10.1093/emboj/20.5.1184
Ganguly A, Rajdev P, Williams SM, Chatterji D (2012) Nonspecific interaction between DNA and protein allows for cooperativity: a case study with mycobacterium DNA binding protein. J Phys Chem B 116(1):621–632. https://doi.org/10.1021/jp209423n
Gao JL, Lu Y, Browne G, Yap BC, Trewhella J, Hunter N, Nguyen KA (2012) The role of heme binding by DNA-protective protein from starved cells (Dps) in the Tolerance of Porphyromonas gingivalis to heme toxicity. J Biol Chem 287(50):42243–42258. https://doi.org/10.1074/jbc.M112.392787
Gaupp R, Ledala N, Somerville GA (2012) Staphylococcal response to oxidative stress. Front Cell Infect Microbiol 2:33. https://doi.org/10.3389/fcimb.2012.00033
Ghatak P, Karmakar K, Kasetty S, Chatterji D (2011) Unveiling the role of Dps in the organization of mycobacterial nucleoid. PLoS ONE 6(1):e16019. https://doi.org/10.1371/journal.pone.0016019
Grant RA, Filman DJ, Finkel SE, Kolter R, Hogle JM (1998) The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat Struct Biol 5(4):294–303. https://doi.org/10.1038/nsb0498-294
Gupta S, Chatterji D (2003) Bimodal protection of DNA by Mycobacterium smegmatis DNA-binding protein from stationary phase cells. J Biol Chem 278(7):5235–5241. https://doi.org/10.1074/jbc.M208825200
Haikarainen T, Thanassoulas A, Stavros P, Nounesis G, Haataja S, Papageorgiou AC (2011) Structural and thermodynamic characterization of metal ion binding in Streptococcus suis Dpr. J Mol Biol 405(2):448–460. https://doi.org/10.1016/j.jmb.2010.10.058
Haikarainen T, Tsou CC, Wu JJ, Papageorgiou AC (2010) Structural characterization and biological implications of di-zinc binding in the ferroxidase center of Streptococcus pyogenes Dpr. Biochem Biophys Res Commun 398(3):361–365. https://doi.org/10.1016/j.bbrc.2010.06.071
Havukainen H, Haataja S, Kauko A, Pulliainen AT, Salminen A, Haikarainen T, Finne J, Papageorgiou AC (2008) Structural basis of the zinc- and terbium-mediated inhibition of ferroxidase activity in Dps ferritin-like proteins. Protein Sci 17(9):1513–1521. https://doi.org/10.1110/ps.036236.108
Hayden JA, Hendrich MP (2010) EPR spectroscopy and catalase activity of manganese-bound DNA-binding protein from nutrient starved cells. J Biol Inorg Chem 15(5):729–736. https://doi.org/10.1007/s00775-010-0640-3
He D, Marles-Wright J (2015) Ferritin family proteins and their use in bionanotechnology. N Biotechnol 32(6):651–657. https://doi.org/10.1016/j.nbt.2014.12.006
Hong Y, Wang G, Maier RJ (2006) Helicobacter hepaticus Dps protein plays an important role in protecting DNA from oxidative damage. Free Radic Res 40(6):597–605. https://doi.org/10.1080/10715760600618882
Howe C, Ho F, Nenninger A, Raleiras P, Stensjo K (2018) Differential biochemical properties of three canonical Dps proteins from the cyanobacterium Nostoc punctiforme suggest distinct cellular functions. J Biol Chem 293(43):16635–16646. https://doi.org/10.1074/jbc.RA118.002425
Ilari A, Stefanini S, Chiancone E, Tsernoglou D (2000) The dodecameric ferritin from Listeria innocua contains a novel intersubunit iron-binding site. Nat Struct Biol 7(1):38–43. https://doi.org/10.1038/71236
Ishikawa T, Mizunoe Y, Kawabata S, Takade A, Harada M, Wai SN, Yoshida S (2003) The iron-binding protein Dps confers hydrogen peroxide stress resistance to Campylobacter jejuni. J Bacteriol 185(3):1010–1017. https://doi.org/10.1128/jb.185.3.1010-1017.2003
Janissen R, Arens MMA, Vtyurina NN, Rivai Z, Sunday ND, Eslami-Mossallam B, Gritsenko AA, Laan L, de Ridder D, Artsimovitch I, Dekker NH, Abbondanzieri EA, Meyer AS (2018) Global DNA Compaction in Stationary-Phase Bacteria Does Not Affect Transcription. Cell 174 (5):1188–1199 e1114. doi:https://doi.org/10.1016/j.cell.2018.06.049
Jeong KC, Hung KF, Baumler DJ, Byrd JJ, Kaspar CW (2008) Acid stress damage of DNA is prevented by Dps binding in Escherichia coli O157:H7. BMC Microbiol 8:181. https://doi.org/10.1186/1471-2180-8-181
Kamyshinsky R, Chesnokov Y, Dadinova L, Mozhaev A, Orlov I, Petoukhov M, Orekhov A, Shtykova E, Vasiliev A (2019) Polymorphic Protective Dps-DNA Co-Crystals by Cryo Electron Tomography and Small Angle X-Ray Scattering. Biomolecules 10 (1). doi:https://doi.org/10.3390/biom10010039
Kanekiyo M, Wei CJ, Yassine HM, McTamney PM, Boyington JC, Whittle JR, Rao SS, Kong WP, Wang L, Nabel GJ (2013) Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 499(7456):102–106. https://doi.org/10.1038/nature12202
Kauko A, Pulliainen AT, Haataja S, Meyer-Klaucke W, Finne J, Papageorgiou AC (2006) Iron incorporation in Streptococcus suis Dps-like peroxide resistance protein Dpr requires mobility in the ferroxidase center and leads to the formation of a ferrihydrite-like core. J Mol Biol 364(1):97–109. https://doi.org/10.1016/j.jmb.2006.08.061
Kim H-J, Oh E, Lee J et al (2011) Synthesis of carbon nanotubes with catalytic iron-containing proteins. Carbon 49(12):3717–3722. https://doi.org/10.1016/j.carbon.2011.04.037
Kottakis F, Papadopoulos G, Pappa EV, Cordopatis P, Pentas S, Choli-Papadopoulou T (2008) Helicobacter pylori neutrophil-activating protein activates neutrophils by its C-terminal region even without dodecamer formation, which is a prerequisite for DNA protection–novel approaches against Helicobacter pylori inflammation. FEBS J 275(2):302–317. https://doi.org/10.1111/j.1742-4658.2007.06201.x
Li X, Pal U, Ramamoorthi N, Liu X, Desrosiers DC, Eggers CH, Anderson JF, Radolf JD, Fikrig E (2007) The Lyme disease agent Borrelia burgdorferi requires BB0690, a Dps homologue, to persist within ticks. Mol Microbiol 63(3):694–710. https://doi.org/10.1111/j.1365-2958.2006.05550.x
Linder MC (2013) Mobilization of stored iron in mammals: a review. Nutrients 5(10):4022–4050. https://doi.org/10.3390/nu5104022
Luijsterburg MS, Noom MC, Wuite GJ, Dame RT (2006) The architectural role of nucleoid-associated proteins in the organization of bacterial chromatin: a molecular perspective. J Struct Biol 156(2):262–272. https://doi.org/10.1016/j.jsb.2006.05.006
Maham A, Tang Z, Wu H, Wang J, Lin Y (2009) Protein-based nanomedicine platforms for drug delivery. Small 5(15):1706–1721. https://doi.org/10.1002/smll.200801602
Maham A, Wu H, Wang J, Kang X, Zhang Y, Lin Y (2011) Apoferritin-based nanomedicine platform for drug delivery: equilibrium binding study of daunomycin with DNA. J Mater Chem 21:8700. https://doi.org/10.1039/c0jm04321d
Martinez A, Kolter R (1997) Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J Bacteriol 179(16):5188–5194. https://doi.org/10.1128/jb.179.16.5188-5194.1997
Morikawa K, Ohniwa RL, Kim J, Maruyama A, Ohta T, Takeyasu K (2006) Bacterial nucleoid dynamics: oxidative stress response in Staphylococcus aureus. Genes Cells 11(4):409–423. https://doi.org/10.1111/j.1365-2443.2006.00949.x
Nair S, Finkel SE (2004) Dps protects cells against multiple stresses during stationary phase. J Bacteriol 186(13):4192–4198. https://doi.org/10.1128/JB.186.13.4192-4198.2004
Narayan OP, Kumari N, Rai LC (2010) Heterologous expression of Anabaena PCC 7120 all3940 (a Dps family gene) protects Escherichia coli from nutrient limitation and abiotic stresses. Biochem Biophys Res Commun 394(1):163–169. https://doi.org/10.1016/j.bbrc.2010.02.135
Nicodeme M, Perrin C, Hols P, Bracquart P, Gaillard JL (2004) Identification of an iron-binding protein of the Dps family expressed by Streptococcus thermophilus. Curr Microbiol 48(1):51–56. https://doi.org/10.1007/s00284-003-4116-3
Okuda M, Kobayashi Y, Suzuki K, Sonoda K, Kondoh T, Wagawa A, Kondo A, Yoshimura H (2005) Self-organized inorganic nanoparticle arrays on protein lattices. Nano Lett 5(5):991–993. https://doi.org/10.1021/nl050556q
Pacello F, Ceci P, Ammendola S, Pasquali P, Chiancone E, Battistoni A (2008) Periplasmic Cu, Zn superoxide dismutase and cytoplasmic Dps concur in protecting Salmonella enterica serovar Typhimurium from extracellular reactive oxygen species. Biochim Biophys Acta 1780(2):226–232. https://doi.org/10.1016/j.bbagen.2007.12.001
Papinutto E, Dundon WG, Pitulis N, Battistutta R, Montecucco C, Zanotti G (2002) Structure of two iron-binding proteins from Bacillus anthracis. J Biol Chem 277(17):15093–15098. https://doi.org/10.1074/jbc.M112378200
Park M, Yun ST, Hwang SY, Chun CI, Ahn TI (2006) The dps gene of symbiotic “Candidatus Legionella jeonii” in Amoeba proteus responds to hydrogen peroxide and phagocytosis. J Bacteriol 188(21):7572–7580. https://doi.org/10.1128/JB.00576-06
Pena MM, Bullerjahn GS (1995) The DpsA protein of Synechococcus sp. Strain PCC7942 is a DNA-binding hemoprotein. Linkage of the Dps and bacterioferritin protein families. J Biol Chem 270 (38):22478–22482. doi:https://doi.org/10.1074/jbc.270.38.22478
Pesek J, Buchler R, Albrecht R, Boland W, Zeth K (2011) Structure and mechanism of iron translocation by a Dps protein from Microbacterium arborescens. J Biol Chem 286(40):34872–34882. https://doi.org/10.1074/jbc.M111.246108
Pulliainen AT, Kauko A, Haataja S, Papageorgiou AC, Finne J (2005) Dps/Dpr ferritin-like protein: insights into the mechanism of iron incorporation and evidence for a central role in cellular iron homeostasis in Streptococcus suis. Mol Microbiol 57(4):1086–1100. https://doi.org/10.1111/j.1365-2958.2005.04756.x
Ren B, Tibbelin G, Kajino T, Asami O, Ladenstein R (2003) The multi-layered structure of Dps with a novel di-nuclear ferroxidase center. J Mol Biol 329(3):467–477. https://doi.org/10.1016/s0022-2836(03)00466-2
Reon BJ, Nguyen KH, Bhattacharyya G, Grove A (2012) Functional comparison of Deinococcus radiodurans Dps proteins suggests distinct in vivo roles. Biochem J 447(3):381–391. https://doi.org/10.1042/BJ20120902
Rocha ER, Herren CD, Smalley DJ, Smith CJ (2003) The complex oxidative stress response of Bacteroides fragilis: the role of OxyR in control of gene expression. Anaerobe 9(4):165–173. https://doi.org/10.1016/S1075-9964(03)00118-5
Roy S, Saraswathi R, Gupta S, Sekar K, Chatterji D, Vijayan M (2007) Role of N and C-terminal tails in DNA binding and assembly in Dps: structural studies of Mycobacterium smegmatis Dps deletion mutants. J Mol Biol 370(4):752–767. https://doi.org/10.1016/j.jmb.2007.05.004
Santos SP, Mitchell EP, Franquelim HG, Castanho MA, Abreu IA, Romao CV (2015) Dps from Deinococcus radiodurans: oligomeric forms of Dps1 with distinct cellular functions and Dps2 involved in metal storage. FEBS J 282(22):4307–4327. https://doi.org/10.1111/febs.13420
Schmidt R, Zahn R, Bukau B, Mogk A (2009) ClpS is the recognition component for Escherichia coli substrates of the N-end rule degradation pathway. Mol Microbiol 72(2):506–517. https://doi.org/10.1111/j.1365-2958.2009.06666.x
Schwartz JK, Liu XS, Tosha T, Diebold A, Theil EC, Solomon EI (2010) CD and MCD spectroscopic studies of the two Dps miniferritin proteins from Bacillus anthracis: role of O2 and H2O2 substrates in reactivity of the diiron catalytic centers. Biochemistry 49(49):10516–10525. https://doi.org/10.1021/bi101346c
Shu JC, Soo PC, Chen JC, Hsu SH, Chen LC, Chen CY, Liang SH, Buu LM, Chen CC (2013) Differential regulation and activity against oxidative stress of Dps proteins in Bacillus cereus. Int J Med Microbiol 303(8):662–673. https://doi.org/10.1016/j.ijmm.2013.09.011
Stefanini S, Cavallo S, Montagnini B, Chiancone E (1999) Incorporation of iron by the unusual dodecameric ferritin from Listeria innocua. Biochem J 338(Pt 1):71–75
Stephani K, Weichart D, Hengge R (2003) Dynamic control of Dps protein levels by ClpXP and ClpAP proteases in Escherichia coli. Mol Microbiol 49(6):1605–1614. https://doi.org/10.1046/j.1365-2958.2003.03644.x
Stillman TJ, Upadhyay M, Norte VA, Sedelnikova SE, Carradus M, Tzokov S, Bullough PA, Shearman CA, Gasson MJ, Williams CH, Artymiuk PJ, Green J (2005) The crystal structures of Lactococcus lactis MG1363 Dps proteins reveal the presence of an N-terminal helix that is required for DNA binding. Mol Microbiol 57(4):1101–1112. https://doi.org/10.1111/j.1365-2958.2005.04757.x
Su M, Cavallo S, Stefanini S, Chiancone E, Chasteen ND (2005) The so-called Listeria innocua ferritin is a Dps protein. Iron incorporation, detoxification, and DNA protection properties. Biochemistry 44 (15):5572–5578. doi:https://doi.org/10.1021/bi0472705
Swift J, Wehbi WA, Kelly BD, Stowell XF, Saven JG, Dmochowski IJ (2006) Design of functional ferritin-like proteins with hydrophobic cavities. J Am Chem Soc 128(20):6611–6619. https://doi.org/10.1021/ja057069x
Theoret JR, Cooper KK, Zekarias B, Roland KL, Law BF, Curtiss R 3rd, Joens LA (2012) The Campylobacter jejuni Dps homologue is important for in vitro biofilm formation and cecal colonization of poultry and may serve as a protective antigen for vaccination. Clin Vaccine Immunol 19(9):1426–1431. https://doi.org/10.1128/CVI.00151-12
Travers A, Muskhelishvili G (2005) Bacterial chromatin. Curr Opin Genet Dev 15(5):507–514. https://doi.org/10.1016/j.gde.2005.08.006
Tsou CC, Chiang-Ni C, Lin YS, Chuang WJ, Lin MT, Liu CC, Wu JJ (2008) An iron-binding protein, Dpr, decreases hydrogen peroxide stress and protects Streptococcus pyogenes against multiple stresses. Infect Immun 76(9):4038–4045. https://doi.org/10.1128/IAI.00477-08
Verma D, Gulati N, Kaul S, Mukherjee S, Nagaich U (2018) Protein Based Nanostructures for Drug Delivery. J Pharm (Cairo) 2018:9285854. https://doi.org/10.1155/2018/9285854
Vtyurina NN, Dulin D, Docter MW, Meyer AS, Dekker NH, Abbondanzieri EA (2016) Hysteresis in DNA compaction by Dps is described by an Ising model. Proc Natl Acad Sci U S a 113(18):4982–4987. https://doi.org/10.1073/pnas.1521241113
Wasilewski S, Karelina D, Berriman JA, Rosenthal PB (2012) Automatic magnification determination of electron cryomicroscopy images using apoferritin as a standard. J Struct Biol 180(1):243–248. https://doi.org/10.1016/j.jsb.2012.07.006
Williams SM, Chatterji D (2017) Flexible aspartates propel iron to the ferroxidation sites along pathways stabilized by a conserved arginine in Dps proteins from Mycobacterium smegmatis. Metallomics 9(6):685–698. https://doi.org/10.1039/c7mt00008a
Williams SM, Chandran AV, Prakash S, Vijayan M, Chatterji D (2017) A Mutation Directs the Structural Switch of DNA Binding Proteins under Starvation to a Ferritin-like Protein Cage. Structure 25 (9):1449–1454 e1443. doi:https://doi.org/10.1016/j.str.2017.07.006
Williams SM, Chandran AV, Vijayabaskar MS, Roy S, Balaram H, Vishveshwara S, Vijayan M, Chatterji D (2014) A histidine aspartate ionic lock gates the iron passage in miniferritins from Mycobacterium smegmatis. J Biol Chem 289(16):11042–11058. https://doi.org/10.1074/jbc.M113.524421
Wolf SG, Frenkiel D, Arad T, Finkel SE, Kolter R, Minsky A (1999) DNA protection by stress-induced biocrystallization. Nature 400(6739):83–85. https://doi.org/10.1038/21918
Yamamoto Y, Poole LB, Hantgan RR, Kamio Y (2002) An iron-binding protein, Dpr, from Streptococcus mutans prevents iron-dependent hydroxyl radical formation in vitro. J Bacteriol 184(11):2931–2939. https://doi.org/10.1128/jb.184.11.2931-2939.2002
Yokoyama H, Fujii S (2014) Structures and metal-binding properties of Helicobacter pylori neutrophil-activating protein with a di-nuclear ferroxidase center. Biomolecules 4(3):600–615. https://doi.org/10.3390/biom4030600
Yoshimura H (2006) Protein-assisted nanoparticle synthesis. Colloids Surf a Physicochem Eng Asp 282–283:464–470. https://doi.org/10.1016/j.colsurfa.2006.01.037
Yu MJ, Ren J, Zeng YL, Zhou SN, Lu YJ (2009) The Legionella pneumophila Dps homolog is regulated by iron and involved in multiple stress tolerance. J Basic Microbiol 49(Suppl 1):S79-86. https://doi.org/10.1002/jobm.200800357
Zeth K, Offermann S, Essen LO, Oesterhelt D (2004) Iron-oxo clusters biomineralizing on protein surfaces: structural analysis of Halobacterium salinarum DpsA in its low- and high-iron states. Proc Natl Acad Sci U S a 101(38):13780–13785. https://doi.org/10.1073/pnas.0401821101
Zhao G, Ceci P, Ilari A, Giangiacomo L, Laue TM, Chiancone E, Chasteen ND (2002) Iron and hydrogen peroxide detoxification properties of DNA-binding protein from starved cells. A ferritin-like DNA-binding protein of Escherichia coli. J Biol Chem 277 (31):27689–27696. doi:https://doi.org/10.1074/jbc.M202094200
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Williams, S.M., Chatterji, D. (2021). An Overview of Dps: Dual Acting Nanovehicles in Prokaryotes with DNA Binding and Ferroxidation Properties. In: Harris, J.R., Marles-Wright, J. (eds) Macromolecular Protein Complexes III: Structure and Function. Subcellular Biochemistry, vol 96. Springer, Cham. https://doi.org/10.1007/978-3-030-58971-4_3
Download citation
DOI: https://doi.org/10.1007/978-3-030-58971-4_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-58970-7
Online ISBN: 978-3-030-58971-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)