Abstract
Anthrax toxin is a major virulence factor of Bacillus anthracis, a Gram-positive bacterium which can form highly stable spores that are the causative agents of the disease, anthrax. While chiefly a disease of livestock, spores can be “weaponized” as a bio-terrorist agent, and can be deadly if not recognized and treated early with antibiotics. The intracellular pathways affected by the enzymes are broadly understood and are not discussed here. This chapter focuses on what is known about the assembly of secreted toxins on the host cell surface and how the toxin is delivered into the cytosol. The central component is the “Protective Antigen”, which self-oligomerizes and forms complexes with its pay-load, either Lethal Factor or Edema Factor. It binds a host receptor, CMG2, or a close relative, triggering receptor-mediated endocytosis, and forms a remarkably elegant yet powerful machine that delivers toxic enzymes into the cytosol, powered only by the pH gradient across the membrane. We now have atomic structures of most of the starting, intermediate and final assemblies in the infectious process. Together with a major body of biophysical, mutational and biochemical work, these studies reveal a remarkable story of both how toxin assembly is choreographed in time and space.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Anthrax
- Pathogenesis
- Structure
- Macromolecular assembly
- Conformational change
- Protein translocation
- pH trigger
Introduction
Anthrax toxin is a major virulence factor of Bacillus anthracis, a Gram-positive, non-motile, rod-shaped, encapsulated bacterium, which can also form highly stable spores that are the causative agents of the disease, anthrax. Disease begins by attachment of spores to mucosa-associated lymphoid tissues of mammals, enabling spore germination and the formation of replication-competent “vegetative cells”, initially replicating in the lymphatic system, before migrating to the vasculature, ultimately reaching very high levels that are often lethal if not diagnosed at an early stage (reviewed in Moayeri and Leppla 2004, 2009; Frankel et al. 2009). While chiefly a disease of livestock, spores can be “weaponized” as a bio-terrorist agent.
Two extrachromosomal plasmids, pX01 and pX02, encode the major pathogenic determinants of Bacillus anthracis—the tripartite Anthrax Toxin (Larson et al. 1988) and a poly(γ-D)glutamate capsule—which distinguish it from its harmless relative, Bacillus cereus. The capsule acts as a physical barrier as well as a “cloaking device” against immune surveillance [reviewed in (Moayeri et al. 2015)]. Anthrax Toxin plays critical roles in both early (stifling the innate immune response) and late (destruction of the host vasculature) stages of infection. The intracellular pathways affected by the enzymes are broadly understood, although the details are dependent on host/strain, cell type and disease progression; these are discussed elsewhere [see (Moayeri et al. 2015) and references therein]. This chapter focuses on what we know about the assembly of secreted toxins on the host cell surface and how the toxin is delivered into the cytosol.
The central component of Anthrax Toxin is the “Protective Antigen” (PA) (so-called because it is the major component of anthrax vaccines Gu et al. 1999; Leppla et al. 2002), which self-oligomerizes and forms complexes with its pay-load, either or both of two secreted toxic enzymes, the Lethal Factor (LF) and Edema Factor (EF), all encoded on the pX01 plasmid (Singh et al. 1999). PA also binds its host cell receptor, CMG2 (Bradley et al. 2001) or a close relative (van der Goot and Young 2009), triggering clathrin-dependent receptor-mediated endocytic (Abrami et al. 2003), and ultimately forms a remarkably elegant yet powerful machine that delivers the toxic enzymes into the cytosol, powered only by the pH gradient across the endosomal-cytosolic membrane (reviewed in Young and Collier 2007).
We now have crystal and EM structures of nearly all of the starting, intermediate and final structures in the infectious process, most at atomic or near-atomic resolution. Together with a major body of biophysical, mutational and biochemical work, these studies reveal a remarkable story of both how the toxin assembles, and how assembly, and thus, pathogenesis, may be choreographed in time and space. Here, I will focus on the facts, but identify features of the story that remain unresolved or ambiguous, and which could form the basis for future research.
Anthrax Protective Antigen and Prepore Formation
PA is secreted by B. anthracis as a water-soluble 83 kDa monomeric pro-protein, “PA83”. It is organized into 4 domains (“D1”–“D4”) (Petosa et al. 1997) (Fig. 18.1), and is fully competent to bind its host receptor, CMG2 (see below). However, PA83 cannot oligomerize to form a water-soluble prepore owing to steric hindrance between N-terminal segments of D1 (Petosa et al. 1997). Instead, a host cell-surface furin-like protease must first cleave a loop between two β-strands within the major sheet of D1 (after residue Arg262), creating N-terminal (PA21) and C-terminal (PA63) fragments (Molloy et al. 1992; Gordon et al. 1995). But physical separation of these fragments requires rupture of the major β-sheet of D1. This process is slow in vitro, but can be achieved by passage of “nicked” PA83 over an ion-exchange column, in which case a heptameric species (PA63)7, is the principal product. However, when an excess of LF or EF is included, a functional octameric species, (PA63)8, predominates (Kintzer et al. 2009), that may play a dominant role at late stages of infection. Crystal structures of both oligomers have been determined, and they are functionally and structurally very similar (the difference in packing angle between PA63 protomers in the two oligomers is only ~6°, which can be readily accommodated by small sidechain adjustments), and for the sake of brevity, the following discussion will generally refer to the heptameric species.
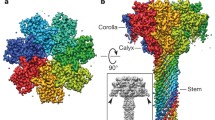
Structure and sequence of PA in its monomeric and prepore forms. a. Structure of full-length PA, with secondary structure colored by domain; except for D2, for which the residues destined to form the central barrel of the pore are colored yellow. A black dashed line indicates where the main sheet of D1 must be torn apart to create PA21 and PA63 following cleavage or “nicking” by furin. The two black balls are Ca2+ ions. b. Sequence of PA83 colored as in a, with secondary structure elements labeled. Residues coordinating the two Ca2+ ions are highlighted in rectangular boxes; non-acidic residues coordinate via their main-chain carbonyls. Orange highlights indicate residues that bridge both Ca2+ ions. c-e Surface representations of the heptameric prepore, colored as in a. c is a side view showing that D1′ is membrane distal and D4 membrane proximal. d is a “top” view revealing the annulus formed by interlocking D1′ domains. D2 is just evident along the channel lumen. E. is a view form the bottom, i.e. the membrane-proximal face. The surface comprises exclusively D2 domains lining the channel lumen, and D4 domains on the periphery
Prepore Structure
A low resolution (4.5 Å resolution) crystal structure of the heptameric (PA63)7 prepore was first described in 1997 (Petosa et al. 1997) (Fig. 18.1c–e). Despite the limited resolution, unbiased phasing (utilizing the heptamer’s non-crystallographic symmetry) demonstrated convincingly that oligomerization comprised essentially a rigid body assembly of PA63 protomers, with a modest shift in one surface loop implicated in LF binding. Later high-resolution (2.8 Å) structures of the heptamer (Lacy et al. 2004) and octamer (Feld et al. 2010) confirmed these features.
The PA63 protomers lie with their longest axes running roughly parallel to the heptamer axis (Fig. 18.1c). The “upper” (i.e., membrane-distal) part of the heptamer is formed by tight self-association of 7 D1′ domains, which form an annulus that defines the top of the prepore (Fig. 18.1d). D3 domains lie just beneath the D1′ domains; they make no contact with each other, but insert one helix (residues 512–517) into the D1′ domain of their counterclockwise neighbor, propping up its surface-exposed β-hairpin (1β12–1β13) by ~5 Å—the largest conformational change upon heptamer formation. D1′ is also stabilized by a pair of Ca2+ ions which share 3 coordinating Asp/Glu ligands, lying just beneath the newly exposed cleaved face of D1′, where they presumably help to stabilize the vestigial “half-domains” (Fig. 18.1a). The D2 domains pack around the heptamer axis and extend along most of its length, tightly to each other (and to the D1′ domains) at the top, but more loosely at the base. The prepore lumen has a tightest constriction of ~20 Å in diameter, near the top of the D2 domains. At the base of heptamer, only 2 domains form the lower (membrane-proximal) surface: the D2 annulus near the center and the D4 domains, which lie distal to the axis. D4 domains make no contacts with each other, but make several important contacts with D2 of their own protomer (see below), as well as forming a cage with D2 of the clockwise protomer which provides a safe haven for the extended protease-sensitive pro-hairpin loop of D2 (Fig. 18.1a), This protection may be critical, since cleavage of one or more of these 7 loops enables prepore but not functional pore formation (Singh et al. 1994).
Nature and Role of the Host Receptor, CMG2
CMG2 is abundant on the surfaces of cells of the innate immune system, as well as endothelial cells lining blood vessel walls, and it is highly conserved among the mammalian targets of B. anthracis. CMG2 has two extracellular domains of similar size, and PA engages the second (membrane distal) of these, one that is highly homologous to integrin “I domains”. Crystal structures of monomeric PA83 bound to a single copy of the CMG2 “I domain” (Santelli et al. 2004) as well as a lower resolution structure of heptameric (PA63)7 bound to 7 molecules of CMG2 display identical interactions (see Fig. 18.6). CMG2 forms a metal-mediated bond to an Asp residue on D4 via its “MIDAS” motif (Emsley et al. 2000), providing a remarkable mimic of an integrin-matrix complex. The interaction site on PA D4 was predicted by mutagenesis (Varughese et al. 1999; Rosovitz et al. 2003); this domain is the most variable of the 4 domains in PA orthologs from other species (which presumably utilize different receptors) (Figs. 18.2, 18.3, 18.4 and 18.5).
LFN bound to a dimeric PA species. a. Two PA protomers are shown as green and cyan surfaces, LFN as yellow cartoon except for the N-terminal segments. In the crystal structure of LFN bound to oligomeric PA63, the α1 helix undergoes a dramatic shift (~40 Å) compared with the structure of the free enzyme, settling into a shallow pocket called the “α-clamp”. The unraveling of the β1 strand and tilting of the α2 helix are consequences of this movement, which positions α1 ~45 Å above the region that will become the Φ-clamp. There are no other significant differences. b. A hypothetical model generated in silico in which the PA21 element of the cyan protomer (counterclockwise from above) has been added by simple overlay of full-length PA onto PA63. PA21 binds without major steric clashes, but only to the “native” conformation of LFN; that is, PA21 binding is predicted to inhibit the first step in LFN activation
Three molecules of full-length LF bound to the PA63 heptamer, derived by cryoEM. a and b Orthogonal views of the hetero-oligomer, with atomic models derived by crystallography fitted into the EM envelope. a The “top” view illustrates that the C-terminal domains are suspended up to 30 Å above the heptamer surface, creating large holes in the complex. b The “side” view illustrates contact points between the N- and C-termini of consecutive LF molecules which may stabilize the heptamer (shown by black arrows). Since LF binds to two consecutive PA protomers, the odd number of protomers creates a “free” N-terminus (N) and an “orphan” PA protomer (O)
Overview of the transition from monomeric PA to the pore conformation. a Monomeric PA oriented so that the invariant D1′ and D3 match those in the pore conformation. An arrows indicates a major point of conformational change: the unraveling of the long b-hairpin (in yellow). b The monomeric pore conformation. The arrow indicates the second point of change: the rotation of the D2 barrel, such that it adopts an upright position, enabling seven protomers to pack tightly across the symmetry axis. D4 is not shown, because it is mobile once the binds to D2 have been broken. c The result of these two reorganizations in the context of the heptamer is the pore conformation. d Overlay of the prepore and pore heptamers, viewed from the “top”. Both the D1′ and D3 domains are essentially invariant, as are the first hairpin and helix of D2, in contrast to the major changes observed in the barrel of D2
A more detailed view of conformation changes on pore formation. At left, a comparison of the PA63 protomers from the pore and prepore, overlaid on the invariant D1′ and D3 domains. The pro-β-barrel segments have been removed to reveal the inward rotation of the D2 domain upon pore (shown in cyan) formation, as well as the stabilization of the loop carrying F427, seven of which coalesce to the form the Φ-clamp (shown at right), which is surrounded by a ring of 14 acidic residues, as shown. The view looks down the axis of the central β-barrel
The bigger surprise was that CMG2 also engages domain D2 of PA, in a region destined to become (the upper) part of the membrane-spanning β-barrel. Thus, by stabilizing this region of D2, as well as cross-linking it to D4, CMG2 acts as a clamp that inhibits conformational changes required for prepore to pore conversion. Indeed, the pH threshold for membrane insertion is reduced from pH 6.0 to 5.0 in the presence of CMG2, suggesting that the receptor discourages premature insertion of the pore into the plasma (Abrami et al. 2004). Pore formation occurs at an earlier point during endosome maturation than protein translocation, which occurs only in late endocytic compartments (Abrami et al. 2004), consistent with the lower pH requirement for the second process. That CMG2 remains bound to PA through the steps of receptor-mediated endocytosis, pore formation and protein translocation (Pilpa et al. 2011), plays a role in the timing of pore formation, and may provide a physical support that holds pore at the correct height for membrane insertion, suggests that it should be considered an integral part of toxin translocation machine.
Loading of the Toxic Enzymes and Insights into Prepore Assembly
A high-resolution crystal structure of the octameric prepore bound to four copies of the N-terminal domain of LF (LFN) has been obtained (Feld et al. 2010), as well as a lower resolution EM structure of the heptameric prepore bound to 3 copies of full-length LF (Fabre et al. 2016). These stoichiometries are consistent with prior mutational and biochemical studies by Collier and co-workers, which showed that the footprint of each LF molecule spans two adjacent protomers of PA (Mogridge et al. 2002). Moreover, their experiments demonstrated that LF did not bind stably to a mutant “nicked” PA83 that could only form monomers; that nicked PA83 did not form dimers; but that mutant pairs with the potential to form only dimers formed a stable complex, a ternary (PA63)2LF entity. In a separate set of experiments, it was concluded that PA oligomerization is driven by dimeric (toxin-laden) receptor intermediates on the cell surface (Kintzer et al. 2010). So if (CMG2)2(PA63)2LF is indeed a stable assembly intermediate for wild-type PA, then it immediately suggests how loading of toxic enzymes onto PA is linked to cell entry. In this scenario, a PA83 monomer binds a receptor and is then nicked by a cell-surface protease; this can now binds LF, which promotes dimerization with a second PA-laden receptor, which in turn promotes oligomer formation and endocytosis.
In fact, the crystal structures reveal that the footprints of LF and PA21 on PA63 are almost distinct (i.e., there is only a small degree of overlap between them), which would provide a low energy kinetic pathway for LF to rapidly displace PA21, and thereby ensure that unladen PA oligomers are not (wastefully) endocytosed. The ternary intermediate, which should be favored by high concentrations of toxic enzyme, also provides a satisfactory rationale for the existence of the octameric species (via the coalescence of four (PA63)2LF intermediates).
Structures of Toxin-Laden PA
In the crystal structure of (PA63)8(LFN)4 (Feld et al. 2010), LFN perches on the outer rim of the heptamer formed by the exposed β-strands of PA D1′, and does indeed straddle two PA63 protomers. The major surprise was a major refolding of the N-terminal β-strand and helix (H1) of LFN to create a new interface (in the counterclockwise protomer). H1 sits in a new site (the “α-clamp”), promoting a “translocation-primed” conformation (Fig. 18.2a), which involves a movement of ~40 Å of helix 1α1 of LFN. Upstream of 1α1 are a further 30 residues that are disordered in the structure.
The structure of the PA63 heptamer bound to three full-length LF molecules (Fabre et al. 2016) confirmed the binding mode to the heptamer, which is mediated entirely by the N-terminal domains. The three LF molecules bind in the conformation observed in the free molecules; i.e., they dock essentially as essentially as rigid bodies. Intriguingly, the C-terminal tails bind to an N-terminal domain of their clockwise neighbors, most likely stabilizing the overall assembly, with the C-terminal domains suspended high (30–40 Å) above the heptamer. The odd number of protomers results in a single “orphan” PA63 that makes no contacts with LF, so that the arrangement creates a large head-to-tail unclosed “horseshoe”, with three large holes in the structure (Fig. 18.3).
More recently, structures of full-length EF (which utilizes a homologous domain, EFN) bound to the pore have been determined by cryoEM at medium resolution (Zhou et al. 2020). These structures confirm that EFN binds in an identical fashion as LFN binds to the prepore. This was expected, but also confirms that the prepore to pore transition, per se, does not trigger a change in binding of the toxic enzymes. One surprise was that the C-terminal domains of EF are oriented in the opposite (counterclockwise) direction to those of LF, but still make stabilizing contacts with the neighboring EFN domain (but on the opposite face). The structures are consistent with a maximum of 3 molecules of EF bound to heptameric pore/prepore,Footnote 1 as well as mixed EF/LF complexes (Pimental et al. 2004).
A Potential Role for PA83/PA21 in Toxin Assembly
There is tantalizing evidence for a role for PA83 (and thus PA21), either nicked or intact, in toxin assembly and prepore stability in some instances. For example, one report provides evidence for “mixed” heptamers containing both PA63 and PA83 that form functional pores in vivo when furin concentrations are limiting (Chekanov et al. 2006), and another report indicates that PA21 binds directly to LF in vitro (Chvyrkova et al. 2007). Indeed, simple in silico rigid-body modeling suggests that, while a single cleavage event must occur in PA to create an LF binding site, there are no steric impediments to binding of PA83 on either the clockwise or counterclockwise sides of the PA63:LF entity. In the latter case (which would create a ternary PA63:LF:PA83 moiety similar to the PA63:LF:PA63 assembly intermediate described above), modeling predicts that the PA21 moiety of PA83 would fit snugly against the N-terminal domain of LF bound to PA63, as well as providing support for LF’s C-terminal domains (and plugging the holes in the heptameric complex). However, binding can occur only to the native conformation of LF, since PA21 occupies the space that includes the α-clamp (Fig. 18.2b). Thus, PA83 (either intact or nicked) could provide additional stabilization for the ternary intermediate as well as the final oligomer, at the same time inhibiting the first step of LF/EF activation (and thus the timing of translocation) in the context of the fully-laden PA heptamer. Subsequent proteolytic/ejection events would be necessary to activate the toxin-laden pore. While speculative, these issues should be readily testable.
The Prepore-Pore Transition
The general nature of this remarkable conformational switch was predicted by Dr. Carlo Petosa more than 20 years ago (Petosa et al. 1997), and was subsequently supported by mutational, biophysical and, ultimately, a high resolution structure of the pore (Jiang et al. 2015).
The structure of the PA63 heptameric pore was determined by single particle cryoEM methods in the presence of lipids at a resolution of 3.5 Å, sufficient for a complete trace of the polypeptide chain (Jiang et al. 2015) (Fig. 18.4c). Remarkably, the structure revealed that conformational changes are almost entirely restricted to D2 (although D4, once freed from its shackles to D2, becomes quite mobile, but still retains its MIDAS bond to the receptor). In contrast, D1′ and D3 remain essentially static: thus, an overlay of all Cα atoms within D1′ and D3 (as well as the connecting elements of D2, the N-terminal hairpin-helix segment) of the prepore and pore gives an RMS difference (for the entire heptameric annulus) of 0.68 Å, well within experimental error (Fig. 18.4d). In fact, this D1′-D3 annulus maintains the integrity of its docking platforms for the toxic enzymes throughout the translocation process.
The residues destined to form the membrane-penetrating portion of the central β-barrel comprise the mobile protease sensitive loop (residues 298–327). However, they lie at the top (i.e. membrane-distal) of the two flanking strands (2β2 and 2β3) which will form the upper part of the central β-barrel. These two strands must therefore first must first peel away from their parent domain, breaking countless intradomain H-bonds in the process. And in order for this to happen, D2 must first break its bonds with D4 and with the receptor.
What triggers these large reorganizations remains a matter for debate (see below). But what occurs is clear. The 2β2-2β3 hairpin lies at one end of the D2 barrel (distal to the heptamer axis. Its loss has little effect on the remainder of the D2′ barrel, which rotates essentially as a rigid body ~30° “inward” toward the heptamer axis. Only the final β-hairpin and helix of D2, which pack against D1′, remain static during this process. The rotation has two important effects.
First, the rotation brings the ends of strands 2β1 and 2β4 (which “hold” the ends of new β-hairpin) ~12 Å closer to the heptamer axis (and ~20 Å closer to their symmetry-related counterparts). This closure enables the 7 long β-hairpins to associate into a 14-stranded β-barrel ~100 Å in length, similar in nature to that found in hemolysin, but roughly twice as long (Fig. 18.4c).
Second, the rotation brings the seven D2′ barrels into close apposition, creating a compact interface that forms the remaining elements of the protein translocation machine within the body of the heptamer. In particular, seven Phe427 residues are brought together to form a flexible annulus with a small hole at its center; this is called the “Φ-clamp” and its integrity is absolutely key to successful protein translocation. It is complemented with rings of acidic residues that surround the Φ-clamp on both the endosomal and cytoplasmic sides (Figs. 18.4 and 18.5).
Protein Translocation
An extensive series of studies, mostly by Krantz, Collier, Leppla and colleagues (Wynia-Smith et al. 2012; Krantz et al. 2005, 2006), has elucidated the basic elements of translocation of the toxic enzymes through the PA pore from endosome to cytosol. Briefly, each ~90 kDa enzyme must unfold, starting at its N-terminus (Gupta et al. 2008), and traverse the Φ-clamp and β-barrel as an extended polypeptide, one at a time; and then refold once in the cytosol. Binding of the toxic enzymes to the rim of the heptamer primes them for this process, and their N-terminal rearrangement creates a platform for the N-terminal helix (the α-clamp) of LFN/EFN to bind at a suitable height for its disordered ~30-residue N-terminus to hang down and make first contact with the Φ-clamp, ~45 Å below the α-clamp (Brown et al. 2015).
The Φ-clamp is watertight (when peptide is engaged), but flexible enough to allow positively charged and large aromatic residues (both of which readily interact with the ring of Phe residues via hydrophobic and/or Π-cation forces) to pass through the pore in either direction; while negatively charged residues cannot. However, the pH of the late endosome is just low enough to protonate Asp/Glu (pKa = 4.0–4.4), at least transiently, enabling them to pass through the Φ-clamp and onto the cytoplasmic side, where the much higher pH immediately deprotonates them, effectively trapping them on the distal side. That is, the pore provides a conduit for a single extended polypeptide to traverse, but it is protonation/deprotonation of Asp/Glu residues that is the key process for driving protein translocation from endosome to cytosol (by defining irreversible directionality). The mechanism is typically referred to as a “Brownian ratchet”—imagined as random oscillations of peptide segments lacking acidic residues (back and forth across the clamp), punctuated by periodic, irreversible, unidirectional translocational steps, whenever protonated Asp/Glu are encountered within the polypeptide chain; these have been called the “teeth” of the ratchet.
Trigger for the Prepore to Pore Transition
Finally, what drives the conformational change from pre-pore to pore? We know that, in vitro, PA63 heptamers insert into synthetic bilayers (in the absence of receptor) to form functional pores when the pH is lowered to ~6.0, which strongly implicates the protonation of histidine residues as triggers for the change. The lower pH required for membrane insertion in the presence of the receptor, CMG2 (pH ~ 5) (Scobie et al. 2007) is consistent with additional restrictions imposed by the very strong interactions (Kd ~170 pM) between PA and CMG2 that lock D2 and D4 together, thereby lowering the pKa values of certain histidines. The evidence that pore formation occurs in early endosomal compartments, while protein translocation occurs later, at a lower pH, would also argue against a role for Asp/Glu titration.
Exhaustive point mutational studies of PA63 showed that the substitution of any single histidine (by cysteine) did not impede pore formation, suggesting that protonation of more than one histidine is necessary. Six of the nine histidines in PA63, do not appear to be relevant to the pH switch: thus, His304, His310, His253 and His597 have weak or negligible interactions in the prepore and are unchanged or solvent-exposed in the pore; while His616 is buried in the center of D4, which remains folded following pore formation; and His211 is on the (invariant) surface of D1′ at the top of the heptamer (where it interacts with LF/EF in a pH-independent manner).
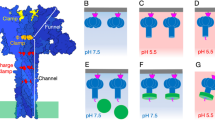
However, there are three histidines (His263, His299 and His336) PA63 that change from buried to an exposed environment upon switching from prepore to pore; moreover, their locations at key points within D2 or at the D2-D4 interface make them plausible candidates for contributing to the pH trigger [Fig. 18.6]. In addition, a conserved residue, His1121, from CMG2, is packed against Arg 659 (D4) and the D2 loop in the receptor complex, and is exposed to solvent following transition to the pore, such that its protonation could also promote the switch.
Potential pH triggers in the context of the PA-CMG2 complex. At right is an overview of the receptor complex, showing the MIDAS-mediated interaction with PA D4, and the interaction with a pro-barrel region of D2, which locks PA in its prepore state. Four histidines are identified that might contribute to the pH trigger: H299, H263 and H336 from D2, and H1121 from CMG2. At left is an expanded view of the environment of two of these histidines. Note how H263 and H299 are held in place by hydrophobic interactions but also form a sandwich around an Arg residue from D3. In addition, H263 inserts into the top of the b-sheet of D4, disrupting interactions that would create a barrel. Protonation of any of these residues is predicted to destabilize an interface, and thereby promote unfolding of the D4 domain
Krantz and colleagues have argued that residues close to the Φ-clamp are somehow involved in the pH trigger, including one Asp and 2 non-titratable residues (Jiang et al. 2015). Their rationale seems to be that these residues form dominant negative mutants, but it is unclear what this has to do with pH-dependent triggers. Notwithstanding, it is clear that identifying such triggers is difficult, and does not readily lend itself to simple mutational approaches.
Notes
- 1.
All of the complexes described here are consistent with each other and with a large body of biochemical and biophysical studies; but they bear no resemblance to earlier EM-derived models of a PA-LF complex (Ren et al. 2004; Tama et al. 2006), which cannot be rationalized in terms of any of the other published complexes, or inferred from translocation intermediates, and so remains enigmatic.
References
Abrami L, Lindsay M, Parton RG, Leppla SH, van der Goot FG (2004) Membrane insertion of anthrax protective antigen and cytoplasmic delivery of lethal factor occur at different stages of the endocytic pathway. J Cell Biol 166(5):645–651. https://doi.org/10.1083/jcb.200312072
Abrami L, Liu S, Cosson P, Leppla SH, van der Goot FG (2003) Anthrax toxin triggers endocytosis of its receptor via a lipid raft-mediated clathrin-dependent process. J Cell Biol 160(3):321–328. https://doi.org/10.1083/jcb.200211018
Bradley KA, Mogridge J, Mourez M, Collier RJ, Young JA (2001) Identification of the cellular receptor for anthrax toxin. Nature 414(6860):225–229. https://doi.org/10.1038/n35101999/n35101999[pii]
Brown MJ, Thoren KL, Krantz BA (2015) Role of the alpha clamp in the protein translocation mechanism of anthrax toxin. J Mol Biol 427(20):3340–3349. https://doi.org/10.1016/j.jmb.2015.08.024
Chekanov AV, Remacle AG, Golubkov VS, Akatov VS, Sikora S, Savinov AY, Fugere M, Day R, Rozanov DV, Strongin AY (2006) Both PA63 and PA83 are endocytosed within an anthrax protective antigen mixed heptamer: a putative mechanism to overcome a furin deficiency. Arch Biochem Biophys 446(1):52–59. https://doi.org/10.1016/j.abb.2005.11.013
Chvyrkova I, Zhang XC, Terzyan S (2007) Lethal factor of anthrax toxin binds monomeric form of protective antigen. Biochem Biophys Res Commun 360(3):690–695. https://doi.org/10.1016/j.bbrc.2007.06.124
Emsley J, Knight CG, Farndale RW, Barnes MJ, Liddington RC (2000) Structural basis of collagen recognition by integrin alpha2beta1. Cell 101(1):47–56. https://doi.org/10.1016/S0092-8674(00)80622-4
Fabre L, Santelli E, Mountassif D, Donoghue A, Biswas A, Blunck R, Hanein D, Volkmann N, Liddington R, Rouiller I (2016) Structure of anthrax lethal toxin prepore complex suggests a pathway for efficient cell entry. J General Physiol 148(4):313–324. https://doi.org/10.1085/jgp.201611617
Feld GK, Thoren KL, Kintzer AF, Sterling HJ, Tang II, Greenberg SG, Williams ER, Krantz BA (2010) Structural basis for the unfolding of anthrax lethal factor by protective antigen oligomers. Nat Struct Mol Biol 17(11):1383–1390. https://doi.org/10.1038/nsmb.1923
Frankel AE, Kuo SR, Dostal D, Watson L, Duesbery NS, Cheng CP, Cheng HJ, Leppla SH (2009) Pathophysiology of anthrax. Front Biosci 14:4516–4524. 3544 [pii]
Gordon VM, Klimpel KR, Arora N, Henderson MA, Leppla SH (1995) Proteolytic activation of bacterial toxins by eukaryotic cells is performed by Furin and by additional cellular proteases. Infect Immun 63(1):82–87
Gu ML, Leppla SH, Klinman DM (1999) Protection against anthrax toxin by vaccination with a DNA plasmid encoding anthrax protective antigen. Vaccine 17 (4):340–344. S0264-410X(98)00210-2 [pii]
Gupta PK, Moayeri M, Crown D, Fattah RJ, Leppla SH (2008) Role of N-terminal amino acids in the potency of anthrax lethal factor. PLoS ONE 3(9):e3130. https://doi.org/10.1371/journal.pone.0003130
Jiang J, Pentelute BL, Collier RJ, Zhou ZH (2015) Atomic structure of anthrax protective antigen pore elucidates toxin translocation. Nature 521(7553):545–549. https://doi.org/10.1038/nature14247
Kintzer AF, Sterling HJ, Tang II, Williams ER, Krantz BA (2010) Anthrax toxin receptor drives protective antigen oligomerization and stabilizes the heptameric and octameric oligomer by a similar mechanism. PLoS ONE 5(11):e13888. https://doi.org/10.1371/journal.pone.0013888
Kintzer AF, Thoren KL, Sterling HJ, Dong KC, Feld GK, Tang II, Zhang TT, Williams ER, Berger JM, Krantz BA (2009) The protective antigen component of anthrax toxin forms functional octameric complexes. J Mol Biol 392(3):614–629. https://doi.org/10.1016/j.jmb.2009.07.037
Krantz BA, Finkelstein A, Collier RJ (2006) Protein translocation through the anthrax toxin transmembrane pore is driven by a proton gradient. J Mol Biol 355(5):968–979. https://doi.org/10.1016/j.jmb.2005.11.030
Krantz BA, Melnyk RA, Zhang S, Juris SJ, Lacy DB, Wu Z, Finkelstein A, Collier RJ (2005) A phenylalanine clamp catalyzes protein translocation through the anthrax toxin pore. Science 309(5735):777–781. https://doi.org/10.1126/science.1113380
Lacy DB, Wigelsworth DJ, Melnyk RA, Harrison SC, Collier RJ (2004) Structure of heptameric protective antigen bound to an anthrax toxin receptor: a role for receptor in pH-dependent pore formation. Proc Natl Acad Sci USA 101(36):13147–13151. https://doi.org/10.1073/pnas.0405405101
Larson DK, Calton GJ, Little SF, Leppla SH, Burnett JW (1988) Separation of three exotoxic factors of Bacillus anthracis by sequential immunosorbent chromatography. Toxicon 26(10):913–921
Leppla SH, Robbins JB, Schneerson R, Shiloach J (2002) Development of an improved vaccine for anthrax. J Clin Invest 110(2):141–144. https://doi.org/10.1172/JCI16204
Moayeri M, Leppla SH (2004) The roles of anthrax toxin in pathogenesis. Curr Opin Microbiol 7(1):19–24. https://doi.org/10.1016/j.mib.2003.12.001/S1369527403001668[pii]
Moayeri M, Leppla SH (2009) Cellular and systemic effects of anthrax lethal toxin and edema toxin. Molecular Aspects Med 30 (6):439–455. S0098-2997(09)00051-X [pii]/10.1016/j.mam.2009.07.003
Moayeri M, Leppla SH, Vrentas C, Pomerantsev AP, Liu S (2015) Anthrax pathogenesis. Annu Rev Microbiol 69:185–208. https://doi.org/10.1146/annurev-micro-091014-104523
Mogridge J, Cunningham K, Lacy DB, Mourez M, Collier RJ (2002) The lethal and edema factors of anthrax toxin bind only to oligomeric forms of the protective antigen. Proc Natl Acad Sci USA 99(10):7045–7048. https://doi.org/10.1073/pnas.052160199
Molloy SS, Bresnahan PA, Leppla SH, Klimpel KR, Thomas G (1992) Human furin is a calcium-dependent serine endoprotease that recognizes the sequence Arg-X-X-Arg and efficiently cleaves anthrax toxin protective antigen. J Biol Chem 267(23):16396–16402
Petosa C, Collier RJ, Klimpel KR, Leppla SH, Liddington RC (1997) Crystal structure of the anthrax toxin protective antigen. Nature 385(6619):833–838. https://doi.org/10.1038/385833a0
Pilpa RM, Bayrhuber M, Marlett JM, Riek R, Young JA (2011) A receptor-based switch that regulates anthrax toxin pore formation. PLoS Pathog 7(12):e1002354. https://doi.org/10.1371/journal.ppat.1002354
Pimental RA, Christensen KA, Krantz BA, Collier RJ (2004) Anthrax toxin complexes: heptameric protective antigen can bind lethal factor and edema factor simultaneously. Biochem Biophys Res Commun 322(1):258–262. https://doi.org/10.1016/j.bbrc.2004.07.105
Ren G, Quispe J, Leppla SH, Mitra AK (2004) Large-scale structural changes accompany binding of lethal factor to anthrax protective antigen: a cryo-electron microscopic study. Structure 12(11):2059–2066. https://doi.org/10.1016/j.str.2004.09.010
Rosovitz MJ, Schuck P, Varughese M, Chopra AP, Mehra V, Singh Y, McGinnis LM, Leppla SH (2003) Alanine-scanning mutations in domain 4 of anthrax toxin protective antigen reveal residues important for binding to the cellular receptor and to a neutralizing monoclonal antibody. J Biol Chem 278(33):30936–30944. https://doi.org/10.1074/jbc.M301154200/M301154200[pii]
Santelli E, Bankston LA, Leppla SH, Liddington RC (2004) Crystal structure of a complex between anthrax toxin and its host cell receptor. Nature 430(7002):905–908. https://doi.org/10.1038/nature02763
Scobie HM, Marlett JM, Rainey GJ, Lacy DB, Collier RJ, Young JA (2007) Anthrax toxin receptor 2 determinants that dictate the pH threshold of toxin pore formation. PLoS ONE 2(3):e329. https://doi.org/10.1371/journal.pone.0000329
Singh Y, Klimpel KR, Arora N, Sharma M, Leppla SH (1994) The chymotrypsin-sensitive site, FFD315, in anthrax toxin protective antigen is required for translocation of lethal factor. J Biol Chem 269(46):29039–29046
Singh Y, Klimpel KR, Goel S, Swain PK, Leppla SH (1999) Oligomerization of anthrax toxin protective antigen and binding of lethal factor during endocytic uptake into mammalian cells. Infect Immun 67(4):1853–1859
Tama F, Ren G, Brooks CL 3rd, Mitra AK (2006) Model of the toxic complex of anthrax: responsive conformational changes in both the lethal factor and the protective antigen heptamer. Protein Sci 15(9):2190–2200. https://doi.org/10.1110/ps.062293906
van der Goot G, Young JA (2009) Receptors of anthrax toxin and cell entry. Mol Aspects Med 30(6):406–412. https://doi.org/10.1016/j.mam.2009.08.007
Varughese M, Teixeira AV, Liu S, Leppla SH (1999) Identification of a receptor-binding region within domain 4 of the protective antigen component of anthrax toxin. Infect Immun 67(4):1860–1865
Wynia-Smith SL, Brown MJ, Chirichella G, Kemalyan G, Krantz BA (2012) Electrostatic ratchet in the protective antigen channel promotes anthrax toxin translocation. J Biol Chem 287(52):43753–43764. https://doi.org/10.1074/jbc.M112.419598
Young JA, Collier RJ (2007) Anthrax toxin: receptor binding, internalization, pore formation, and translocation. Annu Rev Biochem 76:243–265. https://doi.org/10.1146/annurev.biochem.75.103004.142728
Zhou K, Liu S, Hardenbrook NJ, Cui Y, Krantz BA, Zhou ZH (2020) Atomic structures of anthrax prechannel bound with full-length lethal and edema factors. Structure. https://doi.org/10.1016/j.str.2020.05.009
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Liddington, R.C. (2021). Assembly and Function of the Anthrax Toxin Protein Translocation Complex. In: Harris, J.R., Marles-Wright, J. (eds) Macromolecular Protein Complexes III: Structure and Function. Subcellular Biochemistry, vol 96. Springer, Cham. https://doi.org/10.1007/978-3-030-58971-4_18
Download citation
DOI: https://doi.org/10.1007/978-3-030-58971-4_18
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-58970-7
Online ISBN: 978-3-030-58971-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)