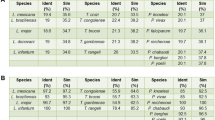
Abstract
Expansion segments (ES) are insertions of a few to hundreds of nucleotides at discrete locations on eukaryotic ribosomal RNA (rRNA) chains. Some cluster around ‘hot spots’ involved in translation regulation and some may participate in biogenesis. Whether ES play the same roles in different organisms is currently unclear, especially since their size may vary dramatically from one species to another and very little is known about their functions. Most likely, ES variation is linked to adaptation to a particular environment. In this chapter, we compare the interaction networks of ES from four kinetoplastid parasites, which have evolved in diverse insect vectors and mammalian hosts: Trypanosoma cruzi, Trypanosoma brucei, Leishmania donovani and Leishmania major. Here, we comparatively analyze ribosome structures from these representative kinetoplastids and ascertain meaningful structural differences from mammalian ribosomes. We base our analysis on sequence alignments and three-dimensional structures of 80S ribosomes solved by cryo-electron microscopy (cryo-EM). Striking differences in size are observed between ribosomes of different parasites, indicating that not all ES are expanded equally. Larger ES are not always matched by large surrounding ES or proteins extensions in their vicinity, a particularity that may lead to clues about their biological function. ES display different species-specific patterns of conservation, which underscore the density of their interaction network at the surface of the ribosome. Making sense of the conservation patterns of ES is part of a global effort to lay the basis for functional studies aimed at discovering unique kinetoplastid-specific sites suitable for therapeutic applications against these human and often animal pathogens.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
Why Expansion Segments?
Because protein synthesis is a universal necessity for all organisms, ribosome function, overall shape and core architecture are conserved across species. Yet, ribosome composition varies considerably among different domains of life. For example, ribosomes from yeast and human are respectively ~40 and ~85% larger than bacterial ribosomes (Melnikov et al. 2012). In addition to comprising more proteins than bacterial ribosomes, eukaryotic ribosomes also contain elongated ribosomal RNAs (rRNA). This ‘extra RNA’ forms expansion segments (ES) (Clark et al. 1984; Hassouna et al. 1984; Ware et al. 1983).
Whether ES are all mere additions that are tolerated by the translation machinery, or whether they have important regulatory roles remains for the most part an open question. Most ES protrude outside of the highly conserved ribosomal core (Bernier et al. 2018; Melnikov et al. 2012). Their size varies across species, although their patterns of insertion are conserved (Gerbi 1996). We know from early structural studies that ES and associated proteins portions comprise somewhat flexible substructures on the solvent-facing side, where they may interact with one another and with proteins to form an outer layer (Anger et al. 2013; Armache et al. 2010; Gao et al. 2005). This web-like organization is more rigid where contacts with the ribosome core are tighter, which argues against a superfluous nature. ES are generally best perceived as peripheral elements that facilitate folding, assembly, or activity. In this view they are to some extent similar to often observed insertions of >200 nt reported in other ribozymes (e.g., group I introns, RNase P, and hammerhead) (Kachouri et al. 2005; Lehnert et al. 1996; Michel and Westhof 1990; Westhof and Massire 2004; Przybilski et al. 2005).
What do we currently know about the function of ES? On the small ribosomal subunit (SSU) of protozoan ribosomes, for instance, a network of three ES (ES3S, ES6S, ES7S) span >150 Å in the vicinity of the mRNA exit channel (Gao et al. 2005; Hashem et al. 2013b). This cluster of ES could to be involved in translation initiation through the regulation of recruitment of initiation factors such as eIF3 (Hashem et al. 2013a, b). We currently hypothesize that this platform is required for translation of protozoan mRNAs, which have particular features compared to most know eukaryotic mRNAs (a hypermodified cap, presence of a 39-nt leader sequence that is trans-spliced to the 5′ UTR of all kinetoplastid mature mRNAs) (Gao et al. 2005; Hashem et al. 2013b). ES may also participate in ribosome biogenesis, as illustrated by the finding that 12 out of the 16 ESs on the large subunit (LSU) of the yeast ribosome are required for optimal growth and correct 25S rRNA processing (Ramesh and Woolford 2016). The roles associated with certain ES in a particular organism may be conserved across various species. For instance, ES27L on the LSU was shown to be consistently alternating between two positions (near L1 and the peptide tunnel exit) from the structures of the human, wheat, fly and yeast ribosomes (Anger et al. 2013; Armache et al. 2010; Beckmann et al. 2001). This observation suggests a similar role, especially since replacing ES27L with that of another species restores function (Sweeney et al. 1994).
In this chapter, we focus on ribosomal ES from the Trypanosoma genus (ES in other eukaryotes have been discussed elsewhere Anger et al. 2013; Li et al. 2017). Tritryps kinetoplastid parasites, i.e. Trypanosoma cruzi, Trypanosoma brucei spp. and Leishmania spp. are human pathogens causing the Chagas disease, African trypanosomiasis and leishmaniasis, respectively. Chagas disease alone affects eight million people in the Americas (PAHO 2017), and due to migration flows it has now emerged in non-endemic countries such as Spain and the United States [>300,000 infected individuals in the US (Bern and Montgomery 2009)] (Gascon et al. 2010). Current treatments rely on toxic drugs that are often effective only during specific stages of the disease, further highlighting the urge to develop new strategies for targeting these parasites (Field et al. 2017). Being highly distinctive from those of mammalian ribosomes (Gao et al. 2005; Hashem et al. 2013b), theses ES in kinetoplastids could potentially be promising targets for therapeutic applications. However, efficiently targeting kinetoplastid-specific ES requires a deep structural understanding of their organization and their function, which is the motivation behind the comparative analysis we present here.
Expansion Segments in Kinetoplastid Ribosomes Are Large but Structurally Organized
Biochemical and structural studies on ribosomes from kinetoplastids pinpointed unique features for two Trypanosoma species. The first structure of the ribosome from T. cruzi was solved at 12 Å resolution (Gao et al. 2005) and that of T. brucei brucei (referred to as T. brucei in the following) at 5.6 Å resolution (Hashem et al. 2013b). We can now compare these structures to higher-resolution structures of the 80S ribosome from L. donovani (Zhang et al. 2016; Shalev-Benami et al. 2016, 2017) and T. cruzi (Liu et al. 2016; Querido et al. 2017), which we revised and/or expanded from the published models (Fig. 13.1a, b). We also include in this analysis a previously unpublished structure of the L. major 80S ribosome (Fig. 13.1c) at ~5 Å resolution (the structure can be shared upon simple request. Images were collected on an FEI Polara electron microscope equipped with a Gatan Summit K2 Camera. After image processing, nearly 150,000 particle images were used in the final reconstruction). Sequence conservation between the L. donovani and L. major ribosomes is >98% (rRNA) and >97% (r-protein; except for eS24 ~ 93%). Our strategy was to model the ES for these four species of kinetoplastids according to rRNA structural alignments that we generated using non-redundant sequences in SSU-Align (Nawrocki 2009), which we manually adjusted in Ugene (Okonechnikov et al. 2012) and validated using R-scape (Rivas et al. 2017). Because of their high level of flexibility, in many cases it was not possible to directly model some ES in their cryo-EM map densities (Fig. 13.1a, b), as had been done previously for other eukaryotic ribosomes (Anger et al. 2013).
Strategy for modeling flexible expansion segments in ribosomes. a Modeling the complete T. cruzi 80S at 4 Å resolution. We modeled expansion segments in the SSU of our previously published model [PDB ID: 5OPT Brito Querido et al. 2017; Pitula et al. 2002; Ayub et al. 2012)]. We also included coordinates from a 2.5 Å-structure of the 60S particle from T. cruzi [PDB ID: 5T5H (Liu et al. 2016)]. RNA and protein elements were added (a total of 1816 nucleotides and 810 aminoacids) are shown in red. Residues built within density limits (~8 to 15 Å) are shown in ribbon mode, while residues beyond are shown as surfaces. Insets illustrate representative regions of the original unfiltered maps. Dashed boxes point to regions located on the side of the ribosome hidden from view. b The quality of the L. donovani electron density map at 2.9 Å resolution [PDB ID: 5T2A (Zhang et al. 2016)] enabled us to add 873 nucleotides and 802 aminoacids to the original model (in red). c The revised model of the L. donovani ribosome was used as the basis for building the structure of the ribosome from L. major at 5 Å resolution. The L. major map and coordinates will be deposited in the PDB
A side-by-side comparison of the ribosomes from T. brucei, L. donovani, L. major, and T. cruzi reveals that many ES are several-fold larger than their counterparts in mammals (Fig. 13.2). Although the 28S rRNA in the LSU of T. brucei and T. cruzi is halfway in size between yeast and human, their 18S in the SSU is >20% longer than their human counterpart (Table 13.1). Most of this extra rRNA sequence is found within ES6S and ES7S that are inserted in helices 21 and 26. ES6S and ES7S are respectively 230 and 17 nucleotides long in H. sapiens, but 429 and 136 and 512 and 164 nucleotides long in L. major and T. cruzi. ES6S, ES7S. It should also be noted that ES9S and ES31L adopt kinetoplastid-specific tertiary structures and they cluster around the mRNA channel exit. In particular, ES6S, ES7S and ES9S are located at a very strategic region where initiation factor 3 is known to bind in mammals, forming bridges between the LSU and SSU that are not found in other eukaryotic ribosomes (Hashem et al. 2013b).
The complex outer layer of expansion segments at the surface of ribosomes from kinetoplastids. a Secondary structure schematic for the LSU and SSU rRNA of kinetoplastids. Expansion segments are highlighted in colors and labeled according to conventional numbering (Gerbi 1996). b Overall structure of the 80S ribosomes from four kinetoplastids, viewed from the mRNA L1-stalk side (left) and SSU beak sides (middle). Ribosomes are shown in surface view, except for ES rendered as ribbons and colored as in panel (a). (Right) Ribosome structures from other organisms are shown from the L1-stalk side as a reference (references in the main text)
From aligning more than 80 sequences for the SSU and 25 for the LSU, we found that all ES from kinetoplastids have a conserved structure, with large extensions for some helical regions (Figs. 13.3 and 13.4). This confirms and expands on prior findings obtained with fewer than ten kinetoplastid sequences for all ES (Hashem et al. 2013b), and with 3000 eukaryotic sequences for ES6S only (Wuyts et al. 2000). The proportion of conserved versus variable regions varies among ES, with ES3S (~200 nt long) being the most conserved. Large size variations are observed even among kinetoplastids. For example, ES6S, ES7S, ES7L, ES19L and ES27L have a comparable size among the genera Angomonas, Herpetomonas and Phytomonas. The same ES are significantly larger in Leishmania and Trypanosoma species, the largest being found in T. cruzi. Although ES3S, ES9S, ES10S and ES31L are all larger in Leishmania spp. and in T. brucei than in T. cruzi, the size differences remain relatively modest (Table 13.1).
Conservation of interaction networks between kinetoplastids. a Interaction network and consensus secondary structure of ES3S. Note that ES2S forms a hairpin at the 5′ of h8. Statistically significant covarying base pairs are shown in green [E-value <0.05 (Rivas et al. 2017)]. This and subsequent covariation models were generated using R2R (Weinberg and Breaker 2011). b Close-up of the left foot of the ribosome from L. donovani, showing the arrangement of several ES and r-proteins. c Conserved binding of KSRP, which interacts with ES3S, ES6S and eS6. d Conserved spatial arrangement of stem Ab of ES3S and r-protein eS8. Sequence alignments for all ES are available directly from the authors upon request
Sequence and structure variability among ES is significant but localized. a Interaction networks and consensus secondary structures of ES6S and ES7S (color-coding as in Fig. 13.3). b Conformational variability as a function of the length of certain ES across kinetoplastids. Top: The size of ES7S barely changes across kinetoplastids, in contrast to that of ES6S. The increasing size of ES6S leads to displacement of ES7S helices. The regions covered by the electron density maps (grey) are shown as ribbons, while the sections modeled beyond the limit of the current experimental maps are shown as surfaces. Bottom: Conformational changes in particular of ES31L occur as the length of the surrounding ES19L and ES27L increases
Expansion Segments Form Kinetoplastid-Specific Interaction Networks
The overall high degree of ES conservation can be explained by interaction networks at the ribosome surface. For example, in all cryo-EM structures of kinetoplastids, ES3S is tightly packed as it forms the left foot of the SSU (Fig. 13.3a). ES3S interacts with ES2S, domains B and D of ES6S (Fig. 13.4a), ES12S (forming the right foot of the SSU) and r-proteins eS4, eS6, eS8 and KSRP (Fig. 13.3a, b). The structure of the pseudoknot formed between ES3S and ES6S is conserved across parasitic and eukaryotic ribosomes of known structures, as reported earlier (Armache et al. 2010; Alkemar and Nygard 2006; Hashem et al. 2013b; Zhang et al. 2016). The kinetoplastid-specific r-protein (KSRP) that we identified in T. cruzi (Brito Querido et al. 2017) is similarly bound to ES3S and ES6S in Trypanosoma and Leishmania, through its two conserved RNA recognition motifs (RRM) and a C-terminal helix that also interacts with eS6 (Fig. 13.3c). The only variable region within ES3S is the Ab stem, which nonetheless conserves its spatial arrangement as it bends around eS8 in the four kinetoplastids models (Fig. 13.3d). It is important to emphasize that ES3S simultaneously displays structural characteristics conserved in all eukaryotes (pseudoknot with ES6S) and unique features only found in kinetoplastids (KSRP binding).
Even for larger and more variable ES such as ES6S and ES7S, their core shows a high degree of conservation (Fig. 13.4a), supporting a conserved topology. ES6S is the largest ES on the SSU (400–500 nt) and its highly conserved domains A–D interact tightly with r-proteins and KSRP at the SSU solvent-exposed surface. ES6S interacts also with ES7S, which is of a similar size in T. cruzi and T. brucei, and ~25% larger in T. cruzi than in L. donovani. In contrast to stem Ab in ES3S, whose length changes only slightly across parasites, stems E1–E3 in ES6S roughly double in size from T. brucei through Leishmania to T. cruzi (Fig. 13.4a, b). As a result, the number of stem-loops protruding into the solvent is different among parasites (Fig. 13.4b). Both the nature and the presence of these stems dictate the positioning of the conserved stems of ES7S, which are possibly more conformationally restricted in Leishmania and T. cruzi than in T. brucei. Note that although the lack of density prevented pinpointing the exact location of the expansions seen in E2 and E3, the interaction between E3 and ES7S seen in T. brucei cannot occur for the other three model parasites, where E3 is much longer (Fig. 13.4b). These structural differences probably reflect the dynamic properties of this area, which is a ‘hotspot’ for translation initiation regulation (Hashem et al. 2013a).
ES 9S, 10S and 12S, on the other hand, are among the smallest SSU ES. ES12S, for example, is an extension of h44 by 9–10 base pairs in all kinetolastids, while ES10S exists as a 5–9 base pair stem only in T. brucei, T. congolense, T. simiae, and T. vivax (Fig. 13.5). The shorter ES seem to follow a similar trend as the longer ES, thus presenting conserved cores and variable stems.
On the LSU, ES are concentrated on two regions, the L1-stalk region, and the solvent-exposed face (Fig. 13.2b). The ES at the solvent-exposed face appear less interconnected compared to the SSU ES. ES7L is isolated from the other sizable LSU ES. It presents a two-pronged fork shape (Figs. 13.6a and 13.7c). Because of its numerous interactions with r-proteins (Fig. 13.6a), its densities are relatively well resolved and present a good local resolution. It forms a pseudoknot with the small ES15L (Fig. 13.6b, d). Another more isolated ES is 5L (also called ES42L) (Fig. 13.6e). Although situated close to ES7L, no direct interactions were observed with the latter. ES42L presents a high level of sequence conservation and interacts with r-proteins eL8, eL15 and uL29 (Fig. 13.6e).
Covariation analysis and interaction networks of LSU expansion segments. a Consensus sequence and secondary structure for ES7L. b Close-up of the pseudoknot formed by ES7L and ES15L in T. cruzi and T. gondii [PDB ID: 5XXB (Li et al. 2017)]. c Close-up of the region at the top of stem A1 of ES7L in L. donovani and H. sapiens. d–f Consensus sequences and secondary structures for ES15L, ES5L (also known as ES42L) and ES19L (interacting partners are not shown for H16–20). g Close-up of the region comprising ES19L and ES31L. h–i Same as (d–f) for ES31L and ES27L
Compensatory interaction networks on the LSU. a Superimposition of the ribosomes from T. cruzi (light shades) and H. sapiens [PDB ID: 4V6X; dark shades (Anger et al. 2013)]. The view is from the top of that shown in panel 22.4b, after removing regions modeled outside of density. b Close-up of the location where eL34 binds ES31L in kinetoplastids, and of the corresponding region in the human ribosome (PDB ID: 4V6X). For kinetoplastids, the number of the last residue modeled in density as well as the total length of the r-protein are indicated. c Close-up of ES7L bound to uL4 in kinetolastids, and of the corresponding region in the T. gondii ribosome [PDB ID: 5XXB (Li et al. 2017)], where an elongated uL4 interacts with uL16
On the other hand, LSU ES near the L1-stalk region are interconnected, where ES19L, ES27L and ES31L are clustered (Figs. 13.2b and 13.6). The length of ES31L varies only within a range of ~20 nucleotides among kinetoplastids, while that of ES19L and ES27L increases by ~50% between L. donovani and T. cruzi (Table 13.1). As a result, stem F of ES31L adopts different orientations in the various structures, where it is constricted to various degrees by nearby ES (Fig. 13.4b). ES31L is highly structured, as it comprises four nested three-way junctions but a high degree of sequence variability (Fig. 13.6h). Although ES31L possesses a complex predicted tertiary structure, its cryo-EM corresponding densities are poorly resolved in all four kinetoplastid model ribosomes, probably because of its high flexibility. Although ES31L interacts with r-proteins eL27 and eL34 (Fig. 13.6h), the bulk of its rRNA is not stabilized by any other r-proteins, which explains its high level of flexibility. The cryo-EM reconstructions from several kinetoplatids strongly suggest its interaction with ES19L and ES27L because of its proximity to the latter ES (Figs. 13.2b and 13.6g). ES19L appears as an elongated stem of a relatively well resolved mass of density that shows its interactions with r-proteins eS27 and uL23, but also with srRNA1 (Fig. 13.6f). It is interesting to note that ES31L and ES19L interaction occurs indirectly through r-protein eL27 (Fig. 13.6f, h). Last but not least, ES27L contains two three-way junctions conferring it an elongated shape with two grafted branches protruding toward the solvent (Fig. 13.6i). It interacts directly with ES31L and r-proteins eL19 and eL22 (Fig. 13.6i). Thus, all three, ES 19L, 27L and 31L form an interconnected bundle of rRNA near the L1-stalk on the LSU (Fig. 13.2).
Finally, it is worth mentioning that expansion segments may have a conserved structure even in the absence of sequence conservation, as exemplified by ES7S (Figs. 13.2 and 13.4b), and subdomain A3 of ES7L (Figs. 13.6a and 13.7c).
Compensatory Interaction Networks
Compensatory interaction networks are widespread on the LSU, as previously observed upon comparing ribosomes from yeast, wheat, fly, human and several parasites (Anger et al. 2013; Armache et al. 2010; Li et al. 2017; Hashem et al. 2013b). For instance, the larger ES31L in kinetoplastids (70 nt in yeast, 85 nt in human, but 220–237 nt in kinetoplastids) fills the space occupied by an extended helix from the 714 nt-long ES27L in humans, without altering the binding mode of eL27 and eL8 (Fig. 13.7a). Another example is the conserved stem A1 of ES7L that packs against eL6, eL32 and eL33 but not eL28, in contrast to ES7L of other eukaryotic ribosomes (Anger et al. 2013; Ben-Shem et al. 2011; Hashem et al. 2013b; Li et al. 2017) (Fig. 13.6a, d). Moreover, although the pseudoknot formed between ES7L and ES15L is similar to that in T. gondii (Fig. 13.6b, d), the long-range interaction involving the shallow groove of ES7L and a loop from ES39L in human is replaced by interactions with srRNA3 (Fig. 13.6c).
Size increase of ES is often accompanied by extensions of r-proteins (Hashem et al. 2013b; Li et al. 2017). For example, the eL34 r-protein is extended in kinetoplastids by ~50 amino acids compared with human eL34, which can be explained by a larger ES31L (Fig. 13.7b). It therefore came as a surprise that the length of uL4 remains the same across kinetoplastids, even though stem A3 of ES7L nearby dramatically varies (Fig. 13.7c). Nevertheless, exceptions can be observed, such as in P. falciparum and T. gondii where ES7L is of a similar size compared to that of T. brucei (~290 nt). However, uL4 is elongated by ~40 amino acids in P. falciparum and T. gondii, so that it interacts with uL16 (Fig. 13.7c), a late binding protein during ribosome assembly in yeast (Li et al. 2017; Ma et al. 2017), with no obvious correlation with the size of ES7L (Fig. 13.7c).
Concluding Remarks
In line with previous work that presented the structures, and underscored the importance of, expansion segments in other species (Armache et al. 2010; Beckmann et al. 2001; Ben-Shem et al. 2011), this chapter illustrates that ES are not random extensions with poor sequence conservation at the surface of ribosomes. Rather, ES are organized so that they tightly interact with core rRNA and r-proteins, recruit kinetoplastid-specific proteins, and act as dynamic gatekeepers. Some of these interaction networks are found in some but not all parasites, suggesting that the precise role of ES may vary even among species from the same phylum. Through highlighting the importance of ES networks in the ribosomes of kinetoplastid parasites, this work expands the chacterization of kinetoplastid-specific features, in the hope of stimulating the development of safer and more specific anti-parasitic therapeutic.
Authors Contributions
QV and AB did the molecular modeling and cryo-EM maps (re)analysis. AJ and YH acquired L. major ribosome particles and processed the cryo-EM data. QV, YH and JF wrote the manuscript with input from all authors. QV and AB did the Fig.s. YH and JF supervised the research.
References
Alkemar G, Nygard O (2006) Probing the secondary structure of expansion segment ES6 in 18S ribosomal RNA. Biochemistry 45(26):8067–8078. https://doi.org/10.1021/bi052149z
Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, Beckmann R (2013) Structures of the human and Drosophila 80S ribosome. Nature 497(7447):80–85. https://doi.org/10.1038/nature12104
Armache JP, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, Marquez V, Mielke T, Thomm M, Berninghausen O, Beatrix B, Soding J, Westhof E, Wilson DN, Beckmann R (2010) Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution. Proc Natl Acad Sci USA 107(46):19748–19753. https://doi.org/10.1073/pnas.1009999107
Ayub AJ, Lapadula WJ, Hoebeke J, Smulski CR (2012) Ribosomes from Trypanosomatids: unique structural and functional properties. In: Biyani M (ed) Cell-free protein synthesis. Intech, pp 3–28
Beckmann R, Spahn CM, Eswar N, Helmers J, Penczek PA, Sali A, Frank J, Blobel G (2001) Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell 107(3):361–372
Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M (2011) The structure of the eukaryotic ribosome at 3.0 A resolution. Science, 334. https://doi.org/10.1126/science.1212642
Bern C, Montgomery SP (2009) An estimate of the burden of Chagas disease in the United States. Clin Infect Dis 49(5):e52–54. https://doi.org/10.1086/605091
Bernier CR, Petrov AS, Kovacs NA, Penev PI, Williams LD (2018) Translation: the universal structural core of life. Mol Biol Evol 35(8):2065–2076. https://doi.org/10.1093/molbev/msy101
Brito Querido J, Mancera-Martínez E, Vicens Q, Bochler A, Chicher J, Simonetti A, Hashem Y (2017) The cryo-EM structure of a novel 40S kinetoplastid-specific ribosomal protein. Structure 25(12):1785–1794.e3. https://doi.org/10.1016/j.str.2017.09.014
Clark CG, Tague BW, Ware VC, Gerbi SA (1984) Xenopus laevis 28S ribosomal RNA: a secondary structure model and its evolutionary and functional implications. Nucleic Acids Res 12(15):6197–6220. https://doi.org/10.1093/nar/12.15.6197
Field MC, Horn D, Fairlamb AH, Ferguson MAJ, Gray DW, Read KD, De Rycker M, Torrie LS, Wyatt PG, Wyllie S, Gilbert IH (2017) Anti-trypanosomatid drug discovery: an ongoing challenge and a continuing need. Nat Rev Microbiol 15(7):447. https://doi.org/10.1038/nrmicro.2017.69
Gao H, Ayub MJ, Levin MJ, Frank J (2005) The structure of the 80S ribosome from Trypanosoma cruzi reveals unique rRNA components. Proc Natl Acad Sci USA 102(29):10206–10211. https://doi.org/10.1073/pnas.0500926102
Gascon J, Bern C, Pinazo MJ (2010) Chagas disease in Spain, The United States and other non-endemic countries. Acta Trop 115(1–2):22–27. https://doi.org/10.1016/j.actatropica.2009.07.019
Gerbi SA (1996) Expansion segments: regions of variable size that interrupt the universal core secondary structure of ribosomal RNA. In: Zimmermann RA, Dahlberg AE (eds) Ribosomal RNA: Structure. Telford - CRC Press, Evolution, processing and function in protein synthesis, pp 71–87
Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Hellen CU, Pestova TV, Frank J (2013a) Structure of the mammalian ribosomal 43S preinitiation complex bound to the scanning factor DHX29. Cell 153(5):1108–1119. https://doi.org/10.1016/j.cell.2013.04.036
Hashem Y, des Georges A, Fu J, Buss SN, Jossinet F, Jobe A, Zhang Q, Liao HY, Grassucci RA, Bajaj C, Westhof E, Madison-Antenucci S, Frank J (2013b) High-resolution cryo-electron microscopy structure of the Trypanosoma brucei ribosome. Nature 494(7437):385–389. https://doi.org/10.1038/nature11872
Hassouna N, Michot B, Bachellerie JP (1984) The complete nucleotide sequence of mouse 28S rRNA gene. Implications for the process of size increase of the large subunit rRNA in higher eukaryotes. Nucleic Acids Res 12(8):3563–3583. https://doi.org/10.1093/nar/12.8.3563
Kachouri R, Stribinskis V, Zhu Y, Ramos KS, Westhof E, Li Y (2005) A surprisingly large RNase P RNA in Candida glabrata. RNA 11(7):1064–1072. https://doi.org/10.1261/rna.2130705
Lehnert V, Jaeger L, Michel F, Westhof E (1996) New loop-loop tertiary interactions in self-splicing introns of subgroup IC and ID: a complete 3D model of the Tetrahymena thermophila ribozyme. Chem Biol 3(12):993–1009
Li Z, Guo Q, Zheng L, Ji Y, Xie YT, Lai DH, Lun ZR, Suo X, Gao N (2017) Cryo-EM structures of the 80S ribosomes from human parasites Trichomonas vaginalis and Toxoplasma gondii. Cell Res 27(10):1275–1288. https://doi.org/10.1038/cr.2017.104
Liu Z, Gutierrez-Vargas C, Wei J, Grassucci RA, Ramesh M, Espina N, Sun M, Tutuncuoglu B, Madison-Antenucci S, Woolford JL Jr, Tong L, Frank J (2016) Structure and assembly model for the Trypanosoma cruzi 60S ribosomal subunit. Proc Natl Acad Sci USA 113(43):12174–12179. https://doi.org/10.1073/pnas.1614594113
Ma C, Wu S, Li N, Chen Y, Yan K, Li Z, Zheng L, Lei J, Woolford JL Jr, Gao N (2017) Structural snapshot of cytoplasmic pre-60S ribosomal particles bound by Nmd3, Lsg1, Tif6 and Reh1. Nat Struct Mol Biol 24:214–220
Melnikov S, Ben-Shem A, Garreau de Loubresse N, Jenner L, Yusupova G, Yusupov M (2012) One core, two shells: bacterial and eukaryotic ribosomes. Nat Struct Mol Biol 19(6):560–567. https://doi.org/10.1038/nsmb.2313
Michel F, Westhof E (1990) Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol 216(3):585–610. https://doi.org/10.1016/0022-2836(90)90386-Z
Nawrocki EP (2009) Structural RNA homology search and alignment using covariance models. Washington University School of Medicine. https://doi.org/10.7936/K78050MP
Okonechnikov K, Golosova O, Fursov M, Team U (2012) Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28(8):1166–1167. https://doi.org/10.1093/bioinformatics/bts091
PAHO (2017) Chagas disease. Pan American Health Organization. https://www.paho.org/hq/index.php?option=com_topics&view=article&id=10&Itemid=40743. Accessed 20 Oct 2017
Pitula J, Ruyechan WT, Williams N (2002) Two novel RNA binding proteins from Trypanosoma brucei are associated with 5S rRNA. Biochem Biophys Res Commun 290(1):569–576. https://doi.org/10.1006/bbrc.2001.6226
Przybilski R, Graf S, Lescoute A, Nellen W, Westhof E, Steger G, Hammann C (2005) Functional hammerhead ribozymes naturally encoded in the genome of Arabidopsis thaliana. Plant Cell 17(7):1877–1885. https://doi.org/10.1105/tpc.105.032730
Ramesh M, Woolford JL Jr (2016) Eukaryote-specific rRNA expansion segments function in ribosome biogenesis. RNA 22(8):1153–1162. https://doi.org/10.1261/rna.056705.116
Rivas E, Clements J, Eddy SR (2017) A statistical test for conserved RNA structure shows lack of evidence for structure in lncRNAs. Nat Methods 14(1):45–48. https://doi.org/10.1038/nmeth.4066
Shalev-Benami M, Zhang Y, Matzov D, Halfon Y, Zackay A, Rozenberg H, Zimmerman E, Bashan A, Jaffe CL, Yonath A, Skiniotis G (2016) 2.8-A Cryo-EM structure of the large ribosomal subunit from the eukaryotic parasite leishmania. Cell Rep 16 (2):288–294. https://doi.org/10.1016/j.celrep.2016.06.014
Shalev-Benami M, Zhang Y, Rozenberg H, Nobe Y, Taoka M, Matzov D, Zimmerman E, Bashan A, Isobe T, Jaffe CL, Yonath A, Skiniotis G (2017) Atomic resolution snapshot of Leishmania ribosome inhibition by the aminoglycoside paromomycin. Nat Commun 8(1):1589. https://doi.org/10.1038/s41467-017-01664-4
Sweeney R, Chen L, Yao MC (1994) An rRNA variable region has an evolutionarily conserved essential role despite sequence divergence. Mol Cell Biol 14(6):4203–4215
Ware VC, Tague BW, Clark CG, Gourse RL, Brand RC, Gerbi SA (1983) Sequence analysis of 28S ribosomal DNA from the amphibian Xenopus laevis. Nucleic Acids Res 11(22):7795–7817. https://doi.org/10.1093/nar/11.22.7795
Weinberg Z, Breaker RR (2011) R2R–software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinf 12:3. https://doi.org/10.1186/1471-2105-12-3
Westhof E, Massire C (2004) Structural biology. Evolution of RNA architecture. Science 306(5693):62–63. https://doi.org/10.1126/science.1104482
Wuyts J, De Rijk P, Van de Peer Y, Pison G, Rousseeuw P, De Wachter R (2000) Comparative analysis of more than 3000 sequences reveals the existence of two pseudoknots in area V4 of eukaryotic small subunit ribosomal RNA. Nucleic Acids Res 28(23):4698–4708
Zhang X, Lai M, Chang W, Yu I, Ding K, Mrazek J, Ng HL, Yang OO, Maslov DA, Zhou ZH (2016) Structures and stabilization of kinetoplastid-specific split rRNAs revealed by comparing leishmanial and human ribosomes. Nat Commun 7:13223. https://doi.org/10.1038/ncomms13223
Acknowledgements
We thank Robert A. Grassucci for assistance in data acquisition of L. major. This work was supported by the ANR grant ANR-14-ACHN-0024 @RAction program ‘‘ANR CryoEM80S’’ and the ERC-2017-STG #759120 “TransTryp” (to Y.H.). J.F. acknowledges support by HHMI and NIH R01 GM29169.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Vicens, Q., Bochler, A., Jobe, A., Frank, J., Hashem, Y. (2021). Interaction Networks of Ribosomal Expansion Segments in Kinetoplastids. In: Harris, J.R., Marles-Wright, J. (eds) Macromolecular Protein Complexes III: Structure and Function. Subcellular Biochemistry, vol 96. Springer, Cham. https://doi.org/10.1007/978-3-030-58971-4_13
Download citation
DOI: https://doi.org/10.1007/978-3-030-58971-4_13
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-58970-7
Online ISBN: 978-3-030-58971-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)