Abstract
The chapter describes the exceptional symbiotic associations formed between the ciliate Paramecium and Holospora, highly infectious bacteria residing in the host nuclei. Holospora and Holospora-like bacteria (Alphaproteobacteria) are characterized by their ability for vertical and horizontal transmission in host populations, a complex biphasic life cycle, and pronounced preference for host species and colonized cell compartment. These bacteria are obligate intracellular parasites; thus, their metabolic repertoire is dramatically reduced. Nevertheless, they perform complex interactions with the host ciliate. We review ongoing efforts to unravel the molecular adaptations of these bacteria to their unusual lifestyle and the host’s employment in the symbiosis. Furthermore, we summarize current knowledge on the genetic and genomic background of Paramecium–Holospora symbiosis and provide insights into the ecological and evolutionary consequences of this interaction. The diversity and occurrence of symbioses between ciliates and Holospora-like bacteria in nature is discussed in connection with transmission modes of symbionts, host specificity and compatibility of the partners. We aim to summarize 50 years of research devoted to these symbiotic systems and conclude trying to predict some perspectives for further studies.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
1 Characteristics of Holospora and Holospora-Like Bacteria
Symbioses and especially intracellular symbioses are drivers of evolution and biological innovation. The interaction between unicellular eukaryotic host and intracellular symbionts is naturally rather intimate as any interruption leading to the decay of the symbiotic system may ultimately cause the death of the host cell itself. In unicellular eukaryotes, we find a tremendous amount of organismal and functional diversity regarding intracellular symbionts. Hence, we can use them as fascinating models to study the mechanisms of bacterial transmission on individual and the host population level, regulation of interactions between eukaryotic and prokaryotic cells, and coevolution of host and symbiont. It is no surprise that symbioses between different groups of protists and prokaryotes continuously attract scientific attention (Gast et al. 2009; Nowack and Melkonian 2010; Dziallas et al. 2012; Edgcomb 2016; Samba-Louaka et al. 2019). Ciliates, one of the most numerous taxa of protists, harbor a plethora of diverse prokaryotes. The first-discovered endosymbionts of ciliates were Holospora, and now these and related Holospora-like bacteria (HLB) are, probably, among the best-studied prokaryotic symbionts of protists.
Holospora and HLB exhibit several fascinating features. The most conspicuous are the occupation of the host’s nucleus (Fig. 4.1), their cellular dimorphism with the eye-catching long infectious form, and their complex life cycle with an infectious stage. Thanks to the prominent localization and the atypical cell shape, these bacteria were reported already in very early microscopic studies devoted to Paramecium (Bütschli 1887). 130 years ago, bacteria in Paramecium nuclei were observed and described as Holospora (“whole spore”) by Wladimir Hafkine (Hafkine 1890), a well-known bacteriologist from the laboratory of Louis Pasteur. His descriptions were confirmed and formalized according to taxonomic rules a century later (Gromov and Ossipov 1981).
Different paramecia infected with various Holospora species. Holospora undulata (a) in the Paramecium caudatum micronucleus and Holospora parva (b) and Holospora curviuscula (c) in the macronuclei of their Paramecium hosts. The nuclei are heavily infected and appear swollen in size. Note the algal symbionts additionally harbored by Paramecium chlorelligerum (b) and Paramecium busaria (c). Living cells observed by differential interference contrast microscopy. Scale bars: 25 μm (a), 10 μm (b, c)
Holospora and HLB form a monophyletic group within the Holosporales (see Sect. 4.2.1). All of them are obligate intranuclear bacteria as they live “inside the control center” of their eukaryotic hosts (Schulz and Horn 2015). They are capable of vertical and horizontal transmission; thus, they are distributed from mother to daughter cells and can also infect new hosts after uptake from the environment (see Sect. 4.1.2).
The only known hosts for Holospora and HLB are ciliates. Relevant features of ciliates for the symbiotic interaction with HLB are their filter-feeding followed by phagocytosis, which provides to bacteria a possibility to enter the cell, and their nuclear dimorphism, as HLB reside in either somatic polyploid macronuclei or in the germline micronuclei and are restricted to one type or the other.
In this review, we will discuss new insights into adaptation, evolution, and host interactions of the following Holospora species: H. undulata (type species), H. obtusa, H. elegans, H. acuminata, H. curviuscula, and “CandidatusFootnote 1 H. parva.” As there has been no update since the most recent reviews on the diversity of Holospora (Fokin and Görtz 2009; Fujishima and Kodama 2012), we will skip H. recta (Fokin 1991), H. curvata (Fokin and Sabaneyeva 1993), and H. bacillata (Fokin 1989). The group here termed HLB includes the following bacteria: “Ca. Preeria caryophila” (Potekhin et al. 2018, basonym: Holospora caryophila), “Ca. Gortzia infectiva” (Boscaro et al. 2013), “Ca. G. shahrazadis” (Serra et al. 2016), “Ca. G. yakutica” (Beliavskaia et al. 2020), and “Ca. Hafkinia simulans” (Fokin et al. 2019).
“Ca. Paraholospora nucleivisitans” (Eschbach et al. 2009) is only distantly related and lacks typical HLB characteristics (see Sect. 4.2.1). Thus, we will not consider it here as HLB.
1.1 Symbioses between Paramecium and Holospora-like bacteria
Holospora are obligate intracellular bacteria, i.e., cultivation attempts on artificial media outside the host have not been successful so far (Fokin and Görtz 2009). Inside Paramecium, they can elicit dramatic alterations of the host’s nuclear structure (Fig. 4.1) and impact host growth and fitness (see Sect. 4.3.2). There are no indications that they are required by their host under any circumstance. Thus, they can be considered parasites. Nevertheless, we use the more general term “symbiont” in this review as Paramecium and Holospora may form an intimate, long-term interaction; hence, the definition of symbiosis according to de Bary, 1878 (de Bary 1879; Oulhen et al. 2016) applies.
All Holospora species are strictly associated with a Paramecium species. This pattern of host specificity indicates that the intranuclear symbionts coevolved with their paramecia hosts. Very rarely Holospora can enter the “wrong” host species and even complete its infection cycle (Fokin et al. 2005), but yet under laboratory conditions such associations are very unstable and quickly disappear.
In the following, we will review publications and ongoing efforts conducted to unravel the molecular adaptations of these bacteria to their unusual lifestyle and the host’s employment in the symbiosis. Furthermore, we aim to provide insights into the ecological and evolutionary consequences of this interaction.
1.2 Infection, Life Cycle, and Cellular Dimorphism
Holospora and all HLB (see Sect. 4.2.3) display a complex life cycle connected to the infection process and are characterized by two different cell morphologies that serve distinct functions (Figs. 4.2 and 4.3). The reproductive forms (RF) are typical bacterial rod-shaped cells (0.4–1.0 × 2.0–4.0 μm). They can be found multiplying inside the host nucleus. At some point, they differentiate into infectious forms (IF). These cells are much longer than RF and can reach up to 20.0 μm. IF shapes can be straight, spindle-shaped, curved, or sigmoidal (Fig. 4.2; reviewed by Fokin and Görtz 2009; Potekhin et al. 2018). For several decades, these differences served as one major diagnostic character for discrimination of Holospora and HLB species (Gromov and Ossipov 1981; Fokin et al. 1996; Görtz and Schmidt 2005; Fokin and Görtz 2009; Schweikert et al. 2013).
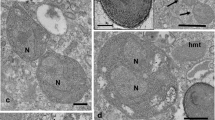
Nuclear infections by Holospora and Holospora-like bacteria. Holospora undulata (a) and Holospora acuminata (b) in the micronucleus (Mic) of their hosts. Infected macronuclei (Mac) harboring Holospora obtusa (c, d), Preeria caryophila (e), and a double infection (f) with H. obtusa (arrow head) and Preeria caryophila (white arrow). The majority of H. obtusa are present as reproductive forms in C, while in D the number of infectious forms has increased. Scale bars: 10 μm
Tripartite compartmentalization of infectious forms. The typical cell structure of infectious forms (IF) comprises a recognition tip (arrow), the periplasmic lumen (P), and the cytoplasm (C). Transmission electron micrographs (a-c) show IF of Holospora obtusa exiting from a food vacuole (a), inside the host cytoplasm (b) and the macronucleus (c). A three-dimensional atomic force microscopy image (d) depicts two IF of Gortzia infectiva. Note a slight depression of the surface of both bacteria at the polar recognition tip (arrow). Scale bars: 2 μm. Images were kindly provided by Dr. Elena Sabaneyeva, St. Petersburg State University (d), and Prof. Sergei Fokin, St. Petersburg State University & University of Pisa (a-c)
On the ultrastructural level (Fig. 4.3), IF are subdivided into recognition tip (also termed infection tip), an enlarged periplasmic lumen, and the remaining condensed cytoplasm (Ossipov 1981; Görtz and Wiemann 1989). The recognition tip plays an important role during the escape of IF from the phagosome (Fig. 4.3a) and in penetration of the nuclear membrane.
IF are the agents of horizontal transmission (Fig. 4.4a). They are released from the nucleus either during cell division (Fig. 4.4b) or at cell death and can persist for a certain time outside a host cell (Fujishima et al. 1991). A new infection cycle starts after phagocytosis of IF by a Paramecium cell. Usually, bacteria inside the phagosome are digested, but IF can avoid this fate by escaping the digestive vacuole with the recognition tip spearheading the exit (Fig. 4.4a). This process is triggered by the acidification of the phagosome. Inhibitors of vacuolar-type ATPases, which block acidification, prevent IF from leaving the vacuole (Fujishima et al. 1997). The importance of acidification for the maturation of IF was also shown by experiments where IF of H. obtusa were microinjected into the macronucleus of Paramecium caudatum bypassing all intermediate stages of the infection cycle. These IF did not form constrictions and failed to differentiate into RF (Skovorodkin et al. 2001). Acidification of isolated IF of H. obtusa induces the production of an IF-specific antigen (Kawai and Fujishima 2000).
Horizontal and vertical transmission of Holospora. (a) Major stages of the infection cycle of Holospora starting with the uptake of an extracellular infectious form (IF) via phagocytosis followed by incorporation in a food vacuole. The IF escapes from the vacuole with recognition tip oriented forward and gradually matures into the activated form. Once in the cytoplasm, the IF recruits the host cytoskeleton for intracellular motility. A perpendicular situated actin comet tail moves the IF to the target nucleus (here the macronucleus) where it penetrates the nuclear membrane again utilizing the recognition tip. Inside the nucleoplasm, the IF constricts and differentiates into reproductive forms (RF). These multiply and can undergo further differentiation from RF into IF. (b) Cell division of Paramecium and division of Holospora-infected nucleus. IF accumulate in the connecting piece, a structure bridging the dividing nuclei, while RF remain attached to the chromatin in the nuclei of daughter cells
It is debated and remains unclear if the phagosome membrane is collapsed in the process of IF escape and the bacteria are naked in the Paramecium cytoplasm (Fujishima 2009) or if the symbionts remain surrounded by remnants of the vacuole membrane (Ossipov 1981; Görtz and Wiemann 1989). Similar to intracellular pathogens like Listeria monocytogenes, Shigella flexneri, and Rickettsia conorii (Stevens et al. 2006; de Souza and Orth 2015), Holospora repurposes the host’s actin cytoskeleton in order to move intracellularly and to reach the target nucleus. The actin tail polymerizes at the side of the IF, occasionally nearly perpendicular to the longitudinal cell axis, and serves as driving force for propulsion through the Paramecium cytoplasm (Sabaneyeva et al. 2009).
Isolated IF can reach the nucleus of a new host within less than 60 min under laboratory conditions. The quick invasion of host nuclei by Holospora is accompanied by extensive ruffling and perturbations of the nuclear envelope (Ossipov 1981; Görtz and Fokin 2009). It implies highly specific recognition mechanisms and bacterial effectors. Indeed, some specific proteins crucial for the escape from the digestive vacuole and invasion of the target nucleus have been biochemically identified (Dohra et al. 1994; Iwatani et al. 2005; Abamo et al. 2008).
Once the nucleus is reached, the IF penetrates the nuclear envelope recognition tip oriented forward. Inside the organelle, IF constrict and differentiate into RF, which will undergo regular bacterial cell divisions until the next differentiation into IF (Fig. 4.4a).
Species-specific targeting to only one type of the host nuclei is another major feature used for Holospora diagnosis (Fokin and Görtz 2009). Rarely, the nucleus recognition is not precise and symbionts may end up in the nontarget compartment, especially in course of massive infections (Borchsenius et al. 1990; Ossipov et al. 1993). Still, with exception of H. curviuscula, which was able to colonize simultaneously both nuclei and to stay in the nonspecific micronucleus for 3–5 months (Borchsenius et al. 1990), Holospora species stop their proliferation and are very quickly lost from the nontarget nucleus (Ossipov et al. 1993; Lebedeva, Skoblo, Ossipov, pers. comm.).
1.3 Molecular Adaptations to an Intranuclear Lifestyle
The uncommon capability to live and replicate inside a host nucleus is characteristic to Holospora and HLB. Obviously, successful infection of nuclei and stable symbiosis (over numerous cell divisions of the host) requires specific adaptations.
Four Holospora genomes have been sequenced so far (Table 4.1), i.e., the micronucleus-specific species H. undulata (Dohra et al. 2013, 2014) and H. elegans (Dohra et al. 2014) and the macronucleus-specific H. obtusa (Dohra et al. 2014) and H. curviuscula (Garushyants et al. 2018). The latter is a symbiont of Paramecium bursaria, while the other three infect Paramecium caudatum. Furthermore, genome assembly and annotation are in progress for Preeria caryophila (Potekhin, pers. comm). Even though none of the genomes is closed, they are relatively large for obligate intracellular bacteria with draft genome sizes ranging from 1.27 to 1.72 Mb for Holospora (Table 4.1), while Preeria has a smaller genome of ca. 1 Mb (Potekhin, pers. comm). Comparative analysis (Garushyants et al. 2018) of four Holospora draft genomes revealed that all contain a considerable fraction of repetitive DNA (up to 15%), transposases, and phage-related genes.
Holospora rely on their host for energy production and provision of amino acids. The single major metabolic pathway that is almost intact in all Holospora is the fatty acids synthesis (Garushyants et al. 2018). All four sequenced Holospora are unable to synthesize any amino acid and lack the majority of genes involved in energy production, e.g., basically all enzymes for glycolysis, the Entner-Doudoroff pathway, the pentose phosphate pathway, the citric acid cycle, and the components of the F1F0-ATPase (Garushyants et al. 2018). All of them possess the pyruvate dehydrogenase and can generate ATP by converting pyruvate to acetyl-CoA and further to acetoacetyl-CoA and acetoacetate (Garushyants et al. 2018). Noteworthy, all Holospora have a set of ribonucleotide reductases. Hence, given the intranuclear lifestyle, it is conclusive that Holospora might use host-derived nucleotides and/or ribonucleotides as energy source (Garushyants et al. 2018). Not only do they encode ribose transport and nucleotide transport proteins for their uptake (Linka et al. 2003; Garushyants et al. 2018), they are capable to interconvert them. As the intracellular abundance of ribonucleotides is estimated to be significantly higher than that of nucleotides, those have been suggested as preferred energy source for Holospora (Garushyants et al. 2018). At the same time, RNA synthesis in the transcriptionally inert micronucleus is several orders of magnitudes lower than in the somatic macronucleus (Freiburg 1988). This could represent the crucial difference between the two types of nuclei in terms of nucleus preference by Holospora species. Their ability to propagate exclusively in one type is, probably, connected with metabolic peculiarities of the respective species.
Regarding infection-related adaptations, there have been substantial efforts to characterize engaged components both on ultrastructural level and biochemically (for a summary see Fujishima 2009). Probably the best-characterized protein involved in the infection of Holospora is the secreted 89-kDa protein of the recognition tip (Iwatani et al. 2005). It interacts with the phagosome membrane during IF escape and forms a fine fibrous structure between bacterial and vacuolar membranes. Comparative genomics revealed that the corresponding gene is conserved in all four Holospora genomes (Garushyants et al. 2018). Another interesting, interspecies-conserved gene encodes the 5.4-kDa periplasm-specific peptide, which has been described as a major protein of the IF periplasmic region likely playing a crucial role in the differentiation from RF to IF (Dohra et al. 1997). Genome mining (Garushyants et al. 2018) in Holospora genomes revealed about fifty proteins either containing transmembrane helixes or predicted to be secreted. Among them is the outer membrane protein A (OmpA), which is encoded in multiple copies in all analyzed Holospora species. OmpA interacts with host glycoproteins and is required for efficient entry into the host cell in some Rickettsiales (Ojogun et al. 2012), well-studied obligate intracellular pathogens, which are related to Holospora (Fig. 4.5). However, almost none of the predicted secreted or extracellular proteins have orthologs with known function in other bacteria. Interestingly, Holospora seem to lack protein secretion systems besides the complete Sec system (Garushyants et al. 2018). In particular, no type IV VirB secretion system was found, which plays an important role in host interaction in many Rickettsiales (Gillespie et al. 2015). Glycosylation of surface structures is likely employed by Holospora, as a loss of IF infectivity was observed after exposure to alpha-mannosidase (Fujishima et al. 1991). As many glycoproteins are associated with virulence factors of medically significant pathogens (Schmidt et al. 2003), glycosylation of outer membrane components of Holospora might serve specific functions in infection and horizontal transmission.
Phylogenetic reconstruction of the order Holosporales demonstrating that Holospora and Holospora-like bacteria (HLB) share a common ancestor. Maximum likelihood tree was calculated with IQ-TREE based on an alignment of 94 16S rRNA gene sequences obtained from GenBank comprising1377 characters. The applied best fit evolutionary model is TVMe+R5. Numbers near nodes indicated Ultrafast Bootstrap support values of IQ-TREE. Numbers in brackets indicate sequences in collapsed groups. Symbols depict occupied intracellular localization (black circle – micronucleus; black ellipse – macronucleus; white ellipse – occasionally macronucleus; white ellipse with bars – close to macronucleus; no symbol – cytoplasm). Scale bar corresponds to 0.07 sequence divergence. Ca. stands for Candidatus
As mentioned before, nonmotile Holospora use the host’s cytoskeleton for intracellular movement (Sabaneyeva et al. 2009) by employing actin polymerization as a driving force for propulsion through Paramecium cytoplasm. The actin tail, which in case of Holospora is not localized at the cell pole as, e.g., in Listeria (Lambrechts et al. 2008) but on its side, is composed of closely packed parallel microfilaments. Treatment with nocodazole, which interferes with the polymerization of microtubules, blocks the transport of IF to the nucleus and indicates that Paramecium microtubules are required as well in bacterial invasion of the nucleus (Sabaneyeva et al. 2005).
An intriguing and still open question is the ability of Holospora to discriminate between the host micro- and macronucleus. The difference between the nuclear envelope markers of Paramecium nuclei is still elusive, although the pore complexes have been proposed as nucleus-specific (Iwamoto et al. 2017). Various physical and chemical treatments (e.g., pH, temperature, detergents, etc.) revealed no effect on IF recognition and infection abilities. Thus, it was speculated that bacterial surface proteins might not play a crucial role in organelle targeting (Fujishima et al. 1991).
Once Holospora are inside their target nucleus and differentiated into RF, they exhibit a strong affinity to host chromatin that IF lack (Görtz and Wiemann 1989; Fokin et al. 1996). This difference between RF and IF is important for their vertical transmission (see Sect. 4.1.5). It is intriguing to speculate about additional interactions of Holospora with components of the host nucleus and their potential outcome, e.g., alteration of host gene expression (see Sect. 4.1.4, 4.3.2) or symbiont distribution at host division. Potential factors involved in intranuclear symbiont-host crosstalk might be IF surface proteins 25 kDa and 50 kDa of size that were identified to specifically bind to Paramecium nuclear proteins (Ehrsam and Görtz 1999).
1.4 Host-Symbiont Compatibility and Dissection of the Infection Process
The complex series of events leading to the successful establishment of a symbiotic association can be interrupted at different stages. It is well known that the outcome of an infection depends on the combination of both partners’ genotypes (Lambrechts et al. 2006), which applies also to symbioses between ciliates and Holospora. Some Paramecium strains can be considered as universal recipients for all strains of a certain symbiont species, while others are resistant to infection (Barhey and Gibson 1984; Fujishima and Fujita 1985; Rautian et al. 1990, 1993; Skoblo et al. 1996, 2001; Bella et al. 2016). The molecular mechanisms of crosstalk between Paramecium and Holospora are unknown, though it was shown that H. obtusa alters the expression of multiple host genes after establishing endosymbiosis (Nakamura et al. 2004).
The infection cycle of Holospora species has been studied in great detail (Borchsenius et al. 1992; Rautian et al. 1993; Skoblo and Lebedeva 1993; Skoblo et al. 1996, 2001; Kawai and Fujishima 2000). It comprises following distinct stages (Fig. 4.4): (1) entrance by phagocytosis, (2) escape from the food vacuole, (3) transport to the nuclei and nucleus penetration, (4) differentiation of IF into RF, (5) propagation of RF in the nucleus, and (6) maturation of RF into next generation of IF. Each of these stages can be blocked in certain combinations of partners. The most controversial is selective feeding of paramecia and thereby avoiding the ingestion of Holospora that has not been firmly proven. On the other hand, carbohydrate residues on the IF surface are important for engulfment by Paramecium (Sabaneyeva, pers. comm.) and some strains indeed do not engulf Holospora (Skoblo et al. 1996).
An interesting outcome in some incompatible host–symbiont combinations is the so-called symbiogenic lysis, in which Holospora simultaneously disintegrate in the host nuclei (Ossipov et al. 1993; Skoblo et al. 2001). Symbiont cells swell, their outer and cytoplasmic membranes visibly separate, and ribosomes disappear. Then bacterial outer membranes are disrupted and the protoplasts finally lyse (Ossipov et al. 1993). This phenomenon might be related to an unknown Paramecium defense mechanism or it could be operated by the bacteria themselves. Virus-induced lysis cannot be ruled out but seems unlikely insofar as no viruses have been observed in Holospora by transmission electron microscopy. However, in the genome of H. undulata, a possibly functional prophage is encoded (Garushyants et al. 2018).
Symbiosis establishment between Holospora and Paramecium is a discrete process that can be interrupted at different stages, confirming that these are independently controlled. The regular arresting in a particular combination of partners is a strong evidence of its genetic determination. Thus, the blockages may be considered as phenotypic markers of genes involved in symbiosis control. Genetic analysis of Paramecium bursaria susceptible and resistant to H. curviuscula confirmed that some infection stages are controlled by several host genes (Makarov, Skoblo and Ossipov, unpublished). Transplantation of macronuclear karyoplasm from susceptible Paramecium strains to resistant ones conferred the latter the ability to be infected by Holospora and allowed to deduce at least three Paramecium genes involved in susceptibility to infection (Rautian et al. 1996).
1.5 Holospora-Induced Changes of Host Cellular Machineries
Infections with Holospora (and HLB) comprise numerous consequences for affected Paramecium cells. First we will discuss morphological and ultrastructural alterations, while ecological and evolutionary consequences will be discussed later (see Sect. 4.3).
During various steps of their infection cycle, Holospora interfere with host membranes, the cytoskeleton, and even host chromatin. Most prominent alterations induced by Holospora are changes in size and shape of infected nuclei (Fig. 4.1). Besides, Holospora infection often leads to a complete loss of micronucleus, micronucleus aberrations, or appearance of additional micronuclei in the host (Ossipov 1981).
The nuclei can be completely filled with bacterial cells and enormously swollen in size and volume. In micronuclear infections by H. undulata, the infected organelle can increase its volume up to 80 times (Fig. 4.1a; Ossipov 1981). Astonishingly, paramecia do not necessarily always suffer from such an occupation of their nuclei, but the effects can differ dramatically. Hyperinfections, when the macronucleus occupies the major volume of the cell and is densely packed with IF, almost always end lethally for the host cell in case of H. obtusa or H. curviuscula (Ossipov 1981; Borchsenius et al. 1983). It was also one cause of failure in the formation of a stable symbiotic system between Paramecium strains and Preeria caryophila, while in some other cases paramecia could not survive exposure to P. caryophila at early stages of infection development for unknown reasons (Potekhin et al. 2018).
The universal consequence of Holospora presence in the nuclei is a decrease of DNA content, dispersion of chromatin and nuclear aberrations, even when the infection was cured or disappeared (Ossipov 1981; Rautian et al. 1993). It is unknown how the intranuclear bacteria interact with the genetic material of the ciliate, but they do not cause significant damage to the integrity of the macronuclear genome (Potekhin et al. 1999).
Holospora can impact the regular course of sexual processes in their hosts. These are autogamy, a process of self-fertilization, and conjugation (Mulisch 2003). During conjugation, two ciliate cells adhere to each other and build a temporary cytoplasmic bridge. The micronuclei of each conjugant cell undergo meiosis and then mitosis, and haploid gametic pronuclei are exchanged between the paired cells. In each cell, they fuse to form zygotic nucleus, which divides mitotically. Anlagen of new micronuclei and macronuclei start to develop, while the old macronucleus degrades gradually. Similarly, autogamy involves meiosis and further mitosis of the micronuclei and fusion of haploid pronuclei but in the same cell, followed by development of the new macronucleus and disintegration of the old one. Thus, the intracellular habitat of Holospora species is destroyed at each sexual event, which can occur more or less frequently, depending on the biology of the host species. For example, species of the Paramecium aurelia complex pass autogamy every 25–30 vegetative divisions (Potekhin et al. 2018), while for Paramecium caudatum or Paramecium bursaria autogamy has never been observed and conjugation can be rare. The intranuclear bacteria have evolved different strategies to cope with sexual processes of their host (see Sect. 4.3.2). For example, H. undulata inhibits conjugation as ultimate consequence (Görtz and Fujishima 1983; Fokin and Görtz 2009). Preeria caryophila, instead, does not prevent sexual processes in its host (Potekhin et al. 2018), but reinfects the new macronuclear anlagen (see Sect. 4.2.2). Elimination of the infection with H. undulata resulted in retrieval of host’s ability for sexual processes (Ossipov 1981). Interestingly, H. elegans occasionally produce irreversible changes in the micronucleus that when ciliates were cured of the infection, they could not proceed with regular conjugation (Fujishima and Görtz 1983).
Transmission of Holospora also involves the modification of typical Paramecium cell structures and processes. During its vegetative cell cycle, Paramecium reproduces by binary fission and its two types of nuclei undergo mitosis (micronuclei) or amitotic division (macronuclei). Once the cell divides, the bacterial symbionts are transmitted along with their host organelle. Over the course of host nucleus division, Holospora induce formation of the so-called connecting piece, resulting of IF concentration in a particular median body of the dividing nucleus (Fig. 4.4b). This process has been intensively studied in Holospora and HLB (Fokin et al. 1996; Fokin and Görtz 2009). Holospora IF remain in the connecting piece linking the parts of dividing nucleus. While RF are accumulated in the new nuclei due to their high chromatin affinity and are distributed to the clonal offspring, the IF are collected in the connecting piece in order to maximize their exit from the host and further transmission success (Fig. 4.4b). Indeed, after the karyokinesis, the connecting piece gets in cyclosis and is eventually expelled from the cytoproct, so that IF can start a new infection cycle (Wiemann and Görtz 1989; Fokin et al. 1996). The formation of the connecting piece has been used to differentiate between “classic” Holospora species and other bacteria, here termed HLB (Fokin et al. 1996). None of the latter are able to provoke connection piece formation in their hosts (see Sect. 4.2.2). However, since H. parva, the most recently described Holospora species found in the extremely rare Paramecium chlorelligerum, also does not induce connecting piece formation (Lanzoni et al. 2016), it cannot be considered as an apomorphic feature for all Holospora species.
2 Differences and Similarities between Classic Holospora and Holospora-Like Bacteria
Our understanding of the diversity, occurrence, and phylogeny of symbionts, not only those of ciliates and other protists, is constantly increasing. For Holosporaceae (Fig. 4.5), ten new reports were published recently (Boscaro et al. 2013, 2019; Lanzoni et al. 2016; Serra et al. 2016; Tashyreva et al. 2018; Potekhin et al. 2018; Fokin et al. 2019; Konecka and Olszanowski 2019; Takeshita et al. 2019; Beliavskaia et al. 2020). Characteristic features of HLB are the cellular dimorphism connected to the diphasic infectious life cycle, the special ultrastructural organization of IF shared with Holospora species, and occupancy of the host nucleus as a major niche in the host cell. The question if all these new symbionts should be considered as HLB or if they are simply a group of related bacteria with different characteristics is discussed (see Sect. 4.2.1).
2.1 Evolutionary History and Systematics of Holosporaceae
At the time of its description, the family Holosporaceae (Görtz and Schmidt 2005) was included in the order Rickettsiales within Alphaproteobacteria. Recently, the order Holosporales (Szokoli et al. 2016) was establised as a sister group to Rickettsiales and has been confirmed according to several phylogenetic reconstructions (Boscaro et al. 2019; Castelli et al. 2019; Fokin et al. 2019). This interpretation was then called into question (Muñoz-Gómez et al. 2019). Whether phylogenomics based on increasing data sets affiliates Holosporales with Rickettsiales or Rhodospirillales is awaiting future studies.
Still, Rickettsiales and Holosporales have many features in common. Both contain exclusively intracellular bacteria (with the prominent exception of the epibiotic parasite Deianiraea; Castelli et al. 2019) colonizing hosts from various groups of protists. Holosporales currently includes four families (Fig. 4.5) and all HLB are members of the family Holosporaceae. However, we recommend to avoid using the term HLB synonymously with Holosporaceae. The latter additionally comprises several recently detected symbionts, e.g. Mystax (Korotaev et al. 2020), Nesciobacter (George et al. 2019), Cytomitobacter (Tashyreva et al. 2018), Hydrogenosomobacter (Takeshita et al. 2019), and Fujishimia (Boscaro et al. 2019), which live and replicate within their host’s cytoplasm and apparently are not characterized by two morphological stages and do not clearly exhibit a life cycle with horizontal transmission.
On the other hand, two Holosporaceae members besides HLB show a certain degree of affinity for the host nucleus (Fig. 4.5): Bealeia paramacronuclearis (Szokoli et al. 2016) that generally accumulates in close proximity to the host macronucleus, and, more prominent, Paraholospora nucleivisitans (Eschbach et al. 2009). This symbiont of Paramecium sexaurelia alternates between the cytoplasm and the nucleus but never occupies both subcellular compartments simultaneously. Thus, this symbiont shares certain features associated to HLB but lacks the HLB-typical infectivity and cellular dimorphism. Furthermore, Paraholospora nucleivisitans branches separately in phylogenetic reconstructions (Fig. 4.5). Thus, it should not be considered as HLB.
2.2 Occurence of Holospora-Like Bacteria: Host Range and Cellular Compartments
Potential hosts for Holospora are members of the genus Paramecium, while HLB may also be harbored by other ciliates (reviewed by Fokin and Görtz 2009; Fujishima 2009). Next to IF morphology and occupied host compartment, the host species was used as a pivotal feature for the discrimination between Holospora species. As with the type of host nuclei, each Holospora species can infect only a single Paramecium species (Fokin and Görtz 2009; Fujishima and Kodama 2012). HLB, as in case of Preeria caryophila and potentially Gortzia infectiva, are not restricted to a single host species (Boscaro et al. 2013; Potekhin et al. 2018). Hafkinia simulans can infect hosts other than Paramecium (Fokin et al. 2019).
Interestingly, species infecting the huge polyploid macronucleus are more numerous than those colonizing the much smaller micronucleus (Fig. 4.5) with just H. elegans, H. undulata, and H. acuminata as micronuclear symbionts. It should be mentioned that there is increasing doubt if H. elegans and H. undulata truly represent two distinct species (Garushyants et al. 2018), especially as H. undulata is known for a high degree of morphological plasticity (Skoblo et al. 1996; Lebedeva, pers. comm). All described HLB infect exclusively macronuclei (Figs. 4.2 and 4.5).
The HLB phylogenetically closest to the genus Holospora is Hafkinia simulans. It does not infect Paramecium but has been found in the brackish water ciliate Frontonia salmastra (Fokin et al. 2019). Both Paramecium and Frontonia belong to the order Peniculida. Hafkinia differentiates into RF and IF, the latter showing compartmentalization typical for Holospora. Still, Hafkinia IF differ from Holospora IF as they exhibit ultrastructural variability and present occasionally two recognition tips (Fokin et al. 2019). Furthermore, the IF of Hafkinia are the largest described so far (up to 30 μm). They have a very peculiar spindle form, which strongly resembles in shape and dimensions the diatom Phaeodactylum tricornutum, a prey organism of Frontonia salmastra. This morphology might have evolved to increase the likelihood of phagocytosis and, thus, horizontal transmission success (Fokin et al. 2019).
The only HLB genus yet with more than a single described species is Gortzia. It comprises G. infectiva from Paramecium jenningsi (Boscaro et al. 2013), G. shahrazadis from Paramecium multimicronucleatum (Serra et al. 2016), and G. yakutica from Paramecium putrinum (Beliavskaia et al. 2020). G. infectiva was isolated from a habitat in which its host organism, Paramecium jenningsi, co-occurred with Paramecium quadecaurelia cells. The latter carried G. infectiva in the macronucleus, but when monoclonal strains were established the infection was lost from Paramecium quadecaurelia. Reinfection experiments revealed that the bacteria could enter the nucleus but failed to complete their life cycle (Boscaro et al. 2013). All three Gortzia species infect the macronuclei of their hosts and present two distinct morphologies. Their IF have typical appearance of Holospora IF, as observed by light (Beliavskaia et al. 2020), transmission electron microscopy (Boscaro et al. 2013; Serra et al. 2016), and atomic force microscopy (Fig. 4.3d). A special case is the cytoplasmic extrusion in the periplasmic space observed in IF of G. shahrazadis (Serra et al. 2016).
Preeria caryophila [basonyms: Holospora caryophila; alpha particles] infecting the macronucleus of Paramecium aurelia is known since the 1960s (Preer 1969) and has been recently redescribed as type species of the new genus (Potekhin et al. 2018). Preeria caryophila also alternates tiny RF and short IF (max. 6 μm, Fig. 4.2e) in its life cycle. It exhibits the broadest host range described for HLB comprising at least eleven Paramecium species (Potekhin et al. 2018).
Interestingly, HLB show a higher degree of flexibility not only in regards of ciliate host species but also in the confinement to the nuclear compartment. All HLB have been observed occasionally in the cytoplasm of their hosts (e.g., Preeria caryophila in Fig. 4.6). IF in the cytoplasm might occur as a result of inversion of the infection process, which allows the release of IF from the infected nucleus (Fokin et al. 2019). The latter may facilitate the exit from the ciliate cell for intranuclear symbionts unable to induce the connecting piece formation (Fig. 4.3b). Another, nonexclusive explanation is that occasional, potentially temporary, visits to the cytoplasm are a part of the life cycle of these HLB. Evidence therefore has been obtained in G. shahrazadis and P. caryophila. For G. shahrazadis, numerous IF and even multiplying RF were observed in Paramecium cytoplasm in long-persisting associations (Serra et al. 2016). In case of P. caryophila, singular IF often roam outside the macronucleus as observed during conjugation (Fig. 4.6a) or autogamy (Fig. 4.6b; Potekhin et al. 2018). This ability is likely responsible for the fact that some macronuclear HLB (but not Holospora) can infect Paramecium species that regularly undergo autogamy. Those are for example members of the Paramecium aurelia species complex and Paramecium jenningsi, which can harbor Preeria caryophila and Gortzia infectiva. Some Preeria IF are not enclosed in fragments of the old macronucleus but appear in the cytoplasm. These can immediately reinfect the new macronucleus once it is formed (Potekhin et al. 2018). Thus, with such an apparently effective strategy at hand, it is not surprising that Preeria does not prevent autogamy or conjugation (Fig. 4.6) of infected Paramecium strains (Potekhin et al. 2018).
Behavior of Preeria caryophila during sexual processes of their host. (a) Conjugating couple of Paramecium novaurelia, Preeria caryophila cells are present in both macronuclei (Mac) and some infectious forms (IF) are visible in cytoplasm (white arrows). (b) Postautogamous Paramecium biaurelia cell stained with lacto-aceto-orcein, IF are visible in old macronuclear fragments and in the cytoplasm (black arrows). Bright yellow structures are crystals, typical for paramecia Living (a) and fixed (b; with glutaraldehyde) cells observed by differential interference contrast microscopy. Scale bars: 30 μm (a) and 50 μm (b)
3 Ecological and Evolutionary Consequences of Symbiosis with Holospora and Holospora-Like Bacteria for Paramecium
3.1 Low Frequency of Paramecium–Holospora Symbioses in Nature
The low frequency of associations between Paramecium and their infectious bacterial symbionts in nature is paradoxical. Indeed, Paramecium caudatum, many species of the Paramecium aurelia complex, Paramecium bursaria, and Paramecium multimicronucleatum, the natural hosts of Holospora, Preeria, and Gortzia, are rather common ciliates, and potentially many strains are capable of harboring symbionts. Extensive infection studies (Potekhin et al. 2018) demonstrated that at least 20–30% of Paramecium aurelia strains can host P. caryophila. In parallel, all Holospora and HLB are highly infectious bacteria able to colonize their hosts quickly and efficiently. However, there are only dozens of Paramecium-Holospora and HLB associations known to ever have been isolated from the environment and maintained in laboratory collections. Why are infected ciliates not more prevalent?
One explanation is that symbionts may easily get lost under changing environmental conditions. Indeed, ciliates may face periods of nutrient surplus when they can divide much faster than their symbionts, thus critically diluting the number of intracellular bacteria per host. On the contrary, they may sometimes face starvation. Ciliates carrying parasitic bacteria as a burden would be outcompeted or, possibly, would not be able to supply the symbionts sufficiently with necessary metabolites. At the same time, under constant laboratory conditions H. obtusa can persist in Paramecium caudatum strains for at least 30 months, which corresponds to more than 1000 ciliate vegetative divisions (Ossipov 1981). Moreover, the associations between Paramecium and P. caryophila may last for years in the laboratory, for example Paramecium biaurelia strain 562 maintains P. caryophila already for more than 50 years (Preer 1969; Potekhin et al. 2018).
This discrepancy might be explained by the variation in environmental factors influencing the populations of paramecia carrying Holospora as symbionts. Paramecium caudatum populations became unstable and declined when exposed to variable temperature conditions. Furthermore, the impact of infection by H. undulata was additive and enhanced the overall negative effect of the variable environment on Paramecium (Duncan et al. 2011). Environmental fluctuations also caused a decrease in H. undulata prevalence in the host population (Duncan et al. 2011). Moreover, patterns of temporal and spatial environmental fluctuations can impact parasite spread and host population abundance in nature and should be considered in prediction of parasite transmission and epidemics (Duncan et al. 2013). Clearly, such fluctuations are avoided in laboratory culture maintenance.
Another possible cause of a low prevalence of symbiont-bearing ciliates in nature is that infected cells might face a higher risk of extinction. Misbalanced symbiosis leading to hyperinfection is one cause of death of a ciliate (see Sect. 4.1.5). It was shown that H. undulata infection of Paramecium caudatum frequently leads to karyopyknosis (irreversible condensation of chromatin) and further loss of the micronucleus during the first day of infection, which is not always lethal for a ciliate but decreases its fitness (Ossipov et al. 1983). Similarly, up to 54–90% of Paramecium bursaria cells were losing micronuclei after experimental infection by H. acuminata (Skoblo and Lebedeva 1993). The most likely explanation for this phenomenon is the damage of micronuclear membranes due to multiple events of bacterial penetration in experimental infection conditions. It is important to emphasize that multiple penetrations by Holospora under environmental conditions are rather unlikely according to the assumed low frequency of IF. Thus, any infected ciliate isolated from nature presumably contains a monoclonal strain of symbionts, as infection probably mostly starts with single IF entering the host (Ossipov 1981; Skoblo and Lebedeva 1993).
Finally, infected paramecia may get outcompeted in nature by symbiont-free ciliates, which do not have to share resources with bacterial residents. On the other hand, benefits provided by HLB under certain conditions might balance the cost of infection, e.g., the observed increased exponential growth rate in Paramecium when infected by P. caryophila (Bella et al. 2016) or increased stress tolerance (see Sect. 4.3.2).
Of course, it is also possible that infections with Holospora and HLB are not as rare in nature as perceived. The standard approach to search for bacterial infections is sampling and further isolation and cultivation of ciliates. Infected specimens might get quickly lost or simply overlooked during initial picking cells from environmental samples and introducing them into laboratory maintenance. In this regard, it is worth noting that in water samples collected in the last 7 years from ponds, streams, and ditches of Peterhof, a small suburb of Saint Petersburg, eight Paramecium species and all seven matching Holospora species and P. caryophila were retrieved (Lebedeva, pers. comm.). Continued efforts in the assessment of the diversity and occurrence of symbionts of protists will provide a better insight in this puzzling aspect of HLB epidemiology.
3.2 Interference of Intranuclear Symbionts with the Host Nuclei and Host Stress Response
Symbionts influence individual hosts as well as their populations. One of the most important criteria defining a population is interbreeding of its members. In ciliates, sexual processes result in the complete renovation of the nuclear apparatus in a very short time period, and there is no continuous line of either micronuclei or macronuclei in sexual generations. The micronucleus passes through a series of meiotic and mitotic divisions, while the old macronucleus gets completely demolished at each sexual process and is formed de novo from micronucleus derivatives. Thus, Holospora species either have to prevent conjugation of their host or get lost (Fokin 1998). Still, conjugation in presence of H. elegans was reported (Fujishima and Görtz 1983). Some bacteria managed to remain in the pronuclei, but survival of infected exconjugants was severely reduced compared to aposymbiotic cells as they were not able to form new macronuclei and regenerated the old ones (Fujishima and Görtz 1983).
While micronuclear symbionts may mechanically interfere with meiosis, there is no plausible explanation of inhibition of host conjugation by H. obtusa residing in the macronucleus than its influence on host gene expression. As discussed, Preeria caryophila does not prevent sexual processes in its host and, instead, temporarily escapes from the transforming nuclei into cytoplasm; there are no data concerning conjugation of ciliates infected by Gortzia or Hafkinia.
Holospora are parasites. A heavy infection of the Paramecium macronucleus by different Holospora results in a decreased fission rate of the ciliates (Ossipov 1981; Borchsenius et al. 1983). Only in case of the symbiosis between Paramecium chlorelligerum and H. parva the slow growth of infected cells was consistent with that of uninfected ones (Lanzoni et al. 2016). At the same time, no retarding effects on host divisions rates were reported for other symbionts (Ossipov 1981; Kaltz and Koella 2003; Castelli et al. 2016). Interestingly, if the host culture experiences unfavorable cultivation conditions and thus reduces cell division rate, H. undulata apparently becomes more virulent (Magalon et al. 2010; Dusi et al. 2015). Elevated host division rates, on the other hand, increased the levels of parasite vertical transmission and resulted in a near-complete loss of infectivity (Dusi et al. 2015). Insufficient time for the bacteria to mature into IF could explain at least partially these observations, but, obviously, the balance between host division rate as well as prevalence and infectivity of symbionts is rather delicate.
Paramecium have been shown to acquire heat-shock resistance (Hori and Fujishima 2003) and osmotic shock tolerance (Smurov and Fokin 1998) when infected by Holospora. This was considered as an advantageous effect of the symbiosis (Hori et al. 2008). An increase of Hsp70 expression is also known from other symbiotic systems (Kodama et al. 2014; Grosser et al. 2018). It might be either specifically induced by the symbionts or represent part of the Paramecium stress response to a large-scale infection. Nevertheless, elevated levels of Hsp70 allowed paramecia infected by H. obtusa to survive at nonpermissive temperatures (Hori and Fujishima 2003). Infected cells were able to maintain the ciliary movement and continued active swimming at temperatures above and below the physiological range of Paramecium (Fujishima et al. 2005). Heat resistance was not acquired by Paramecium caudatum infected by H. undulata, but this symbiont conferred osmotic shock tolerance to some strains (Duncan et al. 2010).
3.3 Epidemiology of Paramecium-Holospora Symbioses and Impact of Environmental Factors
Many epidemiological parameters of Paramecium-Holospora associations are still unknown. Success of infection is higher if more bacteria simultaneously enter the target nucleus, as each IF generates several multiplying RF. Even such small initial differences can strongly influence the subsequent intensity of infection (Fels et al. 2008). Interestingly, direct transmission from infected cell to recipient as occurring in nature is at any rate not less efficient than experimental infections utilizing homogenates of heavily infected paramecia. The presence of a single infected donor cell was sufficient to infect a population of naïve paramecia with the same rate and prevalence (Potekhin et al. 2018). Optimal parasite strategies may depend on the balance between local transmission and the capacity to reach new habitats through dispersal (Lion and Boots 2010). Surprisingly, Holospora-bearing ciliates tended to disperse less in interconnected microcosms (Fellous et al. 2011).
In the Paramecium–Holospora interaction, a negative correlation between the growth rate of the host and the parasite’s investment in horizontal transmission has been observed. The results suggest a tradeoff between efficient vertical and horizontal transmission. If conditions for Paramecium replication decline, the symbionts switch to horizontal transmission (Kaltz and Koella 2003). Addressing the effects of early and late stages of infection, parasite load, and food abundance, it was shown that a reduced availability of food and thus a lower division rate of the host correlates with a higher Holospora virulence (Restif and Kaltz 2006).
Paramecium offers sufficient resources to host multiple bacterial infections. Paramecia with double infections by Caedimonas varicaedens (Preer 1969; Schrallhammer et al. 2018) or Megaira polyxenophila (Schrallhammer et al. 2013) with P. caryophila, Megaira polyxenophila, and H. undulata (Lanzoni et al. 2019), and even H. obtusa and P. caryophila (Fig. 4.2f) are rarely but repeatedly detected in environmental samples. Simultaneous infection of both nuclei of Paramecium bursaria with H. curviuscula and H. acuminata was achieved many times (Borchsenius et al. 1983). Similarly, the presence of Caedimonas varicaedens in the macronucleus did not prevent an infection with micronuclear-specific Holospora (Skoblo et al. 1996). Even experimental double infections of naïve paramecia were obtained (Duncan et al. 2018), albeit at rather low frequencies. The most exceptional case was likely that of H. undulata (normally restricted to micronuclei) infecting a macronucleus already inhabited by H. obtusa (Lebedeva et al. 1992). Holospora can even serve as shuttle transporting free-living bacteria to the macronucleus (Fokin et al. 2004).
However, antagonistic interactions between different bacterial symbionts have also been observed. Resident symbionts might prevent the efficient colonization of the same host by other bacteria, even if both occupy different compartments (Fokin et al. 1987; Görtz 1987). Mixed infection experiments showed that competitive exclusion is more common than coexistence (Duncan et al. 2018). It is tempting to speculate that certain symbionts may provide their host colonization resistance against invasion by other, potentially harmful microorganisms (Plotnikov et al. 2019). Bacterial competition for the host cell, interactions of two different symbiont species in one host, and tradeoffs of multiple bacterial symbioses remain to be further studied.
4 Outlook and Perspectives
In this chapter, we aimed to summarize currently available data on the formation and maintenance of very peculiar symbiotic systems, where Holospora and HLB reside directly in the nucleus of their host. This field has experienced tremendous progress in the last decade. The expansion of state-of-the-art technologies, first of all Next-Generation Sequencing together with current microscopy and molecular biology techniques, now opens extremely interesting directions for further studies of Paramecium-Holospora and HLB symbioses.
The genomes of several Paramecium species have been sequenced and are available at ParameciumDB (Arnaiz et al. 2019), and the genomes of several Holospora species are either sequenced (Dohra et al. 2013, 2014; Garushyants et al. 2018) or in progress. These are the prerequisites for in-depth interaction analyses by transcriptomics. Comparative transcriptomics together with genetic dissection of the symbiotic systems will allow to detect the genes of host and symbiont differentially expressed at each stage of symbiosis development and maintenance. Further studies of such genes’ functions will approach the molecular interaction mechanisms of both partners and potentially may lead to the identification of new bacterial effectors.
Even in mutually beneficial symbiotic associations, excessive number of symbionts may become a heavy burden for the host decreasing its fitness and leading to defeat in local competition (Cunning and Baker 2014; Parkinson et al. 2017). In case of Holospora and HLB, which are not mutualistic, this problem of symbiont population control becomes crucial. A possible pathway for the regulation of symbionts could be production of antimicrobial peptides (AMP) by ciliates. AMP are an ancient defensive weapon of the eukaryotic cell (Wollman 2016) and have been reported from Paramecium caudatum (Cui et al. 2016). Examples of AMP targeting bacterial symbionts, not eliminating the microbial population but rather keeping it in check, are known from different host organisms (Mergaert 2018). Quorum sensing (QS) may be part of self-regulation mechanisms of the symbiont’s population in the host. Possibly, some of the numerous short (<100 amino acids) peptides with unknown function encoded in Holospora genomes (Garushyants et al. 2018) might be involved as QS signal.
The intimate localization of HLB in the nuclei of their hosts might offer possibilities for crosskingdom horizontal gene transfer (HGT). As the symbionts live and die in the nucleus, their DNA may occasionally be integrated into the host genome. While symbiont DNA would not be fixed in the somatic macronuclear genome, it can become a part of the generative micronucleus genome, which is a “safe haven” for noncoding DNA (Bétermier and Duharcourt 2014). If then bacterial genes are somehow retained in the developing macronuclear genome, they may get a chance to be expressed. Holospora are deficient for nearly all major pathways due to genome reduction. In addition, up to 15% of their genomes is represented by noncoding sequences (Garushyants et al. 2018). At the same time, Holospora switch between several stages and environments in their life cycle and perform complex interactions with the host during the infection process. Hence, Holospora belong to the same category of obligatory bacterial symbionts whose genomes are irreversibly shrinking (Wernegreen 2017; Husnik and Keeling 2019). Severely limited metabolic capacities put Holospora and HLB in absolute dependence of the host making the search for HGT promising. As Holosporales are considered as close relatives of free-living ancestors of mitochondria (Wang and Wu 2015), insights into Paramecium-Holospora and HLB symbioses might provide clues for initial stages in the transition from symbiont to organelle.
Notes
- 1.
Candidatus indicates that these bacteria cannot be cultivated outside their host and thus are not deposited as a pure culture at two culture collections, preventing their full valid description according to the International Code of Nomenclature of Prokaryotes. For brevity’s sake, we will omit Candidatus further on in the text. Organisms originally described before 1980 were given valid names even when cultivation could not be accomplished.
References
Abamo F, Dohra H, Fujishima M (2008) Fate of the 63-kDa periplasmic protein of the infectious form of the endonuclear symbiotic bacterium Holospora obtusa during the infection process. FEMS Microbiol Lett 280:21–27. https://doi.org/10.1111/j.1574-6968.2007.01023.x
Arnaiz O, Meyer E, Sperling L (2019) ParameciumDB 2019: integrating genomic data across the genus for functional and evolutionary biology. Nucleic Acids Res 48:D599–D605. https://doi.org/10.1093/nar/gkz948
Barhey K, Gibson I (1984) A study on the conditions for infection of Holospora caryophila, a macronuclear symbiont of Paramecium biaurelia. Micron Microsc Acta 15:261–268
Beliavskaia AY, Predeus AV, Garushyants SK et al (2020) New intranuclear symbiotic bacteria from macronucleus of Paramecium putrinum — “Candidatus Gortzia yakutica”. Diversity 12(5):198. https://doi.org/10.3390/d12050198
Bella C, Koehler L, Grosser K et al (2016) Fitness impact of obligate intranuclear bacterial symbionts depends on host growth phase. Front Microbiol 7:2084. https://doi.org/10.3389/fmicb.2016.02084
Bétermier M, Duharcourt S (2014) Programmed rearrangement in ciliates: Paramecium. Microbiol Spectr 2:MDNA3-0035–2014
Borchsenius O, Skoblo I, Lebedeva N, Ossipov D (1990) Occasional appearance and short-term persistence of symbiotic bacteria Holospora acuminata in macronucleus of ciliates Paramecium bursaria. Tsitologiia 32:578–582
Borchsenius O, Skoblo I, Lebedeva NA, Ossipov D (1992) The infectiousness of Holospora acuminata the intranuclear symbiont of the ciliate Paramecium bursaria 2. The impossibility of a symbiont system formation is a result of disturbances occuring at the stage when symbiotic bacterium leaves the. Tsitologiia 34:97
Borchsenius O, Skoblo I, Ossipov D (1983) Holospora curviuscula - a new species of macronuclear symbiotic bacteria of Paramecium bursaria. Tsitologiya 25:91–97
Boscaro V, Fokin SI, Schrallhammer M et al (2013) Revised systematics of Holospora-like bacteria and characterization of “Candidatus Gortzia infectiva”, a novel macronuclear symbiont of Paramecium jenningsi. Microb Ecol 65:255–267. https://doi.org/10.1007/s00248-012-0110-2
Boscaro V, Husnik F, Vannini C, Keeling PJ (2019) Symbionts of the ciliate Euplotes: diversity, patterns and potential as models for bacteria-eukaryote endosymbioses. Proc R Soc B Biol Sci 286:20190693. https://doi.org/10.1098/rspb.2019.0693
Bütschli O (1887) Protozoa. H. G. Bronns Klassen und Ordnungen des Tierreichs. Akadem. Verl.-Ges. Geest & Portig, Leipzig
Castelli M, Lanzoni O, Rossi L et al (2016) Evaluation of enrichment protocols for bacterial endosymbionts of ciliates by real-time PCR. Curr Microbiol 72(6):723–732. https://doi.org/10.1007/s00284-016-1006-z
Castelli M, Sabaneyeva E, Lanzoni O et al (2019) Deianiraea, an extracellular bacterium associated with the ciliate Paramecium, suggests an alternative scenario for the evolution of Rickettsiales. ISME J 13:2280–2294. https://doi.org/10.1038/s41396-019-0433-9
Cui P, Dong Y, Li Z et al (2016) Identification and functional characterization of an uncharacterized antimicrobial peptide from a ciliate Paramecium caudatum. Dev Comp Immunol 60:53–65. https://doi.org/10.1016/j.dci.2016.02.016
Cunning R, Baker AC (2014) Not just who, but how many: the importance of partner abundance in reef coral symbioses. Front Microbiol 5:400
de Bary A (1879) Die Erscheinungen der Symbiose. In: Vers. Deut. Naturforscher und Aerzte zu Cassel 1878. Strassburg: Strassburg
de Souza SM, Orth K (2015) Subversion of the cytoskeleton by intracellular bacteria: lessons from Listeria, Salmonella and Vibrio. Cell Microbiol 17:164–173. https://doi.org/10.1111/cmi.12399
Dohra H, Fujishima M, Hoshide K (1994) Monoclonal antibodies specific for periplasmic materials of the macronuclear specific bacterium Holospora obtusa of the ciliate Paramecium caudatum. Eur J Protistol 30:288–294. https://doi.org/10.1016/S0932-4739(11)80075-1
Dohra H, Suzuki H, Suzuki T et al (2013) Draft genome sequence of Holospora undulata strain HU1, a micronucleus-specific symbiont of the ciliate Paramecium caudatum. Genome Announc 1:1–2. https://doi.org/10.1128/genomeA.00664-13
Dohra H, Tanaka K, Suzuki T et al (2014) Draft genome sequences of three Holospora species (Holospora obtusa, Holospora undulata, and Holospora elegans), endonuclear symbiotic bacteria of the ciliate Paramecium caudatum. FEMS Microbiol Lett 359:16–18. https://doi.org/10.1111/1574-6968.12577
Dohra H, Yamamoto K, Fujishima M, Ishikawa H (1997) Cloning and sequencing of gene coding for a periplasmic 5.4 kDa peptide of the macronucleus-specific symbiont Holospora obtusa of the ciliate Paramecium caudatum. Zool Sci 14:69–75
Duncan AB, Dusi E, Schrallhammer M et al (2018) Population-level dynamics in experimental mixed infections: evidence for competitive exclusion among bacterial parasites of Paramecium caudatum. Oikos 127:1380–1389. https://doi.org/10.1111/oik.05280
Duncan AB, Fellous S, Accot R et al (2010) Parasite-mediated protection against osmotic stress for Paramecium caudatum infected by Holospora undulata is host genotype specific. FEMS Microbiol Ecol 74:353–360. https://doi.org/10.1111/j.1574-6941.2010.00952.x
Duncan AB, Fellous S, Kaltz O (2011) Reverse evolution: selection against costly resistance in disease-free microcosm populations of paramecium caudatum. Evolution (N Y) 65:3462–3474. https://doi.org/10.1111/j.1558-5646.2011.01388.x
Duncan AB, Gonzalez A, Kaltz O (2013) Stochastic environmental fluctuations drive epidemiology in experimental host-parasite metapopulations. Proc Biol Sci 280:20131747. https://doi.org/10.1098/rspb.2013.1747
Dusi E, Gougat-Barbera C, Berendonk TU, Kaltz O (2015) Long-term selection experiment produces breakdown of horizontal transmissibility in parasite with mixed transmission mode. Evolution (N Y) 69:1069–1076. https://doi.org/10.1111/evo.12638
Dziallas C, Allgaier M, Monaghan MT, Grossart H (2012) Act together—implications of symbioses in aquatic ciliates. Front Microbiol 3:1–17. https://doi.org/10.3389/fmicb.2012.00288
Edgcomb VP (2016) Marine protist associations and environmental impacts across trophic levels in the twilight zone and below. Curr Opin Microbiol 31:169–175. https://doi.org/10.1016/j.mib.2016.04.001
Ehrsam E, Görtz H-D (1999) Surface proteins of the gram-negative bacterium Holospora obtusa bind to macronuclear proteins of its host Paramecium caudatum. Eur J Protistol 35:304–308. https://doi.org/10.1016/S0932-4739(99)80008-X
Eschbach E, Pfannkuchen M, Schweikert M et al (2009) “Candidatus Paraholospora nucleivisitans”, an intracellular bacterium in Paramecium sexaurelia shuttles between the cytoplasm and the nucleus of its host. Syst Appl Microbiol 32:490–500. https://doi.org/10.1016/j.syapm.2009.07.004
Fellous S, Quillery E, Duncan AB, Kaltz O (2011) Parasitic infection reduces dispersal of ciliate host. Biol Lett 7:327–329. https://doi.org/10.1098/rsbl.2010.0862
Fels D, Vignon M, Kaltz O (2008) Ecological and genetic determinants of multiple infection and aggregation in a microbial host-parasite system. Parasitology 135:1373–1383. https://doi.org/10.1017/S0031182008004940
Fokin SI (1991) Holospora recta sp. nov. - a micronucleus-specific endobiont of the ciliate Paramecium caudatum. Tsitologiya 33:135–141
Fokin SI (1989) Bacterial endobionts of the ciliate Paramecium woodruffi. I Endobionts of the macronucleus. Tsitologiya 31:839–844
Fokin SI (1998) Strategies of the macronuclear endocytobionts of Paramecium during the sexual process of the host. Symbiosis 25:323–342
Fokin SI, Brigge T, Brenner J, Görtz H-D (1996) Holospora species infecting the nuclei of Paramecium appear to belong into two groups of bacteria. Eur J Protistol 32:19–24. https://doi.org/10.1016/S0932-4739(96)80072-1
Fokin SI, Görtz HD (2009) Diversity of Holospora bacteria in Paramecium and their characterization. In: Fujishima M (ed) Endosymbionts in Paramecium, Microbiology Monographs. Springer, Berlin Heidelberg, pp 161–201
Fokin SI, Ossipov DV, Skoblo I et al (1987) Nonospora macronucleata g. n., sp. n. - a vegetative nucleus symbiont of the ciliate Paramecium caudatum. Tsitologiia 29:963–970
Fokin SI, Sabaneyeva EV (1993) Bacterial endocytobionts of the ciliate Paramecium calkinsi. Eur J Protistol 29:390–395
Fokin SI, Schweikert M, Fujishima M (2005) Recovery of the ciliate Paramecium multimicronucleatum following bacterial infection with Holospora obtusa. Eur J Protistol 41:129–138. https://doi.org/10.1016/j.ejop.2004.11.007
Fokin SI, Serra V, Ferrantini F et al (2019) “Candidatus Hafkinia simulans” gen. nov., sp. nov., a novel Holospora-like bacterium from the macronucleus of the rare brackish water ciliate Frontonia salmastra (Oligohymenophorea, Ciliophora). Microb Ecol 77:1092–1106. https://doi.org/10.1007/s00248-018-1311-0
Fokin SO, Skovorodkin IN, Schweikert M, Görtz H-D (2004) Co-infection of the macronucleus of Paramecium caudatum by free-living bacteria together with the infectious Holospora obtusa. J Eukaryot Microbiol 51:417–424. https://doi.org/10.1111/j.1550-7408.2004.tb00388.x
Freiburg M (1988) Organization and expression of the nuclear genome. In: Görtz H (ed) Paramecium. Springer-Verlag, Berlin, Heidelberg, pp 141–154
Fujishima M (2009) Infection and maintenance of Holospora species in Paramecium caudatum. In: Fujishima M (ed) Endosymbionts in Paramecium. Microbiology Monographs. Springer, Berlin Heidelberg, pp 201–225
Fujishima M, Dohra H, Kawai M (1997) Quantitative changes in periplasmic proteins of the macronucleus-specific bacterium Holospora obtusa in the infection process of the ciliate Paramecium caudatum. J Eukaryot Microbiol 44:636–642
Fujishima M, Fujita M (1985) Infection and maintenance of Holospora obtusa, a macronucleus-specific bacterium of the ciliate Paramecium caudatum. J Cell Sci 76:179–187
Fujishima M, Görtz H-D (1983) Infection of macronuclear anlagen of Paramecium caudatum with the macronucleus-specific symbiont Holospora obtusa. J Cell Sci 64:137–146
Fujishima M, Kawai M, Yamamoto R (2005) Paramecium caudatum acquires heat-shock resistance in ciliary movement by infection with the endonuclear symbiotic bacterium Holospora obtusa. FEMS Microbiol Lett 243:101–105. https://doi.org/10.1016/j.femsle.2004.11.053
Fujishima M, Kodama Y (2012) Endosymbionts in Paramecium. Eur J Protistol 48:124–137. https://doi.org/10.1016/j.ejop.2011.10.002
Fujishima M, Nagahara K, Kojima Y, Sayama Y (1991) Sensitivity of the infectious long form of the macronuclear endosymbiont Holospora obtusa of the ciliate Paramecium caudatum against chemical and physical factors. Eur J Protistol 27:119–126. https://doi.org/10.1016/S0932-4739(11)80333-0
Garushyants SK, Beliavskaia AY, Malko DB et al (2018) Comparative genomic analysis of Holospora spp., intranuclear symbionts of paramecia. Front Microbiol 9:738. https://doi.org/10.3389/fmicb.2018.00738
Gast RJ, Sanders RW, Caron DA (2009) Ecological strategies of protists and their symbiotic relationships with prokaryotic microbes. Trends Microbiol 17:563–569. https://doi.org/10.1016/j.tim.2009.09.001
George EE, Husnik F, Tashyreva D et al (2020) Highly reduced genomes of protist endosymbionts show evolutionary convergence. Curr Biol 30(5):925–933.e3. https://doi.org/10.1016/j.cub.2019.12.070
Gillespie JJ, Kaur SJ, Rahman MS et al (2015) Secretome of obligate intracellular Rickettsia. FEMS Microbiol Rev 39:47–80. https://doi.org/10.1111/1574-6976.12084
Görtz H-D (1987) Different endocytobionts simultaneously colonizing ciliate cells. Ann N Y Acad Sci 503:261–268
Görtz H-D, Fokin SI (2009) Diversity of endosymbiotic bacteria in Paramecium. In: Fujishima M, (ed) Endosymbionts in Paramecium. Microbiology Monographs. Springer, Berlin Heidelberg, pp 131–160
Görtz H-D, Fujishima M (1983) Conjugation and meiosis of Paramecium caudatum infected with the micronucleus-specific bacterium Holospora elegans. Eur J Cell Biol 32:86–91
Görtz H-D, Schmidt H (2005) Family III. Holosporaceae fam. Nov. In: Brenner D, Krieg N, Staley J, Garrity G (eds) Bergey’s manual of systematic bacteriology, 2nd edn. Springer, New York, US, pp 146–149
Görtz H-D, Wiemann M (1989) Route of infection of the bacteria Holospora elegans and Holospora obtusa into the nuclei of Paramecium caudatum. Eur J Protistol 24:101–109
Gromov BV, Ossipov DV (1981) Holospora (ex Hafkine 1890) nom. rev., a genus of bacteria inhabiting the nuclei of paramecia. Int J Syst Bacteriol 31:348–352. https://doi.org/10.1099/00207713-31-3-348
Grosser K, Ramasamy P, Amirabad A et al (2018) More than the “Killer Trait”: Infection with the bacterial endosymbiont Caedibacter taeniospiralis causes transcriptomic modulation in Paramecium host. Genome Biol Evol 10(2):646–656. https://doi.org/10.1093/gbe/evy024
Hafkine WM (1890) Maladies infectieuses des paramecies. Ann Inst Pasteur 4:148–162
Hori M, Fujii K, Fujishima M (2008) Micronucleus-specific bacterium Holospora elegans irreversibly enhances stress gene expression of the host Paramecium caudatum. J Eukaryot Microbiol 55:515–521. https://doi.org/10.1111/j.1550-7408.2008.00352.x
Hori M, Fujishima M (2003) The endosymbiotic bacterium Holospora obtusa enhances heat-shock gene expression of the host Paramecium caudatum. J Eukaryot Microbiol 50:293–298. https://doi.org/10.1111/j.1550-7408.2003.tb00137.x
Husnik F, Keeling PJ (2019) The fate of obligate endosymbionts: reduction, integration, or extinction. Curr Opin Genet Dev 58–59:1–8. https://doi.org/10.1016/j.gde.2019.07.014
Iwamoto M, Osakada H, Mori C et al (2017) Compositionally distinct nuclear pore complexes of functionally distinct dimorphic nuclei in the ciliate Tetrahymena. J Cell Sci 130:1822–1834. https://doi.org/10.1242/jcs.199398
Iwatani K, Dohra H, Lang BF et al (2005) Translocation of an 89-kDa periplasmic protein is associated with Holospora infection. Biochem Biophys Res Commun 337:1198–1205. https://doi.org/10.1016/j.bbrc.2005.09.175
Kaltz O, Koella JC (2003) Host growth conditions regulate the plasticity of horizontal and vertical transmission in Holospora undulata, a bacterial parasite of the protozoan Paramecium caudatum. Evolution (N Y) 57:1535–1542. https://doi.org/10.1111/j.0014-3820.2003.tb00361.x
Kawai M, Fujishima M (2000) Invasion of the macronucleus of Paramecium caudatum by the bacterium Holospora obtusa: fates of the bacteria and timings of invasion steps. Eur J Protistol 36:46–52. https://doi.org/10.1016/S0932-4739(00)80022-X
Kodama Y, Suzuki H, Dohra H et al (2014) Comparison of gene expression of Paramecium bursaria with and without Chlorella variabilis symbionts. BMC Genomics 15:183. https://doi.org/10.1186/1471-2164-15-183
Konecka E, Olszanowski Z (2019) Detection of a new bacterium of the family Holosporaceae (Alphaproteobacteria: Holosporales) associated with the oribatid mite Achipteria coleoptrata. Biologia (Bratisl) 74:1517–1522. https://doi.org/10.2478/s11756-019-00251-w
Korotaev A, Benken K, Sabaneyeva E (2020) “Candidatus Mystax nordicus” aggregates with mitochondria of its host, the ciliate Paramecium nephridiatum. Diversity 12(6):251. https://doi.org/10.3390/d12060251
Lambrechts A, Gevaert K, Cossart P et al (2008) Listeria comet tails: the actin-based motility machinery at work. Trends Cell Biol 18:220–227. https://doi.org/10.1016/j.tcb.2008.03.001
Lambrechts L, Fellous S, Koella JC (2006) Coevolutionary interactions between host and parasite genotypes. Trends Parasitol 22:12–16. https://doi.org/10.1016/j.pt.2005.11.008
Lanzoni O, Fokin SI, Lebedeva N et al (2016) Rare freshwater ciliate Paramecium chlorelligerum Kahl, 1935 and its macronuclear symbiotic bacterium “Candidatus Holospora parva”. PLoS One 11:e0167928. https://doi.org/10.1371/journal.pone.0167928
Lanzoni O, Sabaneyeva E, Modeo L et al (2019) Diversity and environmental distribution of the cosmopolitan endosymbiont “Candidatus Megaira”. Sci Rep 9:1–13. https://doi.org/10.1038/s41598-018-37629-w
Lebedeva N, Rodionova G, Skoblo I (1992) Maintenance of Holospora undulata, micronucleus-specific symbiotic bacterium of Paramecium caudatum, in macronucleus infected by Holospora obtusa. Tsitologiia 34:85
Linka N, Hurka H, Lang BF et al (2003) Phylogenetic relationships of non-mitochondrial nucleotide transport proteins in bacteria and eukaryotes. Gene 306:27–35. https://doi.org/10.1016/S0378-1119(03)00429-3
Lion S, Boots M (2010) Are parasites ‘“prudent”’ in space? Ecol Lett 13:1245–1255. https://doi.org/10.1111/j.1461-0248.2010.01516.x
Magalon H, Nidelet T, Martin G, Kaltz O (2010) Host growth conditions influence experimental evolution of life history and virulence of a parasite with vertical and horizontal transmission. Evolution (N Y) 64:2126–2138. https://doi.org/10.1111/j.1558-5646.2010.00974.x
Mergaert P (2018) Role of antimicrobial peptides in controlling symbiotic bacterial populations. Nat Prod Rep 35:336–356. https://doi.org/10.1039/C7NP00056A
Mulisch M (2003) Nuclei and sexual reprodution. In: Hausmann K, Hülsmann N, Radek R (eds) Protistology, 3rd edn. Schweizerbart Science Publishers, Stuttgart, pp 241–255
Muñoz-Gómez SA, Hess S, Burger G et al (2019) An updated phylogeny of the Alphaproteobacteria reveals that the parasitic Rickettsiales and Holosporales have independent origins. elife 8:e42535. https://doi.org/10.7554/eLife.42535
Nakamura Y, Aki M, Aikawa T et al (2004) Differences in gene expression of the ciliate Paramecium caudatum caused by endonuclear symbiosis with Holospora obtusa, revealed using differential display reverse transcribed PCR. FEMS Microbiol Lett 240:209–213. https://doi.org/10.1016/j.femsle.2004.09.036
Nowack ECM, Melkonian M (2010) Endosymbiotic associations within protists. Philos Trans R Soc B Biol Sci 365:699–712. https://doi.org/10.1098/rstb.2009.0188
Ojogun N, Kahlon A, Ragland SA et al (2012) Anaplasma phagocytophilum outer membrane protein a interacts with sialylated glycoproteins to promote infection of mammalian host cells. Infect Immun 80:3748–3760. https://doi.org/10.1128/IAI.00654-12
Ossipov D (1981) Problems of nuclear heteromorphism in the unicellular organisms. Nauka, Leningrad
Ossipov D, Borchsenius O, Skoblo I (1983) Ultrastructural changes in the nuclear apparatus of Paramecium caudatum induced by symbiotic bacteria Holospora undulata. Tsitologiia 25:33–38
Ossipov DV, Skoblo II, Borchsenius ON, Lebedeva NA (1993) Interactions between Paramecium bursaria (protozoa, Ciliophora, Hymenostomatida) and their nuclear symbionts. I. Phenomenon of symbiogenic lysis of the bacterium Holospora acuminata. Eur J Protistol 29:61–71. https://doi.org/10.1016/S0932-4739(11)80298-1
Oulhen N, Schulz BJ, Carrier TJ (2016) English translation of Heinrich Anton de Bary’s 1878 speech, ‘Die Erscheinung der Symbiose’ (‘De la symbiose’). Symbiosis 69:131–139. https://doi.org/10.1007/s13199-016-0409-8
Parkinson JF, Gobin B, Hughes WOH (2017) The more, the merrier? Obligate symbiont density changes over time under controlled environmental conditions, yet holds no clear fitness consequences. Physiol Entomol 42:163–172. https://doi.org/10.1111/phen.12186
Plotnikov AO, Balkin AS, Gogoleva NE et al (2019) High-throughput sequencing of the 16S rRNA gene as a survey to analyze the microbiomes of free-living ciliates Paramecium. Microb Ecol 78:286–298. https://doi.org/10.1007/s00248-019-01321-x
Potekhin A, Brigge T, Rautian M (1999) Genetic consequences of the intranuclear symbiosis Paramecium-Holospora analyzed by PFGE. In: Wagner E (ed) Symbiosis to Eukaryotism. University Press, Geneva, pp 169–177
Potekhin A, Schweikert M, Nekrasova I et al (2018) Complex life cycle, broad host range and adaptation strategy of the intranuclear Paramecium symbiont Preeria caryophila comb. nov. FEMS Microbiol Ecol 94:fiy076. https://doi.org/10.1093/femsec/fiy076
Preer LB (1969) Alpha, an infectious macronuclear symbiont of Paramecium aurelia. J Protozool 16:570–578
Rautian M, Vishnyakov A, Makarov S, Ossipov D (1996) Transformation of Paramecium caudatum clone resistant to infection by intranuclear symbiotic bacteria of genus Holospora. Eur J Protistol 32:135–140. https://doi.org/10.1016/S0932-4739(96)80090-3
Rautian MS, Skoblo II, Lebedeva NA, Ossipov DV (1993) Genetics of symbiotic interactions between Paramecium bursaria and the intranuclear bacterium Holospora acuminata, natural genetic variability by infectivity and susceptibility. Acta Protozool 32:165–173
Rautian MS, Skoblo II, Lebedeva NA, Ossipov DV (1990) Complementation in the symbiont-host system formation between the clones in the ciliate Paramecium caudatum and the stocks of the bacterium Holospora obtusa. Tsitologiia 32:584–591
Restif O, Kaltz O (2006) Condition-dependent virulence in a horizontally and vertically transmitted bacterial parasite. Oikos 114:148–158. https://doi.org/10.1111/j.2006.0030-1299.14611.x
Sabaneyeva EV, Derkacheva ME, Benken KA et al (2009) Actin-based mechanism of Holospora obtusa trafficking in Paramecium caudatum. Protist 160:205–219. https://doi.org/10.1016/j.protis.2008.11.006
Sabaneyeva EV, Fokin SI, Gavrilova EV, Kornilova ES (2005) Nocodazole inhibits macronuclear infection with Holospora obtusa in Paramecium caudatum. Protoplasma 226:147–153. https://doi.org/10.1007/s00709-005-0121-7
Samba-Louaka A, Delafont V, Rodier M-H et al (2019) Free-living amoebae and squatters in the wild: ecological and molecular features. FEMS Microbiol Rev 43:415–434. https://doi.org/10.1093/femsre/fuz011
Schmidt MA, Riley LW, Benz I (2003) Sweet new world: glycoproteins in bacterial pathogens. Trends Microbiol 11:554–561. https://doi.org/10.1016/j.tim.2003.10.004
Schrallhammer M, Castelli M, Petroni G (2018) Phylogenetic relationships among endosymbiotic R-body producer: bacteria providing their host the killer trait. Syst Appl Microbiol 41(3):213–220. https://doi.org/10.1016/j.syapm.2018.01.005
Schrallhammer M, Ferrantini F, Vannini C et al (2013) “Candidatus Megaira polyxenophila” gen. nov., sp. nov.: considerations on evolutionary history, host range and shift of early divergent rickettsiae. PLoS One 8:e72581. https://doi.org/10.1371/journal.pone.0072581
Schulz F, Horn M (2015) Intranuclear bacteria: inside the cellular control center of eukaryotes. Trends Cell Biol 25:339–346. https://doi.org/10.1016/j.tcb.2015.01.002
Schweikert M, Fujishima M, Görtz H (2013) Symbiotic associations between ciliates and prokaryotes. In: Rosenberg E, DeLong EF, Lory S et al (eds) The prokaryotes. Springer, Berlin, Heidelberg, pp 427–463
Serra V, Fokin S, Castelli M et al (2016) “Candidatus Gortzia shahrazadis”, a novel endosymbiont of Paramecium multimicronucleatum and a revision of the biogeographical distribution of Holospora-like bacteria. Front Microbiol 7:1704. https://doi.org/10.3389/fmicb.2016.01704
Skoblo I, Lebedeva NA (1993) The infectiousness of Holospora acuminata - an intranuclear bacterial symbiont of Paramecium bursaria (Ciliata). I Syngen specificity of infection Tsitologiia 35:92–99
Skoblo I, Makarov S, Ossipov D (2001) The analysis of symbiotic relationships nature diversity in Paramecium bursaria-Holospora curviuscula system. Tsitologiia 43:520–527
Skoblo II, Lebedeva NA, Rodionova GV (1996) The formation of the Paramecium-Holospora symbiotic system: a study of the compatibility of different symbiont isolates and host clones. Eur J Protistol 32:147–152. https://doi.org/10.1016/S0932-4739(96)80092-7
Skovorodkin IN, Fokin SI, Fujishima M (2001) Fates of the endonuclear symbiotic bacteria Holospora obtusa and Holospora undulata injected into the macronucleus of Paramecium caudatum. Eur J Protistol 37:137–145. https://doi.org/10.1078/0932-4739-00801
Smurov AO, Fokin SI (1998) Resistance of Paramecium caudatum infected with endonuclear bacteria Holospora against salinity impact. Proc Zool RAS 276:175–178
Stevens JM, Galyov EE, Stevens MP (2006) Actin-dependent movement of bacterial pathogens. Nat Rev Microbiol 4:91–101. https://doi.org/10.1038/nrmicro1320
Szokoli F, Castelli M, Sabaneyeva E et al (2016) Disentangling the taxonomy of Rickettsiales and description of two novel symbionts (“Candidatus Bealeia paramacronuclearis” and “Candidatus Fokinia cryptica”) sharing the cytoplasm of the ciliate protist Paramecium biaurelia. Appl Environ Microbiol 82:7236–7247. https://doi.org/10.1128/AEM.02284-16
Takeshita K, Yamada T, Kawahara Y et al (2019) Tripartite symbiosis of an anaerobic scuticociliate with two hydrogenosome-associated endosymbionts, a Holospora-related Alphaproteobacterium and a methanogenic archaeon. Appl Env Microbiol 85:e00854–e00819. https://doi.org/10.1128/AEM.00854-19
Tashyreva D, Prokopchuk G, Votýpka J et al (2018) Life cycle, ultrastructure, and phylogeny of new diplonemids and their endosymbiotic bacteria. MBio 9:1–20. https://doi.org/10.1128/mBio.02447-17
Wang Z, Wu M (2015) An integrated phylogenomic approach toward pinpointing the origin of mitochondria. Sci Rep 5:7949. https://doi.org/10.1038/srep07949
Wernegreen JJ (2017) In it for the long haul: evolutionary consequences of persistent endosymbiosis. Curr Opin Genet Dev 47:83–90. https://doi.org/10.1016/j.gde.2017.08.006
Wiemann M, Görtz H (1989) Release of the endonucleobiotic bacterium Holospora elegans from its host cell Paramecium caudatum. Eur J Protistol 25:100–108
Wollman F-A (2016) An antimicrobial origin of transit peptides accounts for early endosymbiotic events. Traffic 17:1322–1328. https://doi.org/10.1111/tra.12446
Acknowledgments
The research of endosymbiosis is currently supported by Research Innovation Fund of the Albert-Ludwigs University of Freiburg (M.S.) and by Russian Foundation for Basic Research grant 19-04-01256a (A.P.). We are grateful to Dr. E. Sabaneyeva and Prof. S. Fokin for the kind provision of images and to Dr. I. Nekrasova for useful comments and feedback while working on the manuscript. We wish to use an opportunity to thank the founders of Holospora studies, our teachers Prof. Hans-Dieter Görtz and Prof. Dmitry Ossipov, and members of their laboratories, M. Schweikert, T. Brigge, M. Rautian, and I. Skoblo, for introducing us to ciliate-bacterial symbiosis biology; our colleagues and collaborators G. Petroni, O. Kaltz, S. Fokin, E. Sabaneyeva, and N. Lebedeva for inspiring ideas and sharing of knowledge and data; last but not least our current and former groups members and students for stimulating discussions and continuous interest in intranuclear symbiosis.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Schrallhammer, M., Potekhin, A. (2020). Epidemiology of Nucleus-Dwelling Holospora: Infection, Transmission, Adaptation, and Interaction with Paramecium. In: Kloc, M. (eds) Symbiosis: Cellular, Molecular, Medical and Evolutionary Aspects. Results and Problems in Cell Differentiation, vol 69. Springer, Cham. https://doi.org/10.1007/978-3-030-51849-3_4
Download citation
DOI: https://doi.org/10.1007/978-3-030-51849-3_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-51848-6
Online ISBN: 978-3-030-51849-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)