Abstract
The Bdellovibrio and like organisms (BALOs) are obligate predators of gram negative bacteria, for which they have an absolute requirement in order to replicate and complete their life cycle. This peculiar life style profoundly affects their physiology and cellular biology and defines their ecology. BALOs are ubiquitous in soils and in water bodies; while they are not numerically dominant, they can be abundant and diverse. Among the water bodies, BALOs inhabit wastewater treatment plant reactors (WWTP) as well as other schemes where water is recycled. Their capacity to prey upon gram negative bacteria and their semi-generalist feeding suggest that they may play important roles in bacterial biomass turnover and in the reduction of numerous pathogens in these environments. This chapter first introduces the BALO’s life cycle, and features of their ecology and dynamics. It then presents our understanding of BALO community structure and dynamics under varying conditions in WWTPs, detailing latest research studies showing that BALO abundance is greatly influenced by the type of treatment applied to the wastewater. It ends by presenting open questions on our understanding of BALO effects on bacterial communities and by suggesting novels ways to address these questions, considering their capacity to improve wastewater treatment.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
1 Introduction
Bdellovibrio and like organisms (BALOs) are obligate predators of other bacteria. They have an absolute requirement for Gram negative prey in order to replicate and complete their life cycle. This peculiar life style profoundly affects their physiology and cellular biology and defines their ecology. We only partially know the molecular, physiological and structural features enabling this unique life style, and even less about their interactions with, and their effects on, microbial communities and trophic networks in the environment. Nonetheless, thanks to huge technological advances in molecular biology, including molecular ecology and biological computing over the past 15 years, significant strides have been made, providing novel understanding, yielding new concepts and approaches which make it possible to start bridging between cellular features and ecological outputs.
This chapter summarizes the knowledge on BALOs in wastewater treatment plants (WWTPs). Other microbial predators are active in wastewater, mainly protists and phages, and other bacterial predators of bacteria like Myxobacteria (for more details see Chapters by Kuppardt-Kirmse and Chatzinotas “Intraguild Predation: Predatory Networks at the Microbial Scale” and Furness et al. “Predatory Interactions Between Myxobacteria and Their Prey”). These won’t be treated here. Wastewater treatment plants offer “real life” conditions where complex, but mostly limited to the microbial scale, trophic interactions and high microbial diversity combine to provide a highly valuable output for the environment in general and for human communities in particular. Yet, WWTPs are also tractable and controlled engineered environments that can be manipulated and mimicked at various scales, providing unique opportunities to uncover and investigate ecological phenomena at the microbial dimension. In this chapter, we summarize what is known of predatory interactions between bacteria in WWTPs. We will then suggest how novel approaches may bring us closer to understanding their roles in water purification. This, in turn, may help improve WWTP operations by increasing ecological stability, remove pathogens, and provide alternatives to their costly implementation in low and medium income countries (LMICs).
2 The Biology of BALOs
The aim of this section is to provide the reader with some basic knowledge on the phylogeny, and life cycle of BALOs.
2.1 BALO Phylogeny, Distribution in the Environment, and Prey Range
BALO phylogeny and distribution. BALOs belong to the Proteobacteria. Until recently, most were affiliated to the δ-proteobacteria (Rotem et al. 2014). The discovery and isolation of novel bacterial strains has led to a reconsideration of their phylogeny, and to the creation of the class Oligoflexia that includes them (Hahn et al. 2017). Within the Oligoflexia, BALOs form the orders Bdellovibrionales and Bacteriovoracales. The former includes the family Bdellovibrionaceae and the latter the families Bacteriovoracaceae and the Halobacteriovoraceae (Koval et al. 2015; Hahn et al. 2017). In addition, a new family of predators, the Pseudobacteriovoracaceae of which only the type strain Pseudobacteriovorax antillogorgiicola is known is placed in the order Oligoflexales (McCauley et al. 2015; Hahn et al. 2017). As P. antillogorgiicola was isolated from a gorgonian octocoral on marine agar, it is not an obligate predator. Its predation mode (epibiotic or periplasmic, see below) is not known.
The only genus within the Bdellovibrionaceae is Bdellovibrio (Davidov and Jurkevitch 2004), of which two species have been defined: B. bacteriovorus and B. exovorus. B. bacteriovorus is found in soil, and associated with plant roots (Klein and Casida Jr. 1967; Uematsu 1980; Jurkevitch et al. 2000; Oyedara et al. 2016), and in freshwater, e.g. rivers and lakes (Davidov and Jurkevitch 2004; Hobley et al. 2012a; Li and Williams 2015). Both B. bacteriovorus and B. exovorus are found in wastewater, the latter has so far only been found in this habitat (Chanyi et al. 2013). B. bacteriovorus is a periplasmic predator: it invades the space between the cytoplasmic membrane and the outer membrane of its prey. In contrast, B. exovorus is epibiotic, meaning it remains attached to the outer side of its prey, feeding on it from the outside (Koval et al. 2012; Chanyi et al. 2013).
Bacteriovoracales are all periplasmic predators. Bacteriovorax stolpii is a freshwater (including wastewater) and soil bacterium within the Bacteriovoracaceae, a monophyletic offshoot of the Bdellovibrionaceae (Koval et al. 2015). Peredibacter starrii is also found in freshwater and soil environments (Davidov and Jurkevitch 2004). As the family name Peredibacteraceae is deemed illegitimate, P. starrii is classified as a genus within the Bacteriovoracaceae (LPSN Bacterio.net, http://www.bacterio.net/peredibacteraceae.html, May 13, 2019). Lately, it was found that Peredibacter sp. was the most abundant BALO predator in the upper layers of perialpine lakes while Bdellovibrionaceae and Bacteriovoracaceae were proportionally more abundant at greater depths (Paix et al. 2019). The second recognized family in the order Bacteriovoracales is Halobacteriovoraceae. Two species have been defined, i.e. H. marinus and H. litoralis. They form different clusters which preferentially populate estuarine or marine waters, and are apparently selected by salinity levels (Pineiro et al. 2013). They have not been found in freshwater (Koval et al. 2015). A few isolates have been retrieved from salt lakes (Pineiro et al. 2004).
Finally, a few isolates classify to the α-proteobacteria. These are Micavibrio aeruginosavorus and M. admirandus, both epibiotic predators isolated from wastewater and soil (Lambina et al. 1982, 1983; Davidov et al. 2006a). Micavibrio forms a deep branch lineage, sister to the Rhodospirillales but distinct from any other major α-proteobacterial groups (Davidov et al. 2006b; Wang et al. 2011).
The known BALO taxa have only been rarely isolated from or detected in terrestrial animals, including humans (Schwudke et al. 2001; Kikuchi et al. 2009; Iebba et al. 2013) but they appear to be more readily associated with aquatic animals (Kelley and Williams 1992; Wen et al. 2009; Cao et al. 2012, 2015; Richards et al. 2012; Welsh et al. 2015).
Prey Range
BALOs have so far been shown to exclusively prey on Gram negative bacteria, both in the planktonic, suspended phase as well as in biofilms (Kadouri et al. 2005, 2007). Moreover, they can destroy the biofilm matrix of Gram positive bacteria without consuming the cells (Im et al. 2018). BALOs are usually isolated and tested for prey range with bacterial strains from laboratory collections, and these may originate from various source (Chanyi et al. 2013; Cao et al. 2015; Enos et al. 2018). However, when tested for prey range using strains isolated from the same environment the BALO came from, the predators appeared to prefer these co-locating strains, possibly as a result of selection for locally prevailing conditions (Rice et al. 1998; Pineiro et al. 2004). Differences in prey range are also observed between strains of BALO predators belonging to the same species (Jurkevitch et al. 2000; Li et al. 2011). Conversely, different prey strains belonging to the same species are differentially “palatable” to a particular BALO (Jurkevitch et al. 2000; Dashiff et al. 2011; Li et al. 2011). BALOs prey equally well on pathogenic and commensal bacterial strains, as well as on bacteria resistant to antimicrobials (Dashiff et al. 2011; Kadouri et al. 2013).
So far, no universal prey, even for a specific habitat, has been found to be consistently more efficient at “baiting” BALOs. This includes Vibrio haemolyticus P5, which has been extensively used for isolating marine BALOs (Schoeffield and Williams 1990). As many strains can be used as prey in the laboratory, and since many prey may not be culturable (Rinke et al. 2013), the true prey range of BALOs under natural conditions is still not known. Importantly, and as known today, BALO phylogeny, prey range and prey phylogeny are unlinked.
2.2 Essentials of the BALOs’ Predatory Life Cycle
As mentioned above, BALOs exhibit a periplasmic or an epibiotic predatory life style. These appear to be fixed, and to not depend upon the prey (Chanyi et al. 2013).
BALOs actively and rapidly swim during a so-called attack phase (AP) using a single, polar flagellum, in search of prey cells. They possess chemotaxis systems, which they use to detect amino acids (LaMarre et al. 1977), high bacterial biomass (Chauhan and Williams 2006) and to a small extend, prey cells (Lambert et al. 2003). As of today, little is known on the biology of the epibiotic predators beyond their phylogeny and the visual description of their life cycle. Upon encounter, epibiotic predators attach to the prey’s cell wall, consume the prey content from the outside, to leave an empty cell, and grow by binary division. During predation, vesicle-like and remnants of lipid structures can be observed within and outside the prey cell (unpublished). Some genetic details are available, showing that epibiotic predators have significantly smaller genomes than periplasmic BALOs, encoding for up to half of the total secreted proteins found in periplasmic BALOs; they generate energy through glycolysis and the tricarboxylic acid cycle and lack biosynthetic pathways for essential amino acids, vitamins, and precursors, similarly to periplasmic predators (Pasternak et al. 2014). In both periplasmic and epibiotic predators, gene expression is largely altered between the AP and the growth phase (GP), with contrasting expression of motility and search genes and chromosome replication, translation, transcription, energy production and cell division genes (Wang et al. 2011; Karunker et al. 2013). For further detail see (Lambert et al. 2010; Wang et al. 2011; Karunker et al. 2013; Pasternak et al. 2013, 2014).
The life cycle of periplasmic predators is known in much finer details. Some of its main features are presented here. Periplasmic BALOs have absolute requirements for type IVa and type IVb pili for prey invasion, as well as for gliding motility (Evans et al. 2007; Mahmoud and Koval 2010; Avidan et al. 2017; Duncan et al. 2019). In order to enter the prey, the predator makes a hole in the prey’s cell wall, and squeezes through it (Kuru et al. 2017). All the while, the prey’s peptidoglycan is extensively remodelled by specific peptidoglycan endopeptidases to prevent invasion by additional predators, as shown in B. bacteriovorus (Lerner et al. 2012), and the predator prevents self-inflicted damage to its own cell wall by using a protective protein (Lambert et al. 2015). The re-shaped prey cell which now contains the predator is called a bdelloplast. Homologous genes for the cell wall modifying machinery were found in the periplasmic H. marinus but they are absent from either of the epibiotic predators M. aeruginosavorus and B. exovorus (Pasternak et al. 2014). The sequence of events starting with penetration defines a transition phase (TP) characterized by a specific pattern of gene expression (Rotem et al. 2015). The TP is followed by the GP, which is promoted by an as yet undefined soluble prey cell fraction. It is thought that this two-step sensing strategy enables the predator to evaluate prey quality (Rotem et al. 2015). During GP, the prey’s macromolecules are sequentially degraded by different types of hydrolytic enzymes (Dori-Bachash et al. 2008; Karunker et al. 2013; Im et al. 2018). The predatory cell grows as an aseptate filament containing multiple nucleoids, with chromosome replication starting at the onset of the growth phase. The final length of the filament depends upon the size of the prey cell, and sets the number of replications, progeny and cycle duration (Kessel and Shilo 1976; Makowski et al. 2019). GP is sustained by a soluble prey cell-derived signal, the depletion of which leads to growth arrest and to cell division (Ruby and Rittenberg 1983). Strikingly, division is a synchronous, multi-site septation process that can yield an odd- or an even number of progeny (Fenton 2010; Makowski et al. 2019). Cell division is not associated with chromosome replication as this terminates shortly before septation (Makowski et al. 2019). Finally, focal lysis of the bdelloplast creates pores through which flagellated AP progeny cells are released (Fenton et al. 2010).
An intriguing aspect of BALOs’ cell cycle is the spontaneous appearance of host-independent (H-I) derivatives that grow in rich medium in the absence of prey (Barel and Jurkevitch 2001; Roschanski et al. 2011). H-I mutants can retain predatory activity, de facto being facultative predators, but very few strains have been isolated from the wild (Hobley et al. 2012a). Primary saprophytic H-I mutants require prey cell extract for growth, while secondary axenic H-I mutants can robustly grow on rich media (Roschanski et al. 2011). While the mutations responsible for these phenotypes have been mapped (Cotter and Thomashow 1992; Roschanski et al. 2011), their relation to the observed phenotypes is not understood. Moreover, H-I mutants appear to be not as effective predators as wild-type strains are and genetic revertants have not been observed. Altogether, how they survive in nature, i.e. what niche they occupy, is enigmatic. The secondary messenger cyclic-di-GMP was shown to play a role in the transition for wild-type to H-I, as mutations in specific diguanylate cyclases differentially prevented wild-type or H-I growth (Hobley et al. 2012b). Cyclic-di-GMP is often involved in regulating phenotype change in bacteria (Jenal et al. 2017) and its signaling networks may be very developed in B. bacteriovorus (Rotem et al. 2016). So far, no H-I mutants have been retrieved from epibiotic predators. H-I mutants are powerful tools for investigating BALO genetics, as mutants in essential predatory functions which are thus lethal in the wild-type strain are viable due to the non-obligate character of H-I variants (Medina et al. 2008; Duncan et al. 2019). An illustration of the life cycle of periplamic, epibiotic and H-I variant BALOs is shown in Fig. 1.
The life cycle of periplasmic BALOs (panel 1), of epibiotic BALOs (left) and of host-independent variants (right) (panel 2). The internal black line represents the peptidoglycan; the red line, the peptidoglycan processed by the predator upon invasion – see main text for details. Vesicle-like bodies and membrane-like remnants are often visible in prey of epibiotic BALOs (unpublished data)
3 BALO Population Dynamics
Here, we explore various aspects of BALO dynamics in controlled laboratory microcosms. These points will be made relevant in the section dedicated to BALOs in WWTPs.
BALOs have generation times of 2.5–4 h on E. coli-sized prey, depending on predator, prey strain, and conditions, and yield 3–6 progeny per prey (Fenton et al. 2010). Growth in liquid cultures in Erlenmeyer flasks, microtiter plates or else are classically started with a high concentration of prey and predatory populations lower by orders of magnitude. Classically, prey and predators are tracked using dilution plating, counting prey and predator with colony forming units (CFU), and plaque forming units (PFU), per milliter, respectively (Jurkevitch 2012). The development of specific 16S rRNA-gene targeted primers now enables determining predators (and prey) without relying on plate counts (Zheng et al. 2008; Van Essche et al. 2009). Furthermore, BALOs engineered to express fluorescent proteins can also be conveniently tracked and quantified (Mukherjee et al. 2016; Sathyamoorthy et al. 2019).
Within 24–30 h, depending on predator and prey strains, and temperature, the inverse composition is achieved, i.e. high and low predator and prey populations, respectively (Sathyamoorthy et al. 2019). In closed vessels, damping predator-prey oscillations can occur (Afinogenova et al. 1977). Few studies have used open vessels to study BALO predatory dynamics, showing that an oscillating predator-prey regime can be achieved dependent upon prey density, dilution rate or nutrient concentration, with stable oscillations achieved at high prey density (Varon 1979). (Whitby 1977) showed that in the system examined (B. bacteriovorus 6-5-S and Aquaspirillum serpens), dilution rates of 0.1–0.3 sustained stable oscillations for periods of up to 1 month, and a maximum growth rate of 0.45 was measured. At a lower dilution rate (0.05) a stable equilibrium was established, and at higher rates, the predator was washed out, becoming extinct.
Prey biofilms are efficiently preyed upon, and destroyed (Kadouri and O’Toole 2005; Kadouri et al. 2007; Kadouri and Tran 2013). So far, no prey that a BALO can exploit has been shown to be resistant to predation when grown as biofilm vs. as suspended cells (Kadouri and O’Toole 2005; Kadouri et al. 2007; Dashiff et al. 2011; Kadouri and Tran 2013). Although many studies have been conducted to compare prey survival in biofilm vs planktonic growth (Chanyi et al. 2016; Feng et al. 2016; Dharani et al. 2017; Sun et al. 2017) most use the colorimetric crystal violet method that only shows the proportion by which the biofilm has been reduced. Only few have used a comparative metric (cell counts) which can reveal differences in sensitivity to predation between life styles. These few studies however, showed that with E. coli, Acinetobacter baumannii and Enterobacter gergoviae biofilm cells were significantly more resistant to B. bacteriovorus predation than their planktonic counter parts (Kadouri and O’Toole 2005; Dashiff et al. 2011). However, both phenotypes of Pseudomonas aeruginosa and Klebsiella pneumoniae prey were equally sensitive to M. aeruginosavorus and to B. bacteriovorus, respectively (Kadouri et al. 2007; Dashiff et al. 2011), suggesting that both the predator and the prey play a role in determining the outcome of predation under these conditions. Prey sensitivity can be measured by the predator’s growth rate and/or by the remaining living cells of the prey population in a predatory culture. This can vary by orders of magnitude between strains (Dashiff et al. 2011). Remaining cells are resistant to predation, albeit resistance is transient, i.e. plastic, and disappears as the population grows back when nutrients are present (Shemesh and Jurkevitch 2004). While it may be hypothesized that when existing, the differential sensitivity to predation between biofilm and planktonic fractions may stem from intrinsic differences in resistance to predator attachment/penetration due to changes in prey physiology or from effects of the biofilm matrix, the phenomenon is not understood in neither growth phenotypes. Additionally, the predator may also actively affect prey susceptibility, as knockout mutations in nuclease genes in B. bacteriovorus resulted in increased predation in biofilms (Lambert and Sockett 2013).
Under natural conditions, communities are complex, and multiple predators and prey may encounter each other. This issue has barely been researched. Results of experiments conducted with a mixture of prey and a single predator showed that B. bacteriovorus reduces prey in multispecies cultures as efficiently as in single-species cultures, in suspended cells and in biofilms (Loozen et al. 2015; Im et al. 2017). The predator also exhibits prey preferences which can be expressed as differences in remaining prey levels, and faster attachment to a preferred prey (Rogosky et al. 2006).
Finally, temporal predatory dynamics are largely affected by spatial structure. (Hol et al. 2016) grew dual cultures of B. bacteriovorus and E. coli as prey in a micro-chip array composed of connected patches or in a large patch of the total same volume. The prey population drastically declined in the latter, but both the predator and the prey persisted in the former. (Dattner et al. 2017) further showed that in soil, the spatial heterogeneity of the soil matrix enabled co-existence of a viable, slowly declining B. bacteriovorus population over a week (the time frame of the experiment) possibly by providing refuge to a Burkholderia stabilis prey. Under such settings, organic and inorganic particles may act as decoy particles which may further affect predator-prey dynamics (Wilkinson 2001; Hobley et al. 2006). For details on predator-prey dynamics and modelling, and for multi-level predatory interactions and community stability, see the Chapter by Kuppardt-Kirmse and Chatzinotas “Intraguild Predation: Predatory Networks at the Microbial Scale”.
4 BALOs in Wastewater Treatment Plants (WWTPs)
4.1 A Primer on Wastewater Treatment
The basic function of the wastewater treatment plant (WWTP) is to speed up the natural processes by which water purifies itself. A first main goal is to reduce biological oxygen demand (BOD) to low levels (within a few tens of mg per litre) in the effluent released to the environment. BOD is a measure of the amount of oxygen required for microbial metabolism of organic compounds and of ammonia in water. The second main goal is to drastically limit the pathogens present in the effluent as to reduce risks of contamination. Additional goals, which are not always part of WWT include the removal of nutrients (the influent WW is nutrient-rich) to curtail the deleterious effects of increased high nitrogen and high phosphorus concentrations in the environment, such as the eutrophication of aquatic ecosystems, and the removal of inorganic and synthetic organic chemicals, including contaminants of emerging concern, such as pharmaceuticals, hormones and pesticides. Such requirements necessitate further treatment steps and more sophisticated processes upon the classical goals of the WWTP. Lately, due the rapid increase in antibiotic resistance in the clinic and in the community (Berendonk et al. 2015), the role of WWTP in spreading antimicrobial resistance (AMR) and antibiotic resistance genes (ARG) through selection and genetic exchanges between microorganisms has become a major topic of investigation. Dealing with these various demands, which usually are not part of the basic scheme of a WWT, is a challenging task.
The “Classical” WWTP
Wastewater (WW) is collected through the sewage system, flows to the WWTP where it enters primary treatment as influent, often preceded by a preliminary step to remove large floating objects. Primary treatment removes coarse solids by settling, which can be complemented by a sedimentation step for finer particles. The influent then flows to secondary treatment which constitutes the “heart” of the WWTP, where the organic matter is broken down, removing 90% or more of it. The secondary reactor is essentially a microbial digester where myriad biochemical reactions and interactions are maintained by the most diverse microbial community found in man-made systems. Recent studies have shown that microbial communities include over 2000 operational taxonomic units (OTUs, a similarity-based grouping of sequence reads of the same allele, usually the 16S rRNA gene in bacteria) of bacteria and 1000 OTUs of micro-eukaryotes (Semblante et al. 2017; Cohen et al. 2019). These, along with archaea, remove organic carbon and nitrogen to CO2 and ammonium, respectively. Further oxidation of the latter through nitrification yields nitrate. As of today, the most commonly applied secondary treatment is the conventional activated sludge (CAS) process. It is the main focus of this chapter. However, as membrane bioreactors are becoming increasingly popular, the application of BALOs in these systems is also reviewed below. CAS is effected by a suspended growth process in which microbes colonize a mixed liquor consisting of water and of suspended organic matter (flocs). The mixture requires oxygen which is provided by mechanical means or by the injection of pressurized air. Alternatively, wastewater is treated by flowing along with air through a trickling filter (so-called attached growth processes) made of minerals (slag, stones) or plastic materials. Trickling filters provide a large, aerated surface area upon which mixed microbial biofilms develop. In both the suspended and the trickling filter approaches, organic matter mineralization not only supports a large microbial diversity but also a large biomass, called the sludge. In an additional step, the sludge may be separated from the effluent by settling in a clarifier basin. In suspended growth processes, the activated sludge is returned in part to the secondary reactor, enabling continuous operation. Although the suspended sludge is well mixed, the organic flocs, and the water liquor are hosts to sympatrically-segregated bacterial and micro-eukaryotic populations. They also differ in dynamics, as the microbial composition of the liquor fluctuate more strongly and more rapidly than that of the flocs (Cohen et al. 2019). A drawback of this technology are the costs incurred by the need for aeration of the reactor, and often more important, for the disposal and/or treatment of excess sludge for downstream applications, to the extent of up to 50% or more of total operational expenses (Wendland and Ozoguz 2005). Accordingly, improvements upon the existing technologies and practices that can lead to a decrease in operational cost e.g. through sludge reduction, are sorely needed.
Various technologies have been developed to replace or to complement activated sludge-based processes. They include land treatment, constructed wetlands, anaerobic digestion, membrane-based filters and others which are out of the scope of this review.
Things WWTPs Don’t Do – Or Don’t Do Too Well
In addition to degradable organic matter, WW carries refractory contaminants that are only partially, poorly or not degraded in WWTPs (Kümmerer et al. 2018). Among them, pharmaceutically active compounds, personal care products, artificial sweeteners, and endocrine disrupting chemicals are found in the influent and in treated effluents at concentrations ranging from ng.L−1 to μg.L−1 (Tran et al. 2018). A major source of concern are antibiotics and other antimicrobial compounds. Antibiotics are used at large scales in medicine and agriculture. Fifty to ninety percent of the consumed antibiotics or their degradation products are excreted and thus discharged into the environment where they may deleteriously affect aquatic ecosystems (Kümmerer 2009). The large distribution of these compounds at detectable concentrations may be an important factor driving the increase in antibiotic resistance (AR) that finally also impacts upon the clinic (Berendonk et al. 2015). Although discharged concentrations of antibiotics are well below the minimal inhibitory concentration (MIC), they still can drive selection for increased resistance (Negri et al. 2002; Gullberg et al. 2011, 2014). Moreover, mixtures of compounds e.g. antibiotics and heavy metals, or other chemicals can further lower minimal selective concentrations, enhancing multidrug resistance (Gullberg et al. 2014). Accordingly, WWTPs are environments that may promote selection for AR. It has recently been shown that selection also occurs at very low antibiotic concentrations in the complex microbial communities found in WW (Murray et al. 2018). Mobile genetic elements such as plasmids, conjugative transposons and integrons may facilitate the horizontal dissemination of antibiotic resistance genes between bacterial species (von Wintersdorff et al. 2016). The distribution of mobile elements and of the ARGs carried by them largely varies between habitats (Gatica et al. 2019), between regions and even within a WWTP, as seen between sludge and effluent (Gatica et al. 2016) suggesting that dissemination of and selection for ARGs involve complex biotic and abiotic (and their interactions) factors. One such factor can be predation of ARG-carrying bacteria. In that case, ARGs are certainly digested, as the rest of the DNA is (Monnappa et al. 2013).
4.2 Tracking Predator-Prey Interactions In-Situ
Tracking BALO predator-prey interactions in pure culture, i.e. the growth of a predatory strain and the concomitant consumption (and then decrease) of a prey is rather straightforward. Population sizes of the predator and of the prey can be measured by dilution plating, counting colonies of the prey and plaques of the predator (Jurkevitch 2012). Optical density also comes in handy: as BALOs cells are small, they absorb little light, enabling one to follow the decrease in optical density of the prey culture as their cells are lysed by the predator (Jurkevitch 2012). As mentioned above, the ability to express fluorescent proteins in B. bacteriovorus has made direct tracking of the growth of the predator possible and precise (Mukherjee et al. 2016; Sathyamoorthy et al. 2019).
Natural and other complex samples can be addressed by high throughput sequencing. The large number of reads obtained per sample uncovers non-dominant populations such as BALOs, the population sizes of which can then be estimated in terms of relative abundance. The rapid expansion of the application of these technologies has already yielded an understanding that BALOs are ubiquitous in WWT, and that they may play a significant role in bacterial turnover, at least under some conditions and microhabitats as described below. Sequencing also exposes the diversity of BALOs, including hitherto uncultured ones (Kandel et al. 2014). The first studies with quantitative PCR targeting BALOs aimed at assessing the specificity of the primers but they also showed that in a seawater sample (Zheng et al. 2008) and in a freshwater sample (Van Essche et al. 2009), BALO concentration was higher by two orders of magnitude than that detected by plaque counts, providing the first evidence for much larger BALO abundances than previously thought.
4.3 Wastewater BALO Communities and Their Dynamics
From Then Onward
Bdellovibrio bacteriovorus was the first BALO to be isolated, from soil (Stolp and Petzold 1962). As the interest in predatory bacteria grew, they were searched for in various environments, including wastewater (sludge) (Dias and Baht 1965). In this first study, which of course relied on the isolation and the counting of plaques on specific prey (Pseudomonas fluorescens, Salmonella paratyphi), BALOs appeared to be an extremely rare type of bacterium in wastewater, averaging less than 300 cells per ml for the highest counts, and they also seemingly were unaffected by sludge processes. It was concluded that they were not active during sludge treatment. Few studies followed; three studies by (Staples and Fry 1973) and (Fry and Staples 1974, 1976) showed that larger numbers of Bdellovibrio spp. were present in all the WWTPs examined, and that their numbers increased between inflow and effluent. However, the BALO predators still constituted at most 0.01% of the total heterotrophic bacterial community. BALOs were not retrieved from settling sludge, arguably because they cannot withstand anaerobic conditions (Fry and Staples 1976). The largest concentration was measured in effluents where BALOs reached 0.2% of the cultured bacteria (about 105.pfu.ml−1 and 5.107 cfu.ml−1, respectively). However, the authors concluded that the predators (then called ‘parasites’) did not reduce the number of bacteria spilled into the river, the temperature of which (8-13 °C) was shown to prevent their growth.
As wastewater often contains toxic compounds, the sensitivity of BALOs to pollutants was explored. (Varon and Shilo 1981; Cho et al. 2019) showed that the growth B. bacteriovorus was strongly reduced in the presence of organic and inorganic chemicals, including heavy metals. Most of the compounds affecting the predators also affected the prey but a few (cadmium, copper, sodium laureth sulfate) were more potent on the predators. Similar results were obtained by (Markelova 2002; Cho et al. 2019) who showed prey and predator inhibition by 0.1% and 0.01% urea and phenol respectively. However, survival was higher when predators were associated with biofilms, which contained higher proportions of bdelloplasts, suggesting that the predator was shielded (Sanchez-Amat and Torrella 1990).
Thus, BALOs were thought to represent a rather minor fraction of the wastewater bacterial community, susceptible to environmental insults. As with environmental and ecological microbiology at large, culture-independent, DNA-based technologies proved to be a game-changer for asserting BALOs and their function in this environment.
In contrast to the results discussed above, not all pollutants appear to have a similar effect on BALOs: (Chen et al. 2014) using denaturing gradient gel electrophoresis (DGGE), a sequence-based analytic method, found B. bacteriovorus to be an important component of the microbial community of a combined photoreactor and a packed bed bioreactor used for the removal of the triphenylmethane dye ethyl violet. The microbial degraders appeared to be various Ralstonia, Stenotrophomonas, Comamonas and Delftia, with the three later species known to be potential BALO preys (Chanyi et al. 2013). A limitation of these findings is that DGGE cannot provide reliable assessments of relative or absolute abundance.
The developments of BALO-targeted (quantitative)PCR and 16S rRNA-gene sequencing enabled (Kandel et al. 2014) to use culture-independent approaches to quantify BALOs in zero discharge systems (ZDS) in which fish are grown at high density (Shnel et al. 2002; Cytryn et al. 2005). ZDSs are closed water systems. They usually include a nitrification loop (e.g. a trickling filter), a denitrification and an organic matter digester loop, complemented with a sulphide-removal reactor such as a fluidized bed reactor, resulting in the main water contaminants being converted to gases (Shnel et al. 2002; Cytryn et al. 2005). Aquaculture ZDSs sustain large fish yields and can use freshwater as well as seawater (Gelfand et al. 2003; Kandel et al. 2014).
BALOs and the general bacterial populations were analyzed by quantitative PCR (qPCR) over a 7-month period by targeting the Bdellovibrionales and the Bacteriovoracales with taxon-specific and general 16S rRNA gene primers, respectively. It was found that both families of predators co-existed in the different ZDS compartments, in fresh water-based systems as well as in seawater-based systems. Together, the two families of predators constituted 0.13–1.4% of the total Bacteria community. Thus, while BALOs are not a quantitatively major fraction of the community (as expected from obligate predators) they are not so-called “rare populations” (Albertsen et al. 2013). Their relative abundance was highest in the organic matter digester which also sustained the highest bacterial diversity, mostly composed of Gram negative taxa, suggesting a wide range of potential prey and direct coupling between predator and prey abundance. The samples were retrieved from the upper, largely aerobic part and thus whether BALOs can be found (and be active) in settling sludge remained unknown. Yet, and although they are considered aerobic, the presence of cbb3-type oxidases in their genomes suggests that BALOs may colonize oxygen-limited environments such as the upper layers of sediments, where they have previously been found (Williams 1988). (Kadouri and Tran 2013) showed that predatory bacteria preyed upon biofilms in low oxygen conditions but not on planktonic cells. The BALOs were however, not able to prey on biofilms under anoxic conditions. This contradicts a finding by (Monnappa et al. 2013) who found predation albeit limited, under completely anoxic conditions as long as nitrate was present in the medium. Although BALOs do not have bona fide nitrate reductase genes except for Micavibrio aeruginosavorus (Rendulic et al. 2004; Pasternak et al. 2014), they do include a number of nitrite reductases in their genomes. At least one (Bd2203 in B. bacteriovorus HD100) shows homology to nitrate reductases, thereby possibly explaining these results. Noteworthy, facultative predators (mostly Myxococcales) were highly abundant in the systems. As they are Gram negative, they may fall prey to BALOs; the occurrence of such interactions would suggest complex intraguild predation (IGP) networks at the microbial level, including not only phages, and protists, but also facultative and obligate bacterial predators (for more details on IGP, see the Chapter by Kuppardt-Kirmse and Chatzinotas “Intraguild Predation: Predatory Networks at the Microbial Scale”).
4.3.1 BALOs in Advanced WWT Technologies
Effluents from activated sludge bioreactors can be further treated by microfiltration (MF) systems to remove particulate matter, increasing quality, with MF substituting the sludge setting unit and enabling total retention of the suspended solids (Bai and Leow 2002). MF membranes however, foul over time as particulate matter, including microbial cells adhere to them, causing a rapid and continuous reduction of permeation flux with time. In order to test the potential of BALOs to prevent MF membrane fouling, the outcome of predation of an E. coli suspension was evaluated by measuring flux parameters, with B. bacteriovorus predators alone (Kim et al. 2013), in combination with a flocculant (aluminum sulfate, alum) or along with powdered activated carbon (PAC), a material reducing adsorption (Kim et al. 2014). The predator alone treatment efficiently sustained higher membrane fluxes than controls without predators. However, predation led to increased irreversible membrane biofouling, most probably caused by the accumulation of prey cell debris, resulting in pore blockage. The addition of chemical amendments – especially alum- to the predators further increased fluxes over controls, and reduced irreversible fouling. Another lab study was carried out to measure the effect of adding B. bacteriovorus to the membrane filtration process of activated sludge. It used a dead-end reactor with suspended solids of 3–3.5 g.l−1 and a COD of 730–780 mg. l−1, also finding improvements in fluxes (Yılmaz et al. 2014). In summary, BALOs may prove to be a worthwhile additional improvement to ease clogging in microfiltration-based devices.
A series of studies examined various wastewater treatment line architectures containing aerobic, microaerobic or anaerobic side reactors coupled to membrane bioreactors. More specifically, the addition of one or more external microaerobic or anoxic reactors in the return sludge loop of a conventional activated sludge process reduced sludge in large proportions (Semblante et al. 2014). The data supported the idea that the proliferation of slow-growing nitrifiers in the main aerobic sequencing batch reactor, and of hydrolysers and of fermenters causing sludge autolysis in the external oxygen-deficient reactors resulted in sludge reduction (Semblante et al. 2017).
Along those lines, the effects of treatments like hydraulic retention times, side-stream ratio, packing carriers, and ultrasonication on sludge reduction and dewaterability, and pollutant (nitrogen, phosphorus, COD) removal, in the different settings were measured (Cheng et al. 2017, 2018; Zheng et al. 2019). Different combinations of architectures and treatments realized significant improvements over controls (i.e. systems lacking side reactors or packing carriers etc). As an example, micro-aerobic conditions in some treatments favored sludge reduction by enriching for hydrolytic and fermentative bacteria, generating abundant substrates for hydrolysis, bringing about the disintegration of sludge floc structure and contributing to the breakdown of both refractory and biodegradable compounds (Cheng et al. 2018). As another example, packing carriers and ultrasonication applied in an membrane bioreactor (MBR) with an anaerobic side-stream reactor (ASSR-MBR) enriched for hydrolytic bacteria reduced the deterioration of sludge performance caused by a low temperature (Zheng et al. 2019).
In direct relevance to this chapter, it was observed that Bdellovbrio were present under all conditions tested but some led to significant increases in their abundance, with Bdellovibrio populations constituting up to a few percent of the total bacterial community. Microaerobic conditions and high retention times (in some of these systems, hydraulic and solid retention times are similar (Cheng et al. 2017)) promoted high BALO populations, which reached 1.5% of the total Bacteria population; a low side stream ratio or the presence of packing carriers in the ASSR-MBR also significantly increased the BALO community (Cheng et al. 2017; Zheng et al. 2019), albeit to lower levels. Based on these correlative results, it was suggested that along with the hydrolytic populations mentioned above, BALOs contribute to sludge reduction, possibly by the predators affecting turnover of hydrolytic Gram negative populations through predation. Similar results were obtained with other processes aiming at activated sludge reduction based on the insertion of a micro-aerobic or an anoxic tank upstream to an anoxic/aerobic unit containing a feedback loop to both units. These architectures led to an increased abundance of the facultative predators Myxobacteria in studies by both (Zhou et al. 2014) and (Semblante et al. 2017) and in this latter case, also of BALOs. It should be noted that in the Zhou et al. (2014) study, sequencing was performed with the 454 Roche technology which, while enabling long reads, produced relatively low numbers of sequences (a few thousands) per sample. Also worthwhile mentioning, in all the surveyed studies, Micavibrio were absent from the data. Micavibrio strains seem to have a rather restricted prey range compared to most other BALOs (Davidov et al. 2006a; Kadouri et al. 2007; Dashiff et al. 2011), and this property (if true) may restrict their distribution. Nonetheless, a Micavibrio-like bacterium was detected in a sludge incubation experiment in which 13C-labeled bicarbonate was used to monitor the flow of carbon from uncultured nitrifiers to heterotrophs (Dolinšek et al. 2013). The predator was discovered by separating the heavier 13C-labelled nucleic acids, followed by 16S rRNA gene sequencing, and further localized by fluorescent in-situ hybridization (FISH). It was shown to attach to (and seemingly prey on) nitrite-oxidizing sublineage I Nitrospira but not to sublineage II Nitrospira in sludge flocs, suggesting a highly specific interaction.
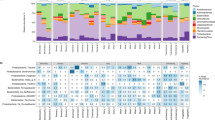
Additional studies experimented with manipulating sludge processes by directly inoculating BALOs into the mixed liquor, demonstrating that BALOs can indeed affect sludge. In their study, (Yu et al. 2017) showed that sludge biolysis increased with the concentration of the introduced predators. It appeared that BALOs promoted bacterial cell lysis resulting in increased sludge disintegration which generally correlated with sludge dewaterability, results that support the role of BALOs in sludge processing. Microscopic observations suggested that BALO-treated flocs were smaller, and had a more porous structure with less connective filaments. Further, the physical state of WW flocs can be manipulated by operational conditions to achieve changes in output parameters (e.g sludge settling time). Under high hydraulic selection pressure that brings about washout of slow settling particles, compact granules containing self-immobilized bacteria in extracellular polymeric substances (EPS) are selected for (Feng et al. 2017; Szabó et al. 2017). The redox status of granules may shift from anaerobic/anoxic in the internal core to aerobic in the granule’s outer layer (de Kreuk et al. 2005). Recently, both Szabó et al. (2017) and Feng et al. (2017) have shown that BALOs populate the granules, inhabiting specific locations within them (Szabó et al. 2017), and altering the structure of their microbial populations (Feng et al. 2017). By applying FISH targeting specific taxa, Szabó et al. (2017) precisely mapped the distribution of various species onto the granules. Bacteria associated with the external layers were also shown to have relatively low retention times suggesting easier washout caused by erosion than internally located microorganisms. Bdellovibrio were found in the inner parts of the granules (Fig. 2) where they actually increased in abundance during the course of the experiment. BALOs may withstand anaerobic conditions, and may even grow under such conditions (see above) but oxygen may still be able to reach these deeper regions (Szabó et al. 2017). One may speculate that predation of -relative to BALOs- large bacteria would increase oxygen diffusion by reducing demand and by creating larger channels. Studies by (Feng et al. 2016, 2017) add to the understanding of BALO-linked processes occurring in the suspended organic fraction in WWTPs. They isolated BALOs and Gram negative bacteria from WWTP, showing that almost all of the latter could be used as prey (Feng et al. 2016), as shown earlier in other aquatic habitats (Rice et al. 1998). Among these potential prey were Bacteroidetes, which are potentially major floc and granule hydrolyzers, and Rhodocyclales, both taxa that had hitherto not been tested as BALO prey. Inoculation of a BALO into suspensions of flocs or of granules, followed by community 16S rRNA gene sequencing showed that the selected predator strain significantly reduced the relative abundances of many taxa, including Bacteroidetes and Rhodocyclales, providing evidence for in situ predation of prey belonging to these genera (Feng et al. 2017). That said, indirect effects brought about by floc/granule structure breakdown due to predation of susceptible strains may release other bacteria to the suspension without predation, as shown by BALOs disrupting biofilms formed by Gram positive bacteria without preying on them (Im et al. 2018). In the Feng et al study (2017), predation led to a remarkable decrease in the floc and in the granule microbial biomass, and in viability by circa 50% and 50-fold, respectively. It can be remarked that Eukarya were also impacted by predation. This indirect effect of bacterial predation further shows the intricate interactions between the various types of predators present.
FISH- confocal laser scanning microscopy images of Bdellovibrio in sludge granules. Cryosections of granules at ×200 magnification and at ×400 magnification. FISH probe BDE-535, according to Mahmoud et al. (2007). Grey, total cells (Syto 40); red stain, Bdellovibrio. (From Szabó et al. (2017) under the terms of the Creative Commons 4.0 International Licence)
Treated wastewater is used to replenish natural habitats and for irrigation, while processed sludge can be used for energy and soil fertilization. Another, complementary approach for using residues of WWT is to develop their added value, for example by producing microbial proteins from sludge, to yield high quality feed and possibly food. Matassa et al. (2016) aerobically converted sludge from a potato-processing plant into protein. The notable feature in relation to microbial predation was the very high proportion (30%) of 16S rRNA reads affiliated to Bdellovibrio, and the high bacterial diversity obtained in a sequencing batch reactor (SBR) operated at low selection pressure, i.e. at high solid retention time, in contrast to a continuous reactor with a short retention time. Explaining how such high relative abundance of predators can be sustained is difficult. One may think that the bacterial (prey) turnover is rapid, and or that a large part of the predatory Bdellovibrio population is actually not predatory but of the “host-independent type” living off the high protein content of the medium. Whether this or that, or any other hypothesis, is valid should be theoretically and empirically tested. If it could be shown that H-I variants grow and take over under the conditions prevailing in the Matassa et al. (2016) potato processing sludge experiment, the selection processes and mechanisms at play would certainly be worthwhile investigating. This would also show that H-I mutants are actually viable in nature and are not mutational dead ends. Another study explored the use of wastewater to produce polyhydroxyalkanoates (PHAs), which are carbon and energy storage compounds of many bacterial strains (Wijeyekoon et al. 2018). PHAs are fully biodegradable and possess thermoplastic properties that make them attractive natural replacements of petroleum-derived plastics. The community of a SBR with a long (4 day) solid retention time was dominated by PHA producing bacteria belonging to the Proteobacteria (73.0%, of which 84% were Rhodocyclaceae) and to the Bacteroidetes (25.2%, of which Saprospiraceae constituted 20.5%). These taxa are dominant in WWTP (Kandel et al. 2014; Semblante et al. 2017; Cohen et al. 2019) and may be preyed upon by BALOs (Feng et al. 2017). The third most abundant taxon in the reactor was Bdellovibrio (3.5%). B. bacteriovorus contains a poly(3-hydroxyalkanoate) depolymerase enabling it to consume medium chain length PHAs (Martínez et al. 2012), conferring energy and an ecological advantage to the predator (Martínez et al. 2013). For more details on biotechnological and industrial applications of BALOs, including applications relevant to PHA production, see the Chapter by Herencias et al. “Emerging Horizons for Industrial Applications of Predatory Bacteria”.
Although still rather limited in scope and number, the studies presented in this and in the above sections indicate that natural BALO populations are an integral part of WWTP reactors and that in contrast to earlier findings, they react dynamically to operational and environmental changes. We would like to tentatively propose that relatively long solid retention times (which controls the concentration of bacteria throughout the system), and the addition of side reactor(s) that operate under various environments (anoxic, microaerobic, aerobic) to a main activated sludge unit promote bacterial diversity and a high abundance of hydrolysers (Bacteroidetes such as Sphingobacteriales) and of Proteobacteria, mainly Rhodocyclales, −that are potentially active in the degradation of organics, in phosphate accumulation and in denitrification-enriching for BALOs and possibly also, Myxobacteria. Hydrolysers, and predators that may prey upon them, may in turn promote floc reduction (Kandel et al. 2014; Zhou et al. 2014; Cheng et al. 2017, 2018; Feng et al. 2017; Semblante et al. 2017; Wijeyekoon et al. 2018) (Fig. 3).
Wastewater treatment schemes that may affect the concentration of predatory bacteria in the system. Side reactors with different operating conditions than the main reactor, and increased solid retention time may positively affect microbial diversity, also increasing Rhodocyclales and Bacteroidetes. The former may degrade organics, denitrification and P accumulation, the latter may increase hydrolysis of flocs, and in concert with increased predators that may also prey upon them, bring about sludge reduction
Two interesting observations will conclude this section. The study by Feng et al. (2017) included a transcriptomics (RNASeq) analysis of flocs and granules exposed or not to B. bacteriovorus. Although community composition and floc structure were altered by the inoculated predators (see above) the expressed functions were not, hinting that predation is either not discriminatory enough in term of taxonomic differences to impact upon the functionality of the community or, alternatively, that functional redundancy and compensation mechanisms are at play. The latter would somewhat be surprising as the large loss in cell viability (50-fold) and biomass (50%) engendered by predation may be defined as a large scale disruption, i.e. a situation thought to bring about functional disruptions. Another study examined the role of the second messenger cyclic di-GMP on the stability of aerobic granules in a sequencing batch reactor (Wan et al. 2013). Cyclic-di-GMP is a cellular signal that strongly affects bacterial phenotypes such as motility, biofilm formation, EPS and cellulose synthesis, virulence, and many other features (Jenal et al. 2017). In B. bacteriovorus, different effectors of cyclic di-GMP metabolism differentially affect H-I formation by preventing it or by making it obligatory; they also affect gliding, progeny exit from the bdelloplast, and attachment to prey (Hobley et al. 2012b; Milner et al. 2014). The addition of manganese to the reactor brought about disintegration of granules, causing a significant decrease in cyclic-di-GMP cellular concentrations of the total bacterial community, leading to a decrease in EPS (Wan et al. 2013). A clone library (therefore restricted in size and coverage as compared high throughput sequencing) showed a high representation of Bdellovibrio 16S rRNA gene inserts in the control treatment (circa 4%). The manganese treated samples still had a rather high (circa 2%) but significantly reduced BALO population. Whether c-di-GMP metabolism plays a role in the association of Bdellovibrio with biofilms (granules are biofilm-like structures) is not known. As with the gene expression changes in the BALOs and in the prey populations in flocs, this interesting question remains to be further investigated.
5 Outlook: Basic Questions, Technological Bridges, and Applications
The information analyzed in this review unequivocally shows that BALOs are almost always present in WWTPs. They usually consist of low abundance but not rare populations and they can significantly increase in size, as a response to biotic and abiotic-driven changes in the environment, showing that BALOs are active members of microbial trophic networks. A key aim is to understand their “space” in the networks, i.e. their effects on the dynamics and stability of microbial ecosystems. This broad aim can be reduced to more focused (yet still broad) questions such as: what is the impact of BALO predation on the community structure and community components thereof; how qualitatively and quantitatively do BALOs directly (by predation) and indirectly (e.g. by breaking down flocs/biofilms) contribute to bacterial mortality and to nutrient release, i.e. to bacterial turnover; what is their relationship to other microbial predators, e.g. other bacterial predators like Myxobacteria, phages and protists? Such knowledge, which can be obtained from experiments under natural and under controlled conditions would be valuable both for theory and for applications. In order to decipher the role and impact of BALOs on the intricate microbial trophic networks of WWTPs, of other microbial ecosystems, and more globally on nutrient flow, precise quantitation and identification of predator and prey interaction dynamics is necessary. It should be remarked that since BALOs require a prey to grow, many BALOs may not be cultured in the laboratory as their prey may by themselves be unculturable. Fortunately, the sequencing revolution has been accompanied by other powerful advances in microbial community analysis, a few of which are presented here.
QPCR based on specific primers can reveal the sizes and fluctuations of specific BALO populations in absolute terms (Zheng et al. 2008; Van Essche et al. 2009) that can also be expressed as relative to the total bacterial population size if this is measured using general primers targeting the 16S rRNA gene (Kandel et al. 2014). Thus, by combining high throughput sequencing and qPCR, it might be possible to track and identify predators and their dynamics in complex samples. Yet, a pertinent question remains: how can the predators’ prey be identified so the impact of predation be quantified in detail? An approach is computational: by statistically correlating fluctuations/co-occurrence in terms of abundance of the Gram negative populations to those of the BALO populations, links may appear (Welsh et al. 2015). These can be characterized and quantified to uncover the potential prey range of the different predators, their impact on the prey population, and possible mechanisms underlying the revealed dynamics. However, empirical approaches that would directly detect such interactions in situ and confirm the computations are forcefully required. Methods could be developed based on existing technologies such as FISH for labelling predators and fluorescence activated cell sorting to obtain bacterial populations interacting with a labelled predator. Sorted samples could be sequenced to reveal the composition of the interacting populations. Emulsion, Paired Isolation and Concatenation (EPIC)-PCR makes use of emulsion PCR to isolate single cells which can be identified and linked to a chosen genomic feature (Spencer et al. 2016). It may therefore be possible to apply it to uncover direct interactions between cells, and obtain comprehensive identification of pairwise interactions between predators and prey. Predatory interactions can also be uncovered and analysed at the metabolic level using stable isotope probing. As shown by (Chauhan et al. 2009) and Dolinšek et al. (2013) using BALOs, nutrient flow from labelled prey to predators can be tracked to identify active predators. In the former case heterotrophic bacteria were labelled using a rich medium, in the latter, autotrophs were labelled with bicarbonate, both using 13C. Predators preying upon these two metabolic classes were then identified by cloning the “heavy”, 13C-labelled DNA. Use of these approaches individually or in conjunction with each other will enable researchers to track and decipher complex interactions in natural ecosystems or in microcosms mimicking them; or in simplified settings aiming a describing in mechanistic details specific interactions between micro-predators and prey under various conditions (Johnke et al. 2017a, b).
These are but a few examples of novel technologies that could in the (hopefully near) future, help solve questions pertaining to the ecological theory of microbial predation, as an example, when using the IGP approach to decipher community interactions. Such approaches would also provide much needed data for modeling predator-prey interactions and understanding how simple, and more complex ecological systems stabilize. Applications could come all along. At the top of the list, WWT could greatly benefit, through the devise of approaches and technologies that improve the ecological stability of WWTPs, by reducing deleterious and operational disruptive fluctuations in community structure and by improving the efficiency of positive processes involving BALOs (e.g. sludge reduction). It might also become possible to envisage small scale, decentralized WWT systems that target microbial biomass, and more specifically pathogen and ARG reduction, helping to reduce their burden on LMICs.
References
Afinogenova AV, Ratner EN, Lambina VA. Oscillations of parasite and host numbers in two-membered bacterial system. Microbiology (Mikrobiologiya). 1977.
Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol. 2013;31:533–8.
Avidan O, Petrenko M, Becker R, Beck S, Linscheid M, Pietrokovski S, Jurkevitch E. Identification and characterization of differentially-regulated type IVb pilin genes necessary for predation in obligate bacterial predators. Sci Rep. 2017;7:1013.
Bai R, Leow HF. Microfiltration of activated sludge wastewater—the effect of system operation parameters. Sep Purif Technol. 2002;29:189–98.
Barel G, Jurkevitch E. Analysis of phenotypic diversity among host-independent mutants of Bdellovibrio bacteriovorus 109J. Arch Microbiol. 2001;176:211–6.
Berendonk TU, Manaia CM, Merlin C, Fatta-Kassinos D, Cytryn E, Walsh F, Bürgmann H, Sørum H, Norström M, Pons M-N, Kreuzinger N, Huovinen P, Stefani S, Schwartz T, Kisand V, Baquero F, Martinez JL. Tackling antibiotic resistance: the environmental framework. Nat Rev Microbiol. 2015;13:310–7.
Cao H, He S, Wang H, Hou S, Lu L, Yang X. Bdellovibrios, potential biocontrol bacteria against pathogenic Aeromonas hydrophila. Vet Microbiol. 2012;154:413–8.
Cao H, An J, Zheng W, He S. Vibrio cholerae pathogen from the freshwater-cultured whiteleg shrimp Penaeus vannamei and control with Bdellovibrio bacteriovorus. J Invertebr Pathol. 2015;130:13–20.
Chanyi RM, Ward C, Pechey A, Koval SF. To invade or not to invade: two approaches to a prokaryotic predatory life cycle. Can J Microbiol. 2013;59:273–9.
Chanyi RM, Koval SF, Brooke JS. Stenotrophomonas maltophilia biofilm reduction by Bdellovibrio exovorus. Environ Microbiol Rep. 2016;8:343–51.
Chauhan A, Williams HN. Response of Bdellovibrio and like organisms (BALOs) to the migration of naturally occurring bacteria to chemoattractants. Curr Microbiol. 2006;53:516–22.
Chauhan A, Cherrier J, Williams HN. Impact of sideways and bottom-up control factors on bacterial community succession over a tidal cycle. Proc Natl Acad Sci U S A. 2009;106:4301–6.
Chen C-Y, Yen S-H, Chung Y-C. Combination of photoreactor and packed bed bioreactor for the removal of ethyl violet from wastewater. Chemosphere. 2014;117:494–501.
Cheng C, Zhou Z, Niu T, An Y, Shen X, Pan W, Chen Z, Liu J. Effects of side-stream ratio on sludge reduction and microbial structures of anaerobic side-stream reactor coupled membrane bioreactors. Bioresour Technol. 2017;234:380–8.
Cheng C, Zhou Z, Pang H, Zheng Y, Chen L, Jiang L-M, Zhao X. Correlation of microbial community structure with pollutants removal, sludge reduction and sludge characteristics in micro-aerobic side-stream reactor coupled membrane bioreactors under different hydraulic retention times. Bioresour Technol. 2018;260:177–85.
Cho G, Kwon J, Soh SM, Jang H, Mitchell RJ. Sensitivity of predatory bacteria to different surfactants and their application to check bacterial predation. Appl Microbiol Biotechnol. 2019;103:8169–78.
Cohen Y, Pasternak Z, Johnke J, Abed-Rabbo A, Kushmaro A, Chatzinotas A, Jurkevitch E. Bacteria and microeukaryotes are differentially segregated in sympatric wastewater microhabitats. Environ Microbiol. 2019;21(5):1757–70.
Cotter T, Thomashow M. Identification of a Bdellovibrio bacteriovorus genetic locus, hit, associated with the host-independent phenotype. J Bacteriol. 1992;174:6018–24.
Cytryn E, Minz D, Gelfand I, Neori A, Gieseke A, de Beer D, van Rijn J. Sulfide-oxidizing activity and bacterial community structure in a fluidized bed reactor from a zero-discharge Mariculture system. Environ Sci Technol. 2005;39:1802–10.
Dashiff A, Junka R, Libera M, Kadouri D. Predation of human pathogens by the predatory bacteria Micavibrio aeruginosavorus and Bdellovibrio bacteriovorus. J Appl Microbiol. 2011;110:431–44.
Dattner I, Miller E, Petrenko M, Kadouri DE, Jurkevitch E, Huppert A. Modelling and parameter inference of predator–prey dynamics in heterogeneous environments using the direct integral approach. J R Soc Interface. 2017;14:20160525.
Davidov Y, Jurkevitch E. Diversity and evolution of Bdellovibrio-and-like organisms (BALOs), reclassification of Bacteriovorax starrii as Peredibacter starrii gen. nov., comb. nov., and description of the Bacteriovorax-Peredibacter clade as Bacteriovoracaceae fam. nov. Int J Syst Evol Microbiol. 2004;54:1439–52.
Davidov Y, Friedjung A, Jurkevitch E. Structure analysis of a soil community of predatory bacteria using culture-dependent and culture-independent methods reveals a hitherto undetected diversity of Bdellovibrio-and-like organisms. Environ Microbiol. 2006a;8:1667–73.
Davidov Y, Huchon D, Koval SF, Jurkevitch E. A new α-proteobacterial clade of Bdellovibrio-like predators: implications for the mitochondrial endosymbiotic theory. Environ Microbiol. 2006b;8:2179–88.
de Kreuk MK, Heijnen JJ, van Loosdrecht MCM. Simultaneous COD, nitrogen, and phosphate removal by aerobic granular sludge. Biotechnol Bioeng. 2005;90:761–9.
Dharani S, Kim DH, Shanks RMQ, Doi Y, Kadouri DE. Susceptibility of colistin-resistant pathogens to predatory bacteria. Res Microbiol. 2017.
Dias FF, Baht JV. Microbial ecology of activated sludge. II. Bacteriophages, Bdellovibrio, coliforms, and other organisms. Appl Microbiol. 1965;13:257–61.
Dolinšek J, Lagkouvardos I, Wanek W, Wagner M, Daims H. Interactions of nitrifying Bacteria and heterotrophs: identification of a Micavibrio-like putative predator of Nitrospira spp. Appl Environ Microbiol. 2013;79:2027–37.
Dori-Bachash M, Dassa B, Pietrokovski S, Jurkevitch E. Proteome-based comparative analyses of growth stages reveal new cell cycle-dependent functions in the predatory bacterium Bdellovibrio bacteriovorus. Appl Environ Microbiol. 2008;74:7152–62.
Duncan MC, Gillette RK, Maglasang MA, Corn EA, Tai AK, Lazinski DW, Shanks RMQ, Kadouri DE, Camilli A. High-throughput analysis of gene function in the bacterial predator Bdellovibrio bacteriovorus. mBio. 2019;10:e01040–19.
Enos BG, Anthony MK, DeGiorgis JA, Williams LE. Prey range and genome evolution of Halobacteriovorax marinus predatory bacteria from an estuary. mSphere. 2018;3:e00508.
Evans KJ, Lambert C, Sockett RE. Predation by Bdellovibrio bacteriovorus HD100 requires type IV Pili. J Bacteriol. 2007;189:4850–9.
Fenton AK, Kanna M, Woods RD, Aizawa SI, Sockett RE. Shadowing the actions of a predator: backlit fluorescent microscopy reveals synchronous nonbinary septation of predatory Bdellovibrio inside prey and exit through discrete bdelloplast pores. J Bacteriol. 2010;192:6329–35.
Feng S, Tan CH, Cohen Y, Rice SA. Isolation of Bdellovibrio bacteriovorus from a tropical wastewater treatment plant and predation of mixed species biofilms assembled by the native community members. Environ Microbiol: n/a-n/a. 2016.
Feng S, Tan CH, Constancias F, Kohli GS, Cohen Y, Rice SA. Predation by Bdellovibrio bacteriovorus significantly reduces viability and alters the microbial community composition of activated sludge flocs and granules. FEMS Microbiol Ecol. 2017;93:fix020.
Fry J, Staples D. The occurrence and role of Bdellovibrio bacteriovorus in a polluted river. Water Res. 1974;8:1029–35.
Fry J, Staples D. Distribution of Bdellovibrio bacteriovorus in sewage works, river water, and sediments. Appl Environ Microbiol. 1976;31:469–74.
Gatica J, Tripathi V, Green S, Manaia CM, Berendonk T, Cacace D, Merlin C, Kreuzinger N, Schwartz T, Fatta-Kassinos D, Rizzo L, Schwermer CU, Garelick H, Jurkevitch E, Cytryn E. High throughput analysis of integron gene cassettes in wastewater environments. Environ Sci Technol. 2016;50:11825–36.
Gatica J, Jurkevitch E, Cytryn E. Comparative metagenomics and network analyses provide novel insights into the scope and distribution of β-lactamase homologs in the environment. Front Microbiol. 2019;10:146.
Gelfand I, Barak Y, Even-Chen Z, Cytryn E, Van Rijn J, Krom MD, Neori A. A novel zero discharge intensive seawater recirculating system for the culture of marine fish. J World Aquacult Soc. 2003;34:344–58.
Gullberg E, Cao S, Berg OG, Ilbäck C, Sandegren L, Hughes D, Andersson DI. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 2011;7:e1002158.
Gullberg E, Albrecht LM, Karlsson C, Sandegren L, Andersson DI. Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. mBio. 2014;5:e01918–4.
Hahn MW, Schmidt J, Koll U, Rohde M, Verbarg S, Pitt A, Nakai R, Naganuma T, Lang E. Silvanigrella aquatica gen. nov., sp. nov., isolated from a freshwater lake, description of Silvanigrellaceae fam. nov. and Silvanigrellales ord. nov., reclassification of the order Bdellovibrionales in the class Oligoflexia, reclassification of the families Bacteriovoracaceae and Halobacteriovoraceae in the new order Bacteriovoracales ord. nov., and reclassification of the family Pseudobacteriovoracaceae in the order Oligoflexales. Int J Syst Evol Microbiol. 2017;67:2555–68.
Hobley L, King JR, Sockett RE. Bdellovibrio predation in the presence of decoys: three-way bacterial interactions revealed by mathematical and experimental analyses. Appl Environ Microbiol. 2006;72:6757–65.
Hobley L, Lerner T, Williams L, Lambert C, Till R, Milner D, Basford S, Capeness M, Fenton A, Atterbury R, Harris M, Sockett RE. Genome analysis of a simultaneously predatory and prey-independent, novel Bdellovibrio bacteriovorus from the River Tiber, supports in silico predictions of both ancient and recent lateral gene transfer from diverse bacteria. BMC Genomics. 2012a;13:670.
Hobley L, Fung RKY, Lambert C, Harris MATS, Dabhi JM, King SS, Basford SM, Uchida K, Till R, Ahmad R, Aizawa S-I, Gomelsky M, Sockett RE. Discrete cyclic di-GMP-dependent control of bacterial predation versus axenic growth in Bdellovibrio bacteriovorus. PLoS Pathog. 2012b;8:e1002493.
Hol FJ, Rotem O, Jurkevitch E, Dekker C, Koster DA. Bacterial predator–prey dynamics in microscale patchy landscapes. Proc R Soc B. 2016;283:20152154.
Iebba V, Santangelo F, Totino V, Nicoletti M, Gagliardi A, De Biase RV, Cucchiara S, Nencioni L, Conte MP, Schippa S. Higher prevalence and abundance of Bdellovibrio bacteriovorus in the human gut of healthy subjects. PLoS One. 2013;8:e61608.
Im H, Choi SY, Son S, Mitchell RJ. Combined application of bacterial predation and violacein to kill polymicrobial pathogenic communities. Sci Rep. 2017;7:14415.
Im H, Dwidar M, Mitchell RJ. Bdellovibrio bacteriovorus HD100, a predator of gram-negative bacteria, benefits energetically from Staphylococcus aureus biofilms without predation. ISME J. 2018;12:2090–5.
Jenal U, Reinders A, Lori C. Cyclic di-GMP: second messenger extraordinaire. Nat Rev Microbiol. 2017;15:271–84.
Johnke J, Boenigk J, Harms H, Chatzinotas A. Killing the killer: predation between protists and predatory bacteria. FEMS Microbiol Lett. 2017a;364:fnx089.
Johnke J, Baron M, de Leeuw M, Kushmaro A, Jurkevitch E, Harms H, Chatzinotas A. A generalist protist predator enables coexistence in multitrophic predator-prey systems containing a phage and the bacterial predator Bdellovibrio. Front Ecol Evol. 2017b;5:124.
Jurkevitch E. Isolation and classification of Bdellovibrio and like organisms. In: Coico R, Kowalik T, Quarles J, Stevenson B, Taylor R, editors. Current protocols in microbiology. New York: Wiley; 2012.
Jurkevitch E, Minz D, Ramati B, Barel G. Prey range characterization, ribotyping, and diversity of soil and rhizosphere Bdellovibrio spp. isolated on phytopathogenic bacteria. Appl Environ Microbiol. 2000;66:2365–71.
Kadouri D, O’Toole GA. Susceptibility of biofilms to Bdellovibrio bacteriovorus attack. Appl Environ Microbiol. 2005;71:4044–51.
Kadouri DE, Tran A. Measurement of predation and biofilm formation under different ambient oxygen conditions using a simple gasbag-based system. Appl Environ Microbiol. 2013;79:5264–71.
Kadouri D, Venzon NC, O’Toole GA. Vulnerability of pathogenic biofilms to Micavibrio aeruginosavorus. Appl Environ Microbiol. 2007;73:605–14.
Kadouri D, O’Toole GA. Susceptibility of biofilms to Bdellovibrio bacteriovorus attack. Appl Environ Microbiol. 2005;71:4044–51.
Kadouri DE, To K, Shanks RMQ, Doi Y. Predatory bacteria: a potential ally against multidrug-resistant gram-negative pathogens. PLoS One. 2013;8:e63397.
Kandel PP, Pasternak Z, van Rijn J, Nahum O, Jurkevitch E. Abundance, diversity and seasonal dynamics of predatory bacteria in aquaculture zero discharge systems. FEMS Microbiol Ecol. 2014;89:149–61.
Karunker I, Rotem O, Dori-Bachash M, Jurkevitch E, Sorek R. A global transcriptional switch between the attack and growth forms of Bdellovibrio bacteriovorus. PLoS One. 2013;8:e61850.
Kelley JI, Williams HN. Bdellovibrios in Callinectus sapidus, the blue crab. Appl Environ Microbiol. 1992;58:1408–10.
Kessel M, Shilo M. Relationship of Bdellovibrio elongation and fission to host cell size. J Bacteriol. 1976;128:477–80.
Kikuchi Y, Bomar L, Graf J. Stratified bacterial community in the bladder of the medicinal leech, Hirudo verbana. Environ Microbiol. 2009;11:2758–70.
Kim E-H, Dwidar M, Mitchell RJ, Kwon Y-N. Assessing the effects of bacterial predation on membrane biofouling. Water Res. 2013;47:6024–32.
Kim E-H, Dwidar M, Kwon Y-N, Mitchell RJ. Pretreatment with alum or powdered activated carbon reduces bacterial predation-associated irreversible fouling of membranes. Biofouling. 2014;30:1225–33.
Klein DA, Casida LE Jr. Occurrence and enumeration of Bdellovibrio bacteriovorus in soil capable of parasitizing Escherichia coli and indigenous soil bacteria. Can J Microbiol. 1967;13:1235–41.
Koval SF, Hynes SH, Flannagan RS, Pasternak Z, Davidov Y, Jurkevitch E. Bdellovibrio exovorus sp. nov., a novel predator of Caulobacter crescentus. Int J Syst Evol Microbiol. 2012.
Koval SF, Williams HN, Stine OC. Reclassification of Bacteriovorax marinus as Halobacteriovorax marinus gen. nov., comb. nov. and Bacteriovorax litoralis as Halobacteriovorax litoralis comb. nov.; description of Halobacteriovoraceae fam. nov. in the class Deltaproteobacteria. Int J Syst Evol Microbiol. 2015;65:593–7.
Kümmerer K. Antibiotics in the aquatic environment – a review – part I. Chemosphere. 2009;75:417–34.
Kümmerer K, Dionysiou DD, Olsson O, Fatta-Kassinos D. A path to clean water. Science. 2018;361:222–4.
Kuru E, Lambert C, Rittichier J, Till R, Ducret A, Derouaux A, Gray J, Biboy J, Vollmer W, VanNieuwenhze M, Brun YV, Sockett RE. Fluorescent D-amino-acids reveal bi-cellular cell wall modifications important for Bdellovibrio bacteriovorus predation. Nat Microbiol. 2017;2:1648–57.
LaMarre AG, Straley SC, Conti SF. Chemotaxis towards amino acids by Bdellovibrio bacteriovorus. J Bacteriol. 1977;131:201–7.
Lambert C, Sockett R. Nucleases in Bdellovibrio bacteriovorus contribute towards efficient self-biofilm formation and eradication of pre-formed prey biofilms. FEMS Microbiol Lett: n/a-n/a. 2013.
Lambert C, Smith M, Sockett R. A novel assay to monitor predator-prey interactions for Bdellovibrio bacteriovorus 109 J reveals a role for methyl-accepting chemotaxis proteins in predation. Environ Microbiol. 2003;5:127–32.
Lambert C, Chang CY, Capeness MJ, Sockett RE. The first bite--profiling the predatosome in the bacterial pathogen Bdellovibrio. PLoS One. 2010;5:e8599.
Lambert C, Cadby IT, Till R, Bui NK, Lerner TR, Hughes WS, Lee DJ, Alderwick LJ, Vollmer W, Sockett ER, Lovering AL. Ankyrin-mediated self-protection during cell invasion by the bacterial predator Bdellovibrio bacteriovorus. Nat Commun. 2015;6:1–10.
Lambina VA, Afinogenova AV, Romai Penabad S, Konovalona SM, PushkarevaA.P. Micavibrio admirandus gen. et sp. nov. Mikrobiologiya. 1982;51:114–7.
Lambina VA, Afinogenova AV, Romay Penabad S, Konovalona SM, Andreev LV. A new species of exoparasitic bacteria from the genus Micavibrio destroying gram-negative bacteria. Mikrobiologiya. 1983;53:777–80.
Lerner TR, Lovering AL, Bui NK, Uchida K, Aizawa S-I, Vollmer W, Sockett RE. Specialized peptidoglycan hydrolases sculpt the intra-bacterial niche of predatory Bdellovibrio and increase population fitness. PLoS Pathog. 2012;8:e1002524.
Li N, Williams H. 454 pyrosequencing reveals diversity of Bdellovibrio and like organisms in fresh and salt water. Antonie Van Leeuwenhoek. 2015;107:305–11.
Li H, Liu C, Chen L, Zhang X, Cai J. Biological characterization of two marine Bdellovibrio-and-like organisms isolated from Daya bay of Shenzhen, China and their application in the elimination of Vibrio parahaemolyticus in oyster. Int J Food Microbiol. 2011;151:36–43.
Loozen G, Boon N, Pauwels M, Slomka V, Rodrigues Herrero E, Quirynen M, Teughels W. Effect of Bdellovibrio bacteriovorus HD100 on multispecies oral communities. Anaerobe. 2015;35(Part A):45–53.
Mahmoud KK, McNeely, D., Elwood, C., Koval, S.F. (2007) Design and performance of a 16S rRNA-targeted oligonucleotide probe for detection of members of the genus Bdellovibrio by fluorescence in situ hybridization. Appl Environ Microbiol 73: 7488–7493.
Mahmoud KK, Koval SF. Characterization of type IV pili in the life cycle of the predator bacterium Bdellovibrio. Microbiology. 2010;156:1040–51.
Makowski Ł, Trojanowski D, Till R, Lambert C, Lowry R, Sockett RE, Zakrzewska-Czerwińska J. Dynamics of chromosome replication and its relationship to predatory attack lifestyles in Bdellovibrio bacteriovorus. Appl Environ Microbiol. 2019;85:00730–19.
Markelova NY. Effect of toxic pollutants on Bdellovibrio. Process Biochem. 2002;37:1177–81.
Martínez V, de la Peña F, García-Hidalgo J, de la Mata I, García JL, Prieto MA. Identification and biochemical evidence of a medium-chain-length Polyhydroxyalkanoate Depolymerase in the Bdellovibrio bacteriovorus predatory hydrolytic arsenal. Appl Environ Microbiol. 2012;78:6017–26.
Martínez V, Jurkevitch E, García JL, Prieto MA. Reward for Bdellovibrio bacteriovorus for preying on a polyhydroxyalkanoate producer. Environ Microbiol: n/a-n/a. 2013.
Matassa S, Verstraete W, Pikaar I, Boon N. Autotrophic nitrogen assimilation and carbon capture for microbial protein production by a novel enrichment of hydrogen-oxidizing bacteria. Water Res. 2016;101:137–46.
McCauley EP, Haltli B, Kerr RG. Description of Pseudobacteriovorax antillogorgiicola gen. nov., sp. nov., a bacterium isolated from the gorgonian octocoral Antillogorgia elisabethae, belonging to the family Pseudobacteriovoracaceae fam. nov., within the order Bdellovibrionales. Int J Syst Evol Microbiol. 2015;65:522–30.
Medina A, Shanks R, Kadouri D. Development of a novel system for isolating genes involved in predator-prey interactions using host independent derivatives of Bdellovibrio bacteriovorus 109J. BMC Microbiol. 2008;8:33.
Milner DS, Till R, Cadby I, Lovering AL, Basford SM, Saxon EB, Liddell S, Williams LE, Sockett RE. Ras GTPase-like protein MglA, a controller of bacterial social-motility in Myxobacteria, has evolved to control bacterial predation by Bdellovibrio. PLoS Genet. 2014;10:e1004253.
Monnappa AK, Dwidar M, Mitchell RJ. Application of bacterial predation to mitigate recombinant bacterial populations and their DNA. Soil Biol Biochem. 2013;57:427–35.
Mukherjee S, Brothers KM, Shanks RMQ, Kadouri DE. Visualizing Bdellovibrio bacteriovorus by using the tdTomato fluorescent protein. Appl Environ Microbiol. 2016;82:1653–61.
Murray AK, Zhang L, Yin X, Zhang T, Buckling A, Snape J, Gaze WH. Novel insights into selection for antibiotic resistance in complex microbial communities. mBio. 2018;9:e00969–18.
Negri M-C, Morosini M-I, Baquero M-R, Campo Rd, Blázquez J, Baquero F. Very low cefotaxime concentrations select for Hypermutable Streptococcus pneumoniae populations. Antimicrob Agents Chemother. 2002;46:528–30.
Oyedara OO, De Luna-Santillana EDJ, Olguin-Rodriguez O, Guo X, Mendoza-Villa MA, Menchaca-Arredondo JL, Elufisan TO, Garza-Hernandez JA, Garcia Leon I, Rodriguez-Perez MA. Isolation of Bdellovibrio sp. from soil samples in Mexico and their potential applications in control of pathogens. Microbiol Open. 2016;5:992–1002.
Paix B, Ezzedine JA, Jacquet S. Diversity, dynamics, and distribution of Bdellovibrio and like organisms in Perialpine Lakes. Appl Environ Microbiol. 2019;85:e02494–18.
Pasternak Z, Pietrokovski S, Rotem O, Gophna U, Lurie-Weinberger MN, Jurkevitch E. By their genes ye shall know them: genomic signatures of predatory bacteria. ISME J. 2013;7:756–69.
Pasternak Z, Njagi M, Shani Y, Chanyi R, Rotem O, Lurie-Weinberger MN, Koval S, Pietrokovski S, Gophna U, Jurkevitch E. In and out: an analysis of epibiotic vs periplasmic bacterial predators. ISME J. 2014;8:625–35.
Pineiro SA, Sahaniuk GE, Romberg E, Williams H. Predation pattern and phylogenetic analysis of Bdellovibrionaceae from the great salt Lake, Utah. Curr Microbiol. 2004;48:113–7.
Pineiro S, Chauhan A, Berhane T-K, Athar R, Zheng G, Wang C, Dickerson T, Liang X, Lymperopoulou D, Chen H, Christman M, Louime C, Babiker W, Stine OC, Williams H. Niche partition of Bacteriovorax operational taxonomic units along salinity and temporal gradients in the Chesapeake Bay reveals distinct estuarine strains. Microb Ecol. 2013;65:652–60.
Rendulic S, Jagtap P, Rosinus A, Eppinger M, Baar C, Lanz C. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science. 2004;303:689–92.
Rice TD, Williams HN, Turng BF. Susceptibility of bacteria in estuarine environments to autochthonous bdellovibrios. Microb Ecol. 1998;35:256–64.
Richards GP, Fay JP, Dickens KA, Parent MA, Soroka DS, Boyd EF. Predatory bacteria as natural modulators of Vibrio parahaemolyticus and Vibrio vulnificus in seawater and oysters. Appl Environ Microbiol. 2012;78:7455–66.
Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J-F, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu W-T, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–7.
Rogosky A, Moak P, Emmert E. Differential predation by Bdellovibrio bacteriovorus 109J. Curr Microbiol. 2006;52:81–5.
Roschanski N, Klages S, Reinhardt R, Linscheid M, Strauch E. Identification of genes essential for prey-independent growth of Bdellovibrio bacteriovorus HD100. J Bacteriol. 2011;193:1745–56.
Rotem O, Pasternak Z, Jurkevitch E. Bdellovibrio and like organisms. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The prokaryotes: deltaproteobacteria and epsilonproteobacteria. Berlin/Heidelberg: Springer; 2014. p. 3–17.
Rotem O, Pasternak Z, Shimoni E, Belausov E, Porat Z, Pietrokovski S, Jurkevitch E. Cell-cycle progress in obligate predatory bacteria is dependent upon sequential sensing of prey recognition and prey quality cues. Proc Natl Acad Sci. 2015;112:E6028–37.
Rotem O, Nesper J, Borovok I, Gorovits R, Kolot M, Pasternak Z, Shin I, Glatter T, Pietrokovski S, Jenal U, Jurkevitch E. An extended cyclic Di-GMP network in the predatory bacterium Bdellovibrio bacteriovorus. J Bacteriol. 2016;198:127–37.
Ruby EG, Rittenberg SC. Differentiation after premature release of intraperiplasmically growing Bdellovibrio bacteriovorus. J Bacteriol. 1983;154:32–40.
Sanchez-Amat A, Torrella F. Formation of stable bdelloplasts as a starvation-survival strategy of marine bdellovibrios. Appl Environ Microbiol. 1990;56:2127–5.
Sathyamoorthy R, Maoz A, Pasternak Z, Im H, Huppert A, Kadouri D, Jurkevitch E. Bacterial predation under changing viscosities. Environ Microbiol. 2019;21:2997–3010.
Schoeffield AJ, Williams HN. Efficiencies of recovery of bdellovibrios from brackish-water environments by using various bacterial species as prey. Appl Environ Microbiol. 1990;56:230–6.
Schwudke D, Strauch E, Krueger M, Appel B. Taxonomic studies of predatory bdellovibrios based on 16S rRNA analysis, ribotyping and the hit locus and characterization of isolates from the gut of animals. Syst Appl Microbiol. 2001;24:385–94.
Semblante GU, Hai FI, Ngo HH, Guo W, You S-J, Price WE, Nghiem LD. Sludge cycling between aerobic, anoxic and anaerobic regimes to reduce sludge production during wastewater treatment: performance, mechanisms, and implications. Bioresour Technol. 2014;155:395–409.
Semblante GU, Phan HV, Hai FI, Xu Z-Q, Price WE, Nghiem LD. The role of microbial diversity and composition in minimizing sludge production in the oxic-settling-anoxic process. Sci Total Environ. 2017;607–608:558–67.
Shemesh Y, Jurkevitch E. Plastic phenotypic resistance to predation by Bdellovibrio and like organisms in bacterial prey. Environ Microbiol. 2004;6:8–12.
Shnel N, Barak Y, Ezer T, Dafni Z, van Rijn J. Design and performance of a zero-discharge tilapia recirculating system. Aquac Eng. 2002;26:191–203.
Spencer SJ, Tamminen MV, Preheim SP, Guo MT, Briggs AW, Brito IL, Weitz DA, Pitkanen LK, Vigneault F, Virta MP, Alm EJ. Massively parallel sequencing of single cells by epicPCR links functional genes with phylogenetic markers. ISME J. 2016;10:427–36.
Staples DG, Fry JC. Factors which influence the enumeration of Bdellovibrio bacteriovorus in sewage and river water. J Appl Bacteriol. 1973;36:1–11.
Stolp H, Petzold H. Untersuchungen über einen obligat parasitischen Mikroorganismus mit lytischer Aktivität für Pseudomonas-Bakterien. J Phytopathol. 1962;45:364–90.
Sun Y, Ye J, Hou Y, Chen H, Cao J, Zhou T. Predation efficacy of Bdellovibrio bacteriovorus on multidrug-resistant clinical pathogens and their corresponding biofilms. Jpn J Infect Dis. 2017;70:485–9.
Szabó E, Liébana R, Hermansson M, Modin O, Persson F, Wilén B-M. Comparison of the bacterial community composition in the granular and the suspended phase of sequencing batch reactors. AMB Express. 2017;7:168.
Tran NH, Reinhard M, Gin KY-H. Occurrence and fate of emerging contaminants in municipal wastewater treatment plants from different geographical regions-a review. Water Res. 2018;133:182–207.
Uematsu T. Ecology of Bdellovibrio parasitic to rice bacterial leaf blight pathogen, Xanthomonas oryzae. Rev Plant Prot Res. 1980;13:12–26.
Van Essche M, Sliepen I, Loozen G, Van Eldere J, Quirynen M, Davidov Y, Jurkevitch E, Boon N, Teughels W. Development and performance of a quantitative PCR for the enumeration of Bdellovibrionaceae. Environ Microbiol Rep. 2009;1:228–33.
Varon M. Selection of predation-resistant bacteria in continuous culture. Nature. 1979;277:386–8.
Varon M, Shilo M. Inhibition of the predatory activity of Bdellovibrio by various environmental pollutants. Microb Ecol. 1981;7:107–11.
von Wintersdorff CJH, Penders J, van Niekerk JM, Mills ND, Majumder S, van Alphen LB, Savelkoul PHM, Wolffs PFG. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front Microbiol. 2016;7:173.
Wan C, Zhang P, Lee D-J, Yang X, Liu X, Sun S, Pan X. Disintegration of aerobic granules: role of second messenger cyclic di-GMP. Bioresour Technol. 2013;146:330–5.
Wang Z, Kadouri D, Wu M. Genomic insights into an obligate epibiotic bacterial predator: Micavibrio aeruginosavorus ARL-13. BMC Genomics. 2011;12:453.
Welsh RM, Zaneveld JR, Rosales SM, Payet JP, Burkepile DE, Thurber RV. Bacterial predation in a marine host-associated microbiome. ISME J. 2015.
Wen CQ, Lai XT, Xue M, Huang YL, Li HX, Zhou SN. Molecular typing and identification of Bdellovibrio-and-like organisms isolated from seawater shrimp ponds and adjacent coastal waters. J Appl Microbiol. 2009;106:1154–62.
Wendland A, Ozoguz Y. Operation costs of wastewater treatment plants. Ahrensburg: Employee of Hamburg Public Sewage Company; 2005.
Whitby GE. Bdellovibrio bacteriovorus 6-5-s and Aquaspirillum serpens VHL in continuous culture; 1977.
Wijeyekoon S, Carere CR, West M, Nath S, Gapes D. Mixed culture polyhydroxyalkanoate (PHA) synthesis from nutrient rich wet oxidation liquors. Water Res. 2018;140:1–11.
Wilkinson MHF. Predation in the presence of decoys: an inhibitory factor on pathogen control by bacteriophages or bdellovibrios in dense and diverse ecosystems. J Theor Biol. 2001;208:27–36.
Williams HN. A study of the distribution of bdellovibrios in estuarine sediment over an annual cycle. Microb Ecol. 1988;15:9–20.
Yılmaz H, Çelik MA, Şengezer Ç, Özkan M. Use of Bdellovibrio bacteriovirus as biological cleaning method for MBRSystems; 2014.
Yu R, Zhang S, Chen Z, Li C. Isolation and application of predatory Bdellovibrio-and-like organisms for municipal waste sludge biolysis and dewaterability enhancement. Front Environ Sci Eng. 2017;11:10.
Zheng G, Wang C, Williams HN, Pineiro SA. Development and evaluation of a quantitative real-time PCR assay for the detection of saltwater Bacteriovorax. Environ Microbiol. 2008;10:2515–26.
Zheng Y, Cheng C, Zhou Z, Pang H, Chen L, Jiang L-M. Insight into the roles of packing carriers and ultrasonication in anaerobic side-stream reactor coupled membrane bioreactors: sludge reduction performance and mechanism. Water Res. 2019;155:310–9.
Zhou Z, Qiao W, Xing C, Shen X, Hu D, Wang L. A micro-aerobic hydrolysis process for sludge in situ reduction: performance and microbial community structure. Bioresour Technol. 2014;173:452–6.
Acknowledgments
I would like to thank Eddie Cytryn (Agricultural Research Organization, Bet Dagan, Israel) for reading the manuscript and for his sharp suggestions that truly improved it.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Jurkevitch, E. (2020). The Ecology of Bdellovibrio and Like Organisms in Wastewater Treatment Plants. In: Jurkevitch, E., Mitchell, R. (eds) The Ecology of Predation at the Microscale. Springer, Cham. https://doi.org/10.1007/978-3-030-45599-6_2
Download citation
DOI: https://doi.org/10.1007/978-3-030-45599-6_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-45598-9
Online ISBN: 978-3-030-45599-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)