Abstract
Periodontitis is a common bacterially induced inflammatory condition that damages the tooth-supporting apparatus and negatively impacts the systemic health. It affects over 700 million people worldwide with an estimated economic burden totaling to $442 billion annually. A bacterial triad in the subgingival niche comprising of Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia is very influential in the development of periodontitis. Significantly, all these three pathogens produce a sialidase enzyme that can cleave terminal sialic acid residue from host-derived sialoglycoproteins, such as present on the surface of oral epithelial cells and in saliva and gingival crevicular fluid. This ability to release and utilize sialic acid from host glycoproteins is crucial for their growth and immune evasion and survival strategies. In addition, sialic acid cleavage can cause immune dysfunction and disruption of tissue integrity and thus exacerbate periodontal inflammation in various ways. Here, we propose that inhibition of pathogen-derived sialidase activity with sialidase-targeting pharmacological drugs may be an attractive adjunct therapy in the treatment of periodontitis.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
Periodontitis (PD) is an inflammatory disease characterized by progressive destruction of the tooth-supporting structures, often leading to tooth loss. It affects over 700 million people worldwide and is one of the major oral conditions globally after caries that is estimated to cost $442 billion per year [1,2,3]. PD is induced by a subgingival polymicrobial community in which a bacterial triad known as the red complex comprising of Porphyromonas gingivalis (Pg), Treponema denticola (Td), and Tannerella forsythia (Tf) is strongly represented. The environmental niche that these bacteria inhabit, as with most human mucosal infections and colonizable surfaces such as the airways, gut, and female reproductive tract, is rich in glycoproteins decorated with N- or O-linked sugar-glycan chains, of which most are capped at the terminal end with the 9-carbon sugar, sialic acid (Neu5Ac) [4]. A wide variety of pathogens from different genera inhabiting a range of niches within the body, including the oral cavity, are known to utilize Neu5Ac as a source of carbon and nitrogen or to cloak their surface to avoid immune attack (please see review [5]). Likewise, the red-complex bacteria have been shown to produce sialidase that cleaves the terminal sialic acid from glycoprotein-linked glycans on the surface of epithelial cells, immune cells, and oral secretions such as gingival crevicular fluid (GCF). In this review, we summarize accumulating evidence demonstrating that sialic acid harvesting by oral pathogens is not only key to their survival in the oral cavity but might also be responsible for immune dysfunction and disruption of tissue integrity observed in periodontitis. Further, we propose that sialidase inhibition with sialidase-targeting pharmacological drugs, such as those currently employed for influenza and also from a range of other sources, may be an attractive adjunct therapy in controlling periodontitis. While sialidase production is a prominent feature of the red-complex bacteria, it is not limited to these species in the oral cavity. For example, several oral commensal and opportunistic organisms such as Streptococci spp. (mitis, oralis, intermedius), Capnocytophaga spp., Actinomyces naeslundii, Actinomyces oris, Prevotella oralis, Bifidobacterium dentium, and Propionibacterium acnes produce related sialidases [6, 7]. It is likely that both the nature of physical niche in the mouth and the physiology of individual species dictate the importance of sialidase activity in a given setting. Among the species that might be considered as early or primary colonizers of tooth surfaces in the mouth such as Streptococci, Actinomyces, Bifidobacterium, and Propionibacterium are saccharolytic organisms where their primary carbon source is often glucose or other dietary sugars, meaning sialidase activity might chiefly be present to access underlying sugars as part of a more versatile catabolic profile. However, as these organisms are not in direct contact with mucosal surfaces, or immune cells, the impact of their sialidases on inflammation is expected to be limited at best.
Moreover, some bacteria in the oral microbiota that do not produce their own sialidase likely benefit from sialidase activity of other bacteria. For example, some Fusobacterium spp. can catabolize exogenous sialic acid for energy or reprocess sialic acid to decorate their surface with the sugar [5]. Still others can also benefit from community sialidase activity, which might allow them to utilize sialic acid or bind to underlying sugars (cryptic epitopes) on glycans, e.g., S. gordonii, S. mutans, S. sanguinis, and S. salivarius [7,8,9,10]. The contribution of sialidase activity in periodontal inflammation has recently come to light from clinical observations of raised levels of sialidase activity in GCF of periodontitis patients [11]. The negative impact of sialidase activity on periodontal disease is heightened since in the context of the subgingival environment of the periodontal pocket, sialidase activity can disrupt the integrity (structure-function) of host glycoproteins, disrupt pattern-recognition receptor (TLRs) signaling in infiltrating immune cells as well as epithelial layers, and promote the survival, persistence, and pathogenesis of periodontal bacteria.
1 Sialic Acid Foraging by Periodontal Pathogens
Current evidence indicates that the key periodontal pathogens P. gingivalis, T. denticola, and T. forsythia strictly rely on host-derived sialic acid for their survival in the oral cavity and virulence. This is unlike many other human pathogens such as Campylobacter jejuni, Neisseria meningitidis, and some Fusobacterium spp. that have dedicated biosynthetic pathways to synthesize their own sialic acid [5]. The work from our groups showed that T. forsythia mutants lacking sialidase and the sialic acid transporter NanT are unable to acquire environmental sialic acid and are severely attenuated in biological activities such as survivability in biofilms on sialoglycoprotein substrates [12] and interactions with and survival on epithelial cells [13, 14]. In addition, other groups have shown that sialidase deletion in a capsulated strain of P. gingivalis results in reduced capsule thickness phenotype [15] and gingipain protease expression [16]. These studies showed that compared to the wild-type capsulated strain, a sialidase-deficient mutant in vitro formed less biofilms and was less resistant to killing by the host complement [15]. Moreover, while the wild-type strain was able to spread to multiple organs and cause mouse mortality following subcutaneous infection, the sialidase-deficient mutant was found to be highly attenuated showing only localized spreading and ineffective in causing mortality in mice. In the case of T. denticola, sialidase deficiency caused an increased surface deposition of complement attack complex and reduced virulence in a mouse model [17]. In addition, though it is yet to be established in the periodontal setting, our groups have preliminary evidence of sialidase-dependent modulation of epithelial and monocyte responses to LPS stimulation, and as such sialic acid scavenging might exacerbate inflammation by affecting TLR signaling. In this scenario, periodontal pathogen-secreted sialidases might contribute to subgingival inflammation through increasing TLR4 responsiveness to LPS as it has been shown that desialylation of TLR4 promotes its dimerization and activation at other human mucosal layers [18]. Moreover, sialidase-mediated desialylation is a mechanism through which one of the immunosuppressive circuits is compromised in immune cells dependent on Siglecs (sialic acid-binding immunoglobulin-type lectins) [19]. In a normal homeostatic state, Siglec-G/10 (G, mouse/10, human) binds a sialylated receptor CD24 and induces an inhibitory circuit that attenuates TLR signaling. Indeed, the data from our lab show that T. forsythia sialidase treatment in vitro can increase TLR4 activation in macrophages (unpublished). Thus, sialic acid utilization by pathogens can take roles in immune evasion and dysregulation, community biofilm development, and pathogen survival. Plausibly, the sialidase activity might also assist periodontal pathogens in mitigating the toxic effects of ROS (reactive oxygen species) in the inflamed periodontal pockets as released sialic acid residues can act as scavengers of peroxide residues [20, 21]. Taken together, these findings demonstrate that sialidase is an important and common virulence determinant that may contribute to the pathogenicity of periodontal pathogens.
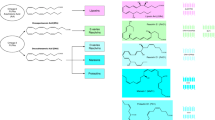
As mentioned above, both P. gingivalis and T. denticola possess sialidases that have been shown to be key to their survival and virulence [15,16,17]. Both P. gingivalis and T. denticola sialidases display a similar domain organization with a C-terminal catalytic domain with homology to glycosyl hydrolase 33 (GH33) family carbohydrate-active enzymes in the CAZy database [22] that is preceded by an N-terminal domain, which in the case of T. denticola is a putative peptidoglycan binding domain [17] while in the case of P. gingivalis shows no homology to any of the known protein domains in the sequence or structural databases (Fig. 12.1). Despite clear data existing that T. denticola seems to require sialidase activity or monomeric sialic acid for growth in serum, its genome sequence lacks any homologs of sialic acid catabolic genes [17], suggesting the existence of novel pathways.
Schematic representation of periodontal pathogen CAzymes. A. sialidases: The GH33 sialidase domains are shown in yellow, with the Asp (D) boxes indicated alongside the catalytic FRIP domains. Signal sequences are indicated in red. N-terminal domains are indicated in gray, with the T. forsythia confirmed carbohydrate-binding domain (CBD); T. denticola putative cell wall anchor (Pfam PF09479) indicated; alongside the P. gingivalis N-terminus, which has no homology in databases
In contrast, biochemical and functional knowledge of the T. forsythia NanH sialidase is well advanced, mostly from work in our teams. As with P. gingivalis and T. denticola, we have revealed that T. forsythia sialidase is key to the bacterium’s microbiology and virulence [5, 12,13,14]. However, unlike P. gingivalis and T. denticola, it expresses its sialidase, NanH, encoded by the nanH gene, as part of a dedicated sialic acid scavenging, transport, and utilization operon (Fig. 12.1). Of note, this operon is present in all of the T. forsythia strains sequenced to date, including the genomes of three clinical isolates recently sequenced by our group [23] and in metagenomic reconstruction from ancient calculus [24]. Notably the nan operon is missing from the related Tannerella BU063 strains isolated to date. NanH is a 62 kDa secreted enzyme again comprising two domains with the C-terminal domain comprising a catalytic domain that is a member of the GH33 family mentioned above. It contains five Asp-box motifs, a conserved catalytic Arginine triad, and FRIP-motif (Fig. 12.1). Once again T. forsythia NanH possesses a 170–180-amino-acid-long, N-terminal domain with no sequence or structural homology in the PDB database outside of sialidase enzymes found within members of the Bacteroidetes. The NanH N-terminal domain has now been characterized by our groups as a novel carbohydrate-binding module (CBM) with broad specificity for host glycans but which prefers sialylated glycans and those with alpha-2,3 glycosidic linkages [25]. As mentioned above, NanH is associated with a novel sialic acid utilization system which contains a novel transporter system (NanOUT) [26,27,28] for the uptake of monomeric sialic acid and a 9-0-sialate-acetylesterase (NanS) [26]. This NanS enzyme is novel in its class as it contains two SGNH-like Sialate-esterase domains and acts to enhance release of Neu5Ac (sialic acid) from host glycoproteins containing diacetylated sialic acids (Neu5,9Ac) that block the action of sialidases (ref). Furthermore, in the nan operon, NanH is followed by a β-hexosaminidase [29], which may act to release subsequent sugars within host glycans [26], and a predicted sialic acid mutarotase that improves utilization of the alpha anomeric form of sialic acid [30].
In addition, our studies have shown that the release, transport, and utilization of sialic acid are critical to the interaction and survival of T. forsythia with epithelial cells as well as bacterium’s growth in glycoprotein-based biofilms [12,13,14, 29]. These data shed light on the ability of the bacterium to cleave and utilize host Neu5Ac as a survival strategy whereby removal of terminal sialic acid residues in host glycoproteins in salivary secretions and on epithelial cells allows the bacterium to colonize and utilize the liberated sialic acid as a source of carbon and nitrogen and possibly as a precursor for the peptidoglycan (PGN) synthesis (Fig. 12.2). This is pertinent as PGN synthesis pathways are notably lacking in T. forsythia, which cannot de novo synthesize the PGN amino sugar N-acetylmuramic acid (MurNAc) from simple nonamino sugars. While it is able to uptake muropeptides released by other bacteria in the oral cavity, e.g., Fusobacterium nucleatum [31], we postulate that the role of T. forsythia sialidase in liberating free monomeric sialic acid might be critical for the bacterium’s survival in the oral cavity as sialic acid could serve as an alternative source for MurNAc synthesis as we have suggested previously [28]. This is significant as the human host is also unable to synthesize and thus provide MurNAc to the bacterium. We postulate that by harvesting sialic acid as an alternative nutritional source, T. forsythia is able to gain a competitive edge over the subgingival cohabiting microbiota. In support of a notion that sialic acid foraging by T. forsythia might be critical in this respect, a community-wide transcriptome analysis of the subgingival microbiome has indicated that the T. forsythia nanH transcript levels likely increase in subjects with periodontitis as compared to healthy controls [32].
Sialic acid scavenging as a means of peptidoglycan biosynthesis and survival by T. forsythia. (a) Genetic organization of sialic acid utilization operon. (b) Putative metabolic pathway of sialic acid utilization to peptidoglycan biosynthesis. Dashed line arrows indicate canonical enzymes involved in the conversion of various sugar intermediates to peptidoglycan have not been found in T. forsythia
One by-product of this use of sialic acid would be a potential increase in availability of peptidoglycan fragments comprising NOD (nucleotide-binding oligomerization domain)-like receptor ligands such as muramyl dipeptide (MDP; NOD2 ligand) and γ-d-glutamyl-meso-diaminopimelic acid (iE-DAP; NOD1 ligand) [33] in the subgingival environment which may heighten NOD-mediated inflammation. Thus, sialic acid scavenging may potentially exacerbate inflammation by affecting TLR signaling as well as putatively increasing NOD-mediated inflammation.
Taken together, sialic acid cleavage by periodontal pathogens contributes to immune dysregulation and evasion, community biofilm development, and pathogen survival (Fig. 12.3). Importantly, support for the notion that sialic acid acquisition by the subgingival microbiome might be important in disease progression comes from our studies demonstrating a heightened expression of microbial sialidase activity in diseased sites compared to healthy sites and the heightened sialidase activity as a predictor of poor standard treatment outcomes [11].
2 Global Sialidase Activity and Impact on Health and Disease
It is important to take into account the contribution of sialidase activity of commensal oral organisms which may be critical for microbial ecology and health or disease in ways that have yet to be fully understood. In this regard, the sialidase activity of commensal bacteria may also be required for their growth/colonization and thus may have roles in maintaining healthy microbiota. The damaging effects of sialidase from commensal bacteria colonizing the supragingival niche might be mitigated both due to sialic acid-rich salivary mucins acting as a coating layer on hard surfaces. The delicate balance between the host and the sialidase activity in health is thus likely maintained by appropriate host barrier functions of salivary proteins including mucins and innate immune responses. This is in contrast to the subgingival niche and inflamed periodontal pockets where sialidases from periodontal pathobionts can directly encounter infiltrating immune cells (monocytes/macrophages) and influence inflammation. In addition, as mentioned above, sialidase activity might promote the survival of pathobionts in the harsh subgingival environment; in the case of T. forsythia, sialidase activity promotes its growth, and for P. gingivalis and T. denticola, sialidase activity is important in protecting these bacteria against the complement attack while it may also allow them easier access to underlying protein substrates that are key for their virulence and nutrition [15, 17]. Thus, the role of sialidase might be context dependent. In a periodontitis setting, inflammation and dysbiosis due to pathobionts P. gingivalis, T. forsythia, and T. denticola and proliferation of commensals might further increase global sialidase activity and thus synergistically exacerbate periodontitis. This notion is supported by observations that sialidase activity is elevated in plaque biofilms [34] and is high in diseased GCF in patients [11].
3 Translational Potential of Sialidase Inhibition
The dependence of key periodontal pathogens on sialic acid for survival, virulence, and immune modulation provides treatment opportunities for periodontitis by targeting sialidase activity with sialidase inhibitors. The potential translatability of blocking periodontal pathogen sialidase activity, heightened by the availability of several current FDA-approved (Tamiflu, Relenza, Peramivir) or novel sialidase inhibitors, to alleviate periodontitis and associated dysbiosis development and inflammation is very promising. Sialidase inhibitors could be used as an adjunct therapy in situations where periodontal pathogens present resistance to antibiotics. This may also minimize the use of antibiotics and development of antibiotic resistance. Our published and preliminary studies have demonstrated that inhibiting sialidase function with pharmacological inhibitors (FDA-approved zanamivir and oseltamivir) can block the ability of T. forsythia to form biofilms and survive on sialoglycoconjugates and epithelial cell monolayers, both in mono-species and mixed-species infections with P. gingivalis and Fusobacterium spp. [12, 14]. Importantly, sialidase inhibition can block the availability of sialic acid for peptidoglycan biosynthesis in T. forsythia, and therefore may reduce any selective advantage T. forsythia might have over other bacteria in biofilms in vivo. In the case of P. gingivalis, lack of sialidase activity in the bacterium results in increasing sensitivity to hydrogen peroxide and reduced gingipain protease activity, suggesting that sialidase activity might be involved in regulating the virulence potential of this keystone pathogen [16]. In a capsulated strain of P. gingivalis, sialidase activity seems to influence capsule formation and confer the bacterium the ability to resist complement attack [15]. Moreover, based on in vivo studies in a mouse model, the role of sialidase activity has been highlighted. It has been shown that while wild-type P. gingivalis has the ability to disseminate to multiple organs following infection, sialidase deficiency abrogates this ability causing only localized spreading around the site of infection [15]. In T. denticola, sialidase activity seems to be responsible for the acquisition of sialic acid as a nutrient as well as for sialic acid modification of surface glycoproteins as a means of blocking deposition of membrane attack complex and killing [17]. An obvious next step is therefore to test the in vivo potential of these synthetic as well as other naturally occurring plant-derived inhibitors (berberine and palmatine) [35] against sialidase-producing pathogens. In this regard, it is tempting to first test the efficacy of sialidase inhibitors/inhibitor cocktails found effective in vitro in conferring protection to pathogen-induced periodontal bone loss in a mouse model, including in polymicrobial situations. The inhibitors might include both those that block the NanH sialidase’s enzymatic activity (Tamiflu) and inhibitors that block NanH’s lectin function (S-Lewis oligosaccharides or glycan mimics). The use of sialidase inhibitors for the treatment of periodontitis is not without risks due to potential off-target effects of these inhibitors on endogenous host sialidases (neuraminidases) playing important roles in the physiology of the host [36,37,38]. However, in principle one can design specific inhibitors that target the bacterial sialidases while not compromising functionality of the host sialidases [39]. We envisage a situation where topical administration of glycosidase inhibitory compounds could be deployed as gels applied as part of nonsurgical root debridement or within mouthwashes.
4 Summary
In this review we provided accumulating evidence demonstrating that sialic acid foraging by oral pathogens is not only key to their survival in the oral cavity but is also responsible for immune dysfunction and dysbiosis observed in periodontitis. We envisage that sialidase neutralization with pharmacological drugs, such as those currently employed for influenza, and also plant derived sialidase inhibitors in development may be an attractive adjunct therapy in controlling periodontitis.
References
Kassebaum NJ, Bernabe E, Dahiya M, Bhandari B, Murray CJ, Marcenes W. Global burden of severe periodontitis in 1990-2010: a systematic review and meta-regression. J Dent Res. 2014;93:1045–53.
Listl S, Galloway J, Mossey PA, Marcenes W. Global economic impact of dental diseases. J Dent Res. 2015;94:1355–61.
Sharma A. Genome functions of Tannerella forsythia in bacterial communities. In: Kolenbrander PE, editor. Oral microbial communities: genome inquiry and interspecies communication. Washington, DC: American Society for Microbiology; 2011. p. 135.
Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta. 1999;1473:4–8.
Stafford G, Roy S, Honma K, Sharma A. Sialic acid, periodontal pathogens and Tannerella forsythia: stick around and enjoy the feast! Mol Oral Microbiol. 2012;27:11–22.
Moncla BJ, Braham P, Hillier SL. Sialidase (neuraminidase) activity among gram-negative anaerobic and capnophilic bacteria. J Clin Microbiol. 1990;28:422–5.
Beighton D, Whiley RA. Sialidase activity of the “Streptococcus milleri group” and other viridans group streptococci. J Clin Microbiol. 1990;28:1431–3.
Byers HL, Homer KA, Beighton D. Utilization of sialic acid by viridans streptococci. J Dent Res. 1996;75:1564–71.
Imaki H, Tomoyasu T, Yamamoto N, Taue C, Masuda S, Takao A, Maeda N, Tabata A, Whiley RA, Nagamune H. Identification and characterization of a novel secreted glycosidase with multiple glycosidase activities in Streptococcus intermedius. J Bacteriol. 2014;196:2817–26.
Wong A, Grau MA, Singh AK, Woodiga SA, King SJ. The role of neuraminidase-producing bacteria in exposing cryptic carbohydrate receptors for Streptococcus gordonii adherence. Infect Immun. 2018;86(7). https://doi.org/10.1128/IAI.00068-18.
Gul SS, Griffiths GS, Stafford GP, Al-Zubidi MI, Rawlinson A, Douglas CWI. Investigation of a novel predictive biomarker profile for the outcome of periodontal treatment. J Periodontol. 2017;88:1135–44.
Roy S, Honma K, Douglas CWI, Sharma A, Stafford GP. Role of sialidase in glycoprotein utilization by Tannerella forsythia. Microbiology. 2011;157:3195–202.
Honma K, Mishima E, Sharma A. Role of Tannerella forsythia NanH sialidase in epithelial cell attachment. Infect Immun. 2011;79:393–401.
Honma K, Ruscitto A, Frey AM, Stafford GP, Sharma A. Sialic acid transporter NanT participates in Tannerella forsythia biofilm formation and survival on epithelial cells. Microb Pathog. 2015;94:12–20.
Li C, Kurniyati HB, Bian J, Sun J, Zhang W, Liu J, Pan Y, Li C. Abrogation of neuraminidase reduces biofilm formation, capsule biosynthesis, and virulence of Porphyromonas gingivalis. Infect Immun. 2012;80:3–13.
Aruni W, Vanterpool E, Osbourne D, Roy F, Muthiah A, Dou Y, Fletcher HM. Sialidase and sialoglycoproteases can modulate virulence in Porphyromonas gingivalis. Infect Immun. 2011;79:2779–91.
Kurniyati K, Zhang W, Zhang K, Li C. A surface-exposed neuraminidase affects complement resistance and virulence of the oral spirochete Treponema denticola. Mol Microbiol. 2013;89:842–56.
Feng C, Stamatos NM, Dragan AI, Medvedev A, Whitford M, Zhang L, Song C, Rallabhandi P, Cole L, Nhu QM, Vogel SN, Geddes CD, Cross AS. Sialyl residues modulate LPS-mediated signaling through the toll-like receptor 4 complex. PLoS One. 2012;7:e32359.
Chen GY, Chen X, King S, Cavassani KA, Cheng J, Zheng X, Cao H, Yu H, Qu J, Fang D, Wu W, Bai XF, Liu JQ, Woodiga SA, Chen C, Sun L, Hogaboam CM, Kunkel SL, Zheng P, Liu Y. Amelioration of sepsis by inhibiting sialidase-mediated disruption of the CD24-SiglecG interaction. Nat Biotechnol. 2011;29:428–35.
Iijima R, Takahashi H, Namme R, Ikegami S, Yamazaki M. Novel biological function of sialic acid (N-acetylneuraminic acid) as a hydrogen peroxide scavenger. FEBS Lett. 2004;561:163–6.
Iijima R, Takahashi H, Ikegami S, Yamazaki M. Characterization of the reaction between sialic acid (N-acetylneuraminic acid) and hydrogen peroxide. Biol Pharm Bull. 2007;30:580–2.
Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–5.
Stafford GP, Chaudhuri RR, Haraszthy V, Friedrich V, Schaffer C, Ruscitto A, Honma K, Sharma A. Draft genome sequences of three clinical isolates of Tannerella forsythia isolated from subgingival plaque from periodontitis patients in the United States. Genome Announc. 2016;4:e01286–16.
Warinner C, Rodrigues JF, Vyas R, Trachsel C, Shved N, Grossmann J, Radini A, Hancock Y, Tito RY, Fiddyment S, Speller C, Hendy J, Charlton S, Luder HU, Salazar-Garcia DC, Eppler E, Seiler R, Hansen LH, Castruita JA, Barkow-Oesterreicher S, Teoh KY, Kelstrup CD, Olsen JV, Nanni P, Kawai T, Willerslev E, von Mering C, Lewis CM Jr, Collins MJ, Gilbert MT, Ruhli F, Cappellini E. Pathogens and host immunity in the ancient human oral cavity. Nat Genet. 2014;46(4):336–44. https://doi.org/10.1038/ng.2906.
Frey AM, Satur MJ, Phansopa C, Parker JL, Bradshaw D, Pratten J, Stafford GP. Evidence for a novel carbohydrate binding module (CBM) of Tannerella forsythia NanH sialidase, key to interactions at the host-pathogen interface. Biochem J. 2018;475(6):1159–76. https://doi.org/10.1042/BCJ20170592.
Phansopa C, Kozak RP, Liew LP, Frey AM, Farmilo T, Parker JL, Kelly DJ, Emery RJ, Thomson RI, Royle L, Gardner RA, Spencer DI, Stafford GP. Characterization of a sialate-O-acetylesterase (NanS) from the oral pathogen Tannerella forsythia that enhances sialic acid release by NanH, its cognate sialidase. Biochem J. 2015;472:157–67.
Phansopa C, Roy S, Rafferty JB, Douglas CW, Pandhal J, Wright PC, Kelly DJ, Stafford GP. Structural and functional characterization of NanU, a novel high-affinity sialic acid-inducible binding protein of oral and gut-dwelling Bacteroidetes species. Biochem J. 2014;458:499–511.
Roy S, Douglas CI, Stafford GP. A novel sialic acid utilisation and uptake system in the periodontal pathogen Tannerella forsythia. J Bacteriol. 2010;192:2285–93.
Roy S, Phansopa C, Stafford P, Honma K, Douglas CW, Sharma A, Stafford GP. Beta-hexosaminidase activity of the oral pathogen Tannerella forsythia influences biofilm formation on glycoprotein substrates. FEMS Immunol Med Microbiol. 2012;65:116–20.
Severi E, Muller A, Potts JR, Leech A, Williamson D, Wilson KS, Thomas GH. Sialic acid mutarotation is catalyzed by the Escherichia coli beta-propeller protein YjhT. J Biol Chem. 2008;283:4841–9.
Ruscitto A, Honma K, Veeramachineni VM, Nishikawa K, Stafford GP, Sharma A. Regulation and molecular basis of environmental muropeptide uptake and utilization in fastidious oral anaerobe Tannerella forsythia. Front Microbiol. 2017;8:648.
Duran-Pinedo AE, Chen T, Teles R, Starr JR, Wang X, Krishnan K, Frias-Lopez J. Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis. ISME J. 2014;8:1659–72.
Caruso R, Warner N, Inohara N, Nunez G. NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity. 2014;41:898–908.
Inui T, Walker LC, Dodds MW, Hanley AB. Extracellular glycoside hydrolase activities in the human oral cavity. Appl Environ Microbiol. 2015;81:5471–6.
Kim JH, Ryu YB, Lee WS, Kim YH. Neuraminidase inhibitory activities of quaternary isoquinoline alkaloids from Corydalis turtschaninovii rhizome. Bioorg Med Chem. 2014;22:6047–52.
Smutova V, Albohy A, Pan X, Korchagina E, Miyagi T, Bovin N, Cairo CW, Pshezhetsky AV. Structural basis for substrate specificity of mammalian neuraminidases. PLoS One. 2014;9:e106320.
Miyagi T, Wada T, Yamaguchi K, Hata K, Shiozaki K. Plasma membrane-associated sialidase as a crucial regulator of transmembrane signalling. J Biochem. 2008;144:279–85.
Kawamura S, Sato I, Wada T, Yamaguchi K, Li Y, Li D, Zhao X, Ueno S, Aoki H, Tochigi T, Kuwahara M, Kitamura T, Takahashi K, Moriya S, Miyagi T. Plasma membrane-associated sialidase (NEU3) regulates progression of prostate cancer to androgen-independent growth through modulation of androgen receptor signaling. Cell Death Differ. 2012;19:170–9.
Khedri Z, Li Y, Cao H, Qu J, Yu H, Muthana MM, Chen X. Synthesis of selective inhibitors against V. cholerae sialidase and human cytosolic sialidase NEU2. Org Biomol Chem. 2012;10:6112–20.
Acknowledgments
The work from AS and coworkers cited in the chapter was supported by grants (DE014749 and DE022870) from the NIDCR.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Stafford, G.P., Sharma, A. (2020). Periodontal Pathogen Sialometabolic Activity in Periodontitis. In: Sahingur, S. (eds) Emerging Therapies in Periodontics. Springer, Cham. https://doi.org/10.1007/978-3-030-42990-4_12
Download citation
DOI: https://doi.org/10.1007/978-3-030-42990-4_12
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-42989-8
Online ISBN: 978-3-030-42990-4
eBook Packages: MedicineMedicine (R0)