Abstract
The aims of this study were to follow up on the establishment of Cotesia vestalis in the coastal regions of Kenya, assessing its contribution in the management of the diamondback moth (Plutella xylostella), parasitism rates, and cultural practices affecting its establishment. Post-release surveys were carried out in five counties in Kenya, Kitui, Kajiado, Kwale, Machakos and Makueni, between 2015 and 2016. The results showed that the overall parasitism rate of C. vestalis in 2015 ranged between 0% and 37.86% while that in 2016 ranged from 0% to 32.19% in the different counties. Farmers carry out routine sprays, either weekly or fortnightly, with different synthetic insecticides. Pyrethroids (60.56%) constituted most of the insecticides used, while only 3.18% of the products used were plant or microorganism- based. Farmers did most of their cabbage production during the rainy season, with production being greatly reduced during the dry seasons. The sampled diamondback moths from Kajiado, Kitui, Makueni, Kwale and Malawi had ≥98% similarity to Cotesia vestalis, as shown by bioinformatics analyses using PCR amplified products of 700 bp, obtained for the mitochondrial COI gene. Alignment showed highly conserved regions, and the phylogenetic analysis revealed two close lineages corresponding to Cotesia vestalis (Genbank accession: FJ154897) and Cotesia spp. (acc. HM430398). Data provide a clear indication that the parasitoid became established in its release sites in Kenya, although parasitism rates are still low probably due to heavy pesticide use and climatic factors.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
10.1 Introduction
Cruciferous vegetables, such as kales and cabbages, are widely grown in Africa for subsistence purposes, as well as for income generation (Ayalew et al. 2002; Lohr and Kfir 2004; Macharia et al. 2005; Grzywacz et al. 2010). The annual global production is estimated at 29 tonnes/ha, while productions in Malawi and Kenya reach approximately 20.7 and 30.9 tonnes/ha, respectively (FAOSTAT 2014).
Crucifer production, worldwide, is often hindered by insect pests which include the diamondback moth (Plutella xylostella L.), cabbage aphids (Brevicoryne brassicae L.), leaf miners (Liriomyza brassicae L.), thrips (Thrips tabaci L.), and cabbageworms (Pieris rapae L.) (Hines and Hutchison 2001; Bjorksten et al. 2005; Munthali 2009). Among these insect pests, P. xylostella is the most destructive one in Eastern Africa (Nyambo and Pekke 1995; Badenes-Perez and Shelton 2006). In instances where no management strategies are put in place, the small larvae of P. xylostella can cause losses between 90% and 98% (Sandur 2004; Macharia et al. 2005). Worldwide losses of crucifer vegetables by P. xylostella are estimated at US$ 1.4 billion (Zalucki et al. 2012), incurring US$ 4–5 billion in management costs (Furlong et al. 2013). In Kenya, annual losses of US$ 7.9 million, due to P. xylostella infestations, have been reported (Macharia et al. 2005).
Management of P. xylostella in Kenya has mainly focused on the use of pesticides, with reported cases of overuse resulting in increases in production costs, health hazards, development of resistance, and destruction of natural enemies (Badenes-Perez and Shelton 2006; Cooper 2009; Macharia et al. 2013). Biological control has thus been promoted as an alternative (Rowell et al. 2005; Kahuthia-Gathu 2012). The parasitoids of P. xylostella mainly found in the East Africa region are Diadegma mollipla Holmgren and Oomyzus sokolowskii Kurdjumov. However, their parasitism rates have been reported to be below 15% in Kenya, Tanzania, Malawi and Ethiopia (Lohr and Kfir 2004). One of the most effective parasitoids used in the management of P. xylostella is Cotesia vestalis Haliday, with reports of 78–88% parasitism rates (Smith and Villet 2001; Rowell et al. 2005). Among all the C. vestalis biotypes, the South African biotype is the most effective, owing to its predominance in both low and high altitudes (Talekar and Shelton 1993; Kfir 1997; Verkerk and Wright 1997; Mosiane et al. 2003), high thermal tolerance (Talekar and Yang 1991), and high parasitism rates (Waladde et al. 2001; Smith 2002).
Once introduced into Uganda, C. vestalis became established (ICIPE, unpublished data). It has been detected on the Kenyan side of Lake Victoria. Additional releases of C. vestalis were carried out in Kajiado, Machakos, Kitui and Makueni counties (Nyambo et al. 2008). However, repeated releases in the eastern region of Kenya resulted in very low parasitism rates (0.5–26.9%) (Nyambo et al. 2008; Kahuthia-Gathu 2012). Although the establishment of C. vestalis at the release sites was confirmed by other studies, most of them relied on morphological identification. A recent release of C. vestalis in Kwale County in 2013 resulted in a very low parasitism rate (unpublished data). This prompted the need to investigate the cultural practices that preclude the successful establishment of the parasitoid in Kenya, particularly in the coastal area. This study also aimed at confirming the molecular identity of the specimens that prevail in those counties and comparing them with those of Southern Africa biotypes, to ascertain their virulence and parasitism rates, in the various regions.
10.2 Materials and Methods
10.2.1 Study Sites
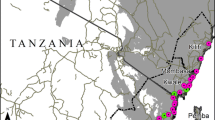
Surveys for the P. xylostella and its parasitoid, C. vestalis, were carried out in five counties: Kwale (Matuga, Diani and Lungalunga), Kajiado, Kitui, Makueni and Machakos (Fig. 10.1). Kajiado, Kitui, Makueni and Machakos counties represented C. vestalis post-release sites in the arid regions at mid-altitude (882–1918 m asl) of Kenya, while Kwale County represented the humid lowlands (3–416 m asl).
The surveys for P. xylostella and C. vestalis in Kwale County were conducted in the Lungalunga, Matuga and Diani regions. Kwale County experiences a bi-modal rainfall distribution, with long rains expected from March to June, and short rains from October to December. The annual rainfall range is 400–1680 mm per year. The soils are generally sandy loam, while some parts are richer in claye and fertile. On the other hand, Kitui, Kajiado, Makueni and Machakos counties are semi-arid areas, which are generally hot and dry. The rainfall distribution is bimodal: long rains are expected between March and May, while short rains are usually expected between October and December. The annual average rainfall ranges between 500 and 1300 mm, and soils are generally sandy with low fertility. Cabbage and kales are the main cruciferous vegetables produced for subsistence use, and for commercial production to a smaller extent.
10.2.2 Sampling for Plutella xylostella and Parasitoids on Cultivated Crucifers
Two surveys were carried out in Kwale County’s during the rainy (June 2015, August 2015) and dry season (January 2016). Two surveys were also carried out in the eastern region, during the rainy (November 2015) and dry season (March 2016). The farms sampled were those on which cabbage is produced, and were at least 1 km apart. Ten (10) farms per region in each county were surveyed during each visit, and the same fields were visited on every survey. In cases where a field was not under crucifer crop production, it was replaced by another farm nearby. However, fewer farms in some regions of Kwale County were surveyed, due to a huge decline in the number of farms on crucifer production because of dry weather conditions in January 2016, since farmers without access to irrigation could not produce vegetables during that period. Ten (10) randomly selected cabbage plants per farm were sampled and thoroughly inspected for P. xylostella larvae, pupa and parasitoid cocoons on the leaves. The samples collected were put into plastic containers fitted with a cloth mesh at the top to allow for ventilation, which were lined with a paper towel at the bottom to prevent condensation. The containers were labelled with the field numbers. The types and numbers of samples collected, other pests found on the crops, field numbers, host plants, and collection dates were recorded and the samples taken to the laboratories. The farms sampled were geo-referenced using a Global Positioning System (GPS, model Magellan® Triton™ 400).
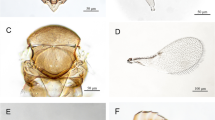
Sample processing was conducted at the International Centre of Insect Physiology and Ecology (ICIPE), Duduville Campus, Kasarani, Nairobi. The samples collected were kept at room temperature (23 ± 2 °C), 50–70% relative humidity, and at a photoperiod of 12:12 h (Light: Darkness). Fresh cabbage leaves were provided daily to the P. xylostella larvae. The emergence of either P. xylostella or parasitoid adults was checked daily until no further emergence was observed. All the P. xylostella and different parasitoid samples that emerged were sexed, identified, and recorded. Identification and sexing of the parasitoids collected was carried out using a Leica EZ4D microscope at a magnification of 10× to enable viewing of the parasitoid genitalia. Moreover, the presence or absence of detailed features that identify C. vestalis were used, based on IntKey (Dallwitz et al. 1999). Parasitoids from each county were reared in Perspex cages measuring 20 × 20 × 20 cm (external dimensions) in the laboratory to obtain a larger number for molecular work. Rearing of C. vestalis was carried out by exposing them to 2nd and 3rd instar larvae of P. xylostella on cabbages for 24 h, removal of the plants and putting the larvae into lunch boxes (11.5 cm diam., 6 cm high), and daily feeding on cabbages until emergence. The C. vestalis were fed daily on honey droplets on a paper strip.
10.2.3 Survey on Pest Management Practices
Household interviews were conducted to collect information on cultural practices on cabbage farming using structured questionnaires (See Appendix A below). The information collected included: the type of cruciferous crop cultivated, variety grown, planting time, harvesting intervals, intercropped farms, soil type, type of manure and fertiliser applied and their application period, irrigation type and rain-fed farms, the pest management strategies applied by farmers, reason for pesticide application, frequency of their application, last application dates, and the change in the frequency on use of the management strategies over time, in addition to the farmers’ knowledge on use of natural enemies in management of P. xylostella.
10.2.4 Molecular Identification of Cotesia vestalis
The Cotesia spp. samples were used for molecular work. Additional samples similar to those released in East Africa (South African biotype) were obtained from the Department of Agricultural Research Services (DARS) in Malawi. All the samples were preserved in 95% ethanol and stored at −20 °C while awaiting genomic DNA extraction. Prior to molecular characterisation, adults were morphologically identified according to the various features used for the identification of Cotesia species of economic importance, as described by IntKey (Dallwitz et al. 1999). Lateral, ventral and dorsal images of the sample specimens were taken with a Leica EZ4D microscope, using LAS EZ software ver. 3.0.0. This was followed by surface sterilisation in 3% sodium hypochlorite, rinsing thrice in distilled water, and placing the samples in labelled 1.5 mL tubes. DNA extraction was performed using ISOLATE II Genomic DNA Kit (Bioline, UK) following the manufacturer’s protocol. Amplification of the target COI gene was done using universal barcode primers; LCO-1490 (5′ GGTCAACAAATCATAAAGATATTGG 3′) and HCO-2198 (5′ TAAACTTCAGGGTGACCAAAAAATA 3′) (Folmer et al. 1994). The PCR was carried out in a total reaction volume of 20 μL containing 5× My Taq Reaction Buffer (5 mM dNTPs, 15 mM MgCl2, stabilisers and enhancers), 10 μmole of each primer, 0.5 mM MgCl2, 0.25 μL My Taq DNA polymerase (Bioline, UK) and 15 ng/μL of DNA template. The reaction was set up in the Nexus Mastercycler gradient (Eppendorf). The following cycling conditions were used: initial denaturation for 2 min at 95 °C, followed by 40 cycles of 30 s at 95 °C, 40 s annealing at 50.6 °C, and 1 min at 72 °C, then a final elongation step of 10 min at 72 °C.
The amplified PCR products were resolved through a 1.2% agarose gel stained with 10 mg/mL ethidium bromide. DNA bands on the gel were analysed and documented using a KETA GL imaging system trans-illuminator (Wealtec Corp). Successively amplified products were excised and purified using Isolate II PCR and Gel Kit (Bioline, UK) following the manufacturer’s instructions. The purified samples were shipped to Macrogen Inc. Europe Laboratory, the Netherlands, for bi-directional sequencing.
10.2.5 Sequence Analysis
The chromatograms were examined with Chromas ver. 2.5.1 (Hall 1999), and when ambiguous sites were found, they were corrected to produce two alternative sequences, corresponding to high and low peaks, respectively, and a sequence was created. The consensus sequences generated from both strands were compared to those available at GenBank, using the built-in BLAST utility (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiple alignment was done with ClustalX version 2.1 (Thompson et al. 1997). jModeltest 2.1.7 program was used to provide an estimate of the best-fit model selection (Darriba et al. 2012). The sequences were then run in RAxML v8.2.0 (Stamatakis 2014) to get the best-scoring Maximum likelihood tree, with a bootstrap test of 1000 replications. The tree was constructed using Fig Tree v. 1.4 (Rambaut 2012).
The sequences obtained were submitted to the Barcode of Life Database (BOLD) and deposited in GenBank (Accession numbers: ABZ6416 and ADL5635). DNA voucher specimens were stored at ICIPE Molecular Pathology Lab., Arthropod Pathology Unit (APU).
10.2.6 Data Analysis
The parasitism rate of P. xylostella by the solitary parasitoid, C. vestalis was calculated as follows:
Calculations of parasitism excluded samples that died before emergence (Nofemela and Kfir 2005). The data on the P. xylostella and C. vestalis densities were checked for normality using the Shapiro-Wilk test, and log-transformed to correct for over-dispersion, while proportional data were arcsine transformed. The data for each survey period were then subjected to Analysis of Variance (ANOVA). Means were separated using Tukey’s Honest Significant Difference (HSD) test at 5% level of significance. R Studio software (ver. 2.15.1) was used for all analyses (http://www.rstudio.org/).
10.3 Results
10.3.1 Incidence of Plutella xylostella and Parasitism Rates by Cotesia vestalis in Kwale County in the Coastal Region
The P. xylostella densities for the June 2015 survey (12.22 ± 1.88 per farm, mean ± SD) was over ten times the densities in Jan 2016 (1.18 ± 0.67 per farm) (Fig. 10.2). On the other hand, the C. vestalis densities for Jan 2016 (0.65 ± 0.37 per farm) were more than twice the densities in June 2015 (0.31 ± 0.11 per farm) (Fig. 10.2). Parasitism rates by C. vestalis in Jan 2016 (7.35 ± 3.523%) were over twice the parasitism in June (2.83 ± 1.02%) (data not shown). The comparison of the P. xylostella densities between the two survey periods showed significant differences (F = 43.11; df = 1,74; P < 0.05), while the C. vestalis densities (F = 0.982; df = 1,74; P = 0.325) were not significantly different.
10.3.2 Incidence of Plutella xylostella and Parasitism Rates by Cotesia vestalis in the Eastern Region
During the November 2015 survey, the density of P. xylostella in Kitui was fivefold (1 ± 0.39 per farm), compared with that recorded in Makueni (0.2 ± 0.13 per farm). In the same survey, the densities of C. vestalis in Makueni were 2.25-fold (1.8 ± 0.80 per farm) higher than in Kitui (0.8 ± 0.33a per farm) (Fig. 10.3). There were no significant differences observed between P. xylostella (F = 3.358; df = 1, 18; p = 0.0835) and C. vestalis densities (F = 0.548; df = 1,18; p = 0.469) in the two counties. The parasitism rates in Makueni (37.86 ± 1 3.88%) were almost twice those in Kitui (20.9 ± 0.92%).
In March 2016, the P. xylostella density in Kitui (2.30 ± 1.76 per farm) was over three times higher than in Makueni (0.6 ± 0.50 per farm) (Fig. 10.3 below). The densities of P. xylostella (F = 1.119; df = 1,18; p = 0.304) and C. vestalis (F = 3.6; df = 1,18; p = 0.074) were not significantly different. The parasitism rate by C. vestalis in Kitui (32.19 ± 12.64%) was more than twice the rate recorded in Kitui (15 ± 10.67%). There was no significant difference between P. xylostella (F = 0.228; df = 1,38; p = 0.636) and C. vestalis (F = 2.896; df = 1,38; p = 0.0969) densities during the 2015 and 2016 surveys, in the Eastern Region.
10.3.3 Survey on Pest Management Practices
A baseline survey of pest management practices indicated that 89.25% of the farm households interviewed at the coastal region were using insecticides. The farmers used a total of 18 different active ingredients of synthetic insecticides under different application regimes. The majority of the insecticides used in the Coastal region were pyrethroids, constituting 75.47% of the total pesticides, followed by neonicotinoids (8.49%) and carbamates (5.66%) (Table 10.1).
Based on the household data collected in Kitui, Makueni and Machakos counties, 78.57% of the farmers were spraying their vegetables with insecticides. The most commonly used insecticides were pyrethroids (46.16%), with a usage threefold higher than that of organophosphates (15.55%). Additionally, only 5.5% of the products used were micro-organism derived, while 1.1% were plant-derived products (Table 10.1).
The proportion of farmers that practised intercropping in the Coastal region ranged between 9.09% and 51.16%, compared to 0–2.5% in the Eastern region, depending on the growing period. On the other hand, the proportion of farms under irrigation in the coastal region was 78.75%, whereas it was 73.12% in the Eastern region (Table 10.2).
The common pests, other than P. xylostella, based on pest densities and frequency of occurrence were cabbage aphids, bollworms, whiteflies, thrips, cabbage loopers, cutworms and leafminers. Bollworms were the most abundant in the Coastal region in both 2015 (65.05%) and 2016 (41.18%). In the Eastern region, cabbage aphids (43.33%) were ranked as the most common pest in 2015, while whiteflies (75.0%) were the most abundant in 2016 (Table 10.3). In regard to farmers’ knowledge on parasitoids, only 41.8% of the farmers in the Coastal region and 30% in Eastern region were well-informed about the use of parasitoids for management of P. xylostella.
10.3.4 Quantification of DNA and PCR Amplification
The purity of the DNA, based on the ratio of absorbance at 260/280nm for all samples, ranged from 1.92 to 2.72, while the nucleic acid concentration of the samples ranged between 30.8 and 146.3 ng/μL. A total of 48 samples (from Kitui, Kwale, Makueni, Kajiado and Malawi) were amplified by PCR, with a band size of approximately 700 base pairs (Fig. 10.4).
10.3.5 Bioinformatics Analysis
BLAST results for the 48 samples from Kajiado, Kitui, Kwale, Makueni and Malawi indicated the complete mitochondrion genome as best hit (E-Value 0.0), with ≥98% similarity to Cotesia vestalis (Genbank accession: FJ154897). The only exception was Kw7 from Kwale that recorded the cytochrome oxidase subunit 1 (COI) gene as best hit (E-value 0.0), with 98% similarity to a Cotesia spp. (Genbank accession: HM430398). All the COI-aligned sequences showed a high degree of conserved residues among the 48 samples. One sample from each group was used to represent samples from the five regions (Fig. 10.5).
Multiple sequence alignment of C. vestalis sequences produced by ClustalX. Conserved regions are marked with asterisks above the sequences. The sequences were sampled from five regions denoted as follows: Kj = Kajiado, Kt = Kitui, Kw = Kwale, Mw = Malawi, Mk = Makueni (see Table 10.4 for samples details)
10.3.6 Phylogenetic Analysis
The C. vestalis sequences grouped into two clusters. The first one had only one sample (Kw7) collected from Kajiado County, which corresponded to Cotesia spp. The second cluster included the remaining samples, which corresponded to Cotesia vestalis. The second cluster was further separated into several groups. The cluster with the highest bootstrap value (99%) had two samples (Mw11 and Mw23) from Malawi. Most of the samples were supported by low bootstrap values.
10.3.7 Genetic Distances and Distance Summary
The divergent distances between the five groups of sequences were estimated by pairwise genetic distances estimated from COI sequences, based on a Kimura-two-parameter algorithm (Tamura et al. 2013). The highest nucleotide distance was between Kw and Mk, with a value of 0.018 (Table 10.5). The distance summary, shown in bold, indicated a within-species mean distance of 23.51%, with maximum distance as 75%, and a minimum distance as 0%. The alignment option used was the Kimura-2-parameter.
10.4 Discussion
The study of P. xylostella dynamics in the semi-arid and coastal areas of Kenya showed much of variability with the planting period in both coastal and eastern regions. Despite several releases, C. vestalis parasitism rates in the coastal region and eastern regions were below 14% and 38%, respectively. Misuse of pesticides could be one of the factors underlying the poor establishment of C. vestalis in both regions. Moreover, farmers continue spraying on a calendar basis, using one or more broad-spectrum synthetic insecticides either weekly or fortnightly, not only to manage P. xylostella, but also other pests including aphids, whiteflies and bollworms. There are reports that the fields with minimum pesticide usage or the organically managed ones have higher densities of parasitoids than those that are frequently sprayed (Ayalew et al. 2002; Kfir 2004; Rowell et al. 2005). Moreover, some products used by farmers, such as spinosad, have detrimental effects on C. vestalis (Oliveira et al. 2011), thus slowing down their increase and reducing the parasitoid efficacy. Overreliance on rainfall for vegetable production necessitated broken and discontinuous productions, and could have been a factor behind the poor establishment of C. vestalis in the release regions. According to the farmers, their frequency of pesticide usage was on the rise, compared with the previous years. They attributed this to the reduced efficacy of the pesticides that they were using. Several studies have shown that P. xylostella developed resistance to abamectin, lufenuron, methomyl and emamectin (Pu et al. 2010; Santos et al. 2011), yet these pesticides are still widely used by farmers.
In the Philippines lowlands, an IPM technology which involved augmentative parasitoid releases and judicious spraying using selective insecticides with strong support from extension efforts, resulted in the successful establishment of C. vestalis, reduced P. xylostella densities, increased yields and reduced production costs (Morallo-Rejesus et al. 1997; Rowell et al. 2005; Jankowski et al. 2007). The same strategy can be adopted in Kenya to increase its efficacy of as a biological control agent. Moreover, stronger links between research and extension would be very helpful in informing the farmers on the use of safe insect management measures against pests and would go a long way in creating farmer awareness of parasitoids. Furthermore, the adapting of IPM programmes that help in management of other common pests such as aphids, bollworms and whiteflies would greatly reduce spraying, since farmers currently continue spraying, even in absence of P. xylostella, and this eventually affect its parasitoids. In a bid to achieve better pest management within the existing cropping system, smallholder farmers need to have access to information on effective IPM technologies through participatory technology transfer approaches. Molecular analysis of the samples collected from Kajiado, Kitui, Kwale, Makueni and Malawi showed that the specimens from the surveyed areas had ≥98% similarity to Cotesia vestalis. The DNA quantification and amplification confirmed that the target gene was present in all samples. Furthermore, bioinformatics showed that all samples had the complete mitochondrion genome as best hit, except one. Some of the samples from different regions clustered together, an indicator that they shared ancestry, while other samples from the same location fell under different clusters, which is an indicator of the occurrence of divergence among the species, over time. The results suggest that there is very close similarity in all the samples, regardless of where they were collected from. This shows that the South African biotype of C. vestalis that was released in the previous years was active in the surveyed areas. The COI gene is highly conserved, as revealed by multiple sequence alignment, and it can be used as a viable marker in confirming Cotesia establishment upon release in the study sites. Similar studies have reported successful use of COI gene in determining the origin of Cotesia flavipes in Ethiopia (Assefa et al. 2008).
In conclusion, this study and the molecular results conclusively show that C. vestalis has established in and around the different release sites in Kenya. However, its contribution in management of P. xylostella is still low, and this could mainly be attributed to the excessive use of broad-spectrum insecticides. Moreover, the COI gene greatly improved the identification of the C. vestalis without much dependence on morphological characters.
References
Assefa, Y., Mitchell, A., Conlong, D. E., & Muirhead, K. A. (2008). Establishment of Cotesia flavipes (Hymenoptera: Braconidae) in sugarcane fields of Ethiopia and origin of founding population. Journal of Economic Entomology, 101, 686–691.
Ayalew, G., Lorh, B., Baumgartner, J., & Ogol, C. (2002). Diamondback moth, Plutella xylostella (L.) (Lepidoptera: Plutellidae) and its parasitoids in Ethiopia. In Proceedings of the international symposium, pp. 21–24.
Badenes-Perez, F. R., & Shelton, A. M. (2006). Pest management and other agricultural practices among farmers growing cruciferous vegetables in the Central and Western highlands of Kenya and the Western Himalayas of India. The International Journal of Pest Management, 52, 303–315.
Bjorksten, T. A., Robinson, M., & La Salle, J. (2005). Species composition and population dynamics of leafmining flies and their parasitoids in Victoria. Australian Journal of Entomology, 44, 186–191.
Cooper, J. (2009). Pest management in horticultural crops; an integrated approach to vegetable pest management with the aim of reducing reliance on pesticides in Kenya. Final technical report.
Dallwitz, M. J., Paine, T. A., & Zurcher, E. J. (1999). User’s guide to the DELTA Editor. https://www.delta-intkey.com/
Darriba, D., Taboada, G. L., Doallo, R., & Posada, D. (2012). jModelTest 2: More models, new heuristics and parallel computing. Nature Methods, 9, 772–772.
de Oliveira, A. C., de Siqueira, H. Á. A., de Oliveira, J. V., da Silva, J. E., & Michereff, F. M. (2011). Resistance of Brazilian diamondback moth populations to insecticides. Science in Agriculture, 68, 154–159.
FAOSTAT. (2014). Food and agriculture organisation, United Nations. http://www.fao.org/faostat.
Folmer, O., Black, M., Hoeh, W., Lutz, R., & Vrijenhoek, R. (1994). DNA primers for amplification of mitochondrial cytochrome C oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology, 3, 294–299.
Furlong, M. J., Wright, D. J., & Dosdall, L. M. (2013). Diamondback moth ecology and management: Problems, progress, and prospects. Annual Review of Entomology, 58, 517–541.
Gichini, G., Löhr, B., Rossbach, A., Nyambo, B., & Gathu, R. (2008). Can low release numbers lead to establishment and spread of an exotic parasitoid: The case of the diamondback moth parasitoid, Diadegma semiclausum (Hellén), in East Africa. Crop Protection, 27, 906–914.
Grzywacz, D., Rossbach, A., Rauf, A., Russell, D. A., Srinivasan, R., & Shelton, A. M. (2010). Current control methods for diamondback moth and other brassica insect pests and the prospects for improved management with lepidopteran-resistant Bt vegetable brassicas in Asia and Africa. Crop Protection, 29, 68–79.
Hall, T. A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. In Nucleic acids symposium series (pp. 95–98).
Hines, R. L., & Hutchison, W. D. (2001). Evaluation of action thresholds and spinosad for lepidopteran pest management in Minnesota cabbage. Journal of Economic Entomology, 94, 190–196.
Jankowski, A., Mithöfer, D., Löhr, B., & Weibel, H. (2007). Economics of biological control in cabbage production in two countries in East Africa. In Utilisation of diversity in land use systems: Sustainable and organic approaches to meet human needs. Proceedings of the Conference on International Agricultural Research for Development ‘Tropentag.
Kahuthia-Gathu, R. (2012). Seasonal incidence of Plutella xylostella (Lepidoptera: Plutellidae) and its associated natural enemies in major crucifer growing areas of Kenya. Journal of Applied Bioscience, 46, 3103–3112.
Kfir, R. (1997). Parasitoids of Plutella xylostella (Lep.: Plutellidae) in South Africa: An annotated list. Entomophaga, 42, 517–523.
Kfir, R. (2004). Effect of parasitoid elimination on populations of diamondback moth in cabbage. In The management of diamondback moth and other crucifer pests: Proceedings of the 4th international workshop (pp. 197–205). Gosford: The Regional Institute Ltd.
Lohr, B., & Kfir, R. (2004). Diamondback moth Plutella xylostella (L.) in Africa. A review with emphasis on biological control. Improving Biocontrol Plutella Xylostella, 2, 5707.
Löhr, B., Gathu, R., Kariuki, C., Obiero, J., & Gichini, G. (2007). Impact of an exotic parasitoid on Plutella xylostella (Lepidoptera: Plutellidae) population dynamics, damage and indigenous natural enemies in Kenya. Bulletin of Entomological Research, 97, 337–350.
Macharia, I., Löhr, B., & De Groote, H. (2005). Assessing the potential impact of biological control of Plutella xylostella (diamondback moth) in cabbage production in Kenya. Crop Protection, 24, 981–989.
Macharia, I., Mithöfer, D., Waibel, H., et al. (2013, September 22–25). Health effects of pesticide use among vegetable farmer in Kenya. In AAAE fourth international conference, Hammamet, Tunisia. African Association of Agricultural Economists (AAAE).
Morallo-Rejesus, B., Inocencio, E., Eusebio, J., & Barroga, S. (1997). Technology transfer of Cotesia-based IPM for diamondback moth on lowland crucifers in the Philippines. In The Management of Diamondback Moth and other crucifer pests. Proceedings of the Third International Workshop (pp. 267–275).
Mosiane, S. M., Kfir, R., & Villet, M. H. (2003). Seasonal phenology of the diamondback moth, Plutella xylostella (L.),(Lepidoptera: Plutellidae), and its parasitoids on canola, Brassica napus (L.), in Gauteng province, South Africa. African Entomology: Journal of the Entomological Society of Southern Africa, 11, 277–285.
Munthali, D. C. (2009). Evaluation of cabbage varieties for resistance to the cabbage aphid. African Entomology: Journal of the Entomological Society of Southern Africa, 17, 1–7.
Nofemela, R. S., & Ktir, R. (2005). The role of parasitoids in suppressing diamondback moth, PIuteIIa xylostella (L.) (Lepidoptera: Plutellidae), populations on unspraved cabbage in the North West Province of South Africa. African Entomology: Journal of the Entomological Society of Southern Africa, 13, 71–83.
Nyambo, B., & Pekke, A. (1995). Brassica planning workshop for East and Southern Africa region, Lilongwe-Malawi. GTZ-IPM Horticulture Nairobi Kenya.
Nyambo, B., Gichini, G., Obiero, J., & Njumwa, G. (2008). Re-distribution of ex-South African strain of Cotesia plutellae Kurdjumov (Hymenoptera, Braconidae) in Africa for control of diamondback moth. In Proceedings of the XXII international congress of entomology, pp. 6–12.
Pu, X., Yang, Y., Wu, S., & Wu, Y. (2010). Characterisation of abamectin resistance in a field-evolved multiresistant population of Plutella xylostella. Pest Management Science, 66, 371–378.
Rambaut, A. (2012). FigTree v1. 4 2006–2012. Institute of Evolutionary Biology, The University of Edinburgh, Edinburgh, UK.
Rowell, B., Bunsong, N., Satthaporn, K., Phithamma, S., & Doungsa-Ard, C. (2005). Hymenopteran parasitoids of diamondback moth (Lepidoptera: Ypeunomutidae) in northern Thailand. Journal of Economic Entomology, 98, 449–456.
Sandur, S. (2004). Implications of Diamondback moth control for Indian cabbage farmers. A report produced for the CIMBAA. http://www.cimbaa.org/downloads/cimbaa_indiandbm_reportfinal-%20apr05.pdf. Last Accessed 17 september 2008.
Santos, V. C., de Siqueira, H. A. A., da Silva, J. E., & de Farias, M. (2011). Insecticide resistance in populations of the diamondback moth, Plutella xylostella (L.) (Lepidoptera: Plutellidae), from the state of Pernambuco, Brazil. Neotropical Entomology, 40, 264–270.
Shi, Z., Guo, S., Lin, W., & Liu, S. (2004). Evaluation of selective toxicity of five pesticides against Plutella xylostella (Lep: Plutellidae) and their side-effects against Cotesia plutellae (Hym: Braconidae) and Oomyzus sokolowskii (Hym: Eulophidae). Pest Management Science, 60, 1213–1219.
Smith, T.J. (2002). The diamondback moth, Plutella xylostella (L.), (Lepidoptera: Plutellidae) and its biological control in the eastern cape province, South Africa.
Smith, T.J., & Villet, M.H. (2001, November 26–29). Parasitoids associated with the diamondback moth, Plutella xylostella (L.), in the Eastern Cape, South Africa. In The management of diamondback moth and other crucifer pests. Proceedings of the fourth international workshop, p. 249.
Stamatakis, A. (2014). RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30, 1312–1313.
Talekar, N. S., & Shelton, A. M. (1993). Biology, ecology, and management of the diamondback moth. Annual Review of Entomology, 38, 275–301.
Talekar, N. S., & Yang, J. C. (1991). Characteristics of parasitism of diamondback moth by two larval parasites [Apanteles plutellae, Diadegma eucerophaga]. Entomophaga, 36, 95.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., & Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30, 2725–2729.
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., & Higgins, D. G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25, 4876–4882.
Verkerk, R. H. J., & Wright, D. J. (1997). Field-based studies with the diamondback moth tritrophic system in Cameron highlands of Malaysia: Implications for pest management. International Journal of Pest Management, 43, 27–33.
Vrijenhoek, R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology, 3, 294–299.
Waladde, S. M., Leutle, M. F., & Villet, M. H. (2001). Parasitism of Plutella xylostella (Lepidoptera, Plutellidae): Field and laboratory observations. South African Journal of Plant and Soil, 18, 32–37.
Zalucki, M. P., Shabbir, A., Silva, R., Adamson, D., Shu-Sheng, L., & Furlong, M. J. (2012). Estimating the economic cost of one of the world’s major insect pests, Plutella xylostella (Lepidoptera: Plutellidae): Just how long is a piece of string? Journal of Economic Entomology, 105, 1115–1129.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Appendix
Appendix
Farmer questionnaire used for Cotesia vestalis post-release Surveys (Kenya- Kajiado, Kitui, Machakos, Makueni).

Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Mukiri, M.J. et al. (2020). Establishment of an Exotic Parasitoid Cotesia vestalis in Coastal Areas of Kenya as Biological Control Agent of Plutella xylostella. In: Niassy, S., Ekesi, S., Migiro, L., Otieno, W. (eds) Sustainable Management of Invasive Pests in Africa. Sustainability in Plant and Crop Protection, vol 14. Springer, Cham. https://doi.org/10.1007/978-3-030-41083-4_10
Download citation
DOI: https://doi.org/10.1007/978-3-030-41083-4_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-41082-7
Online ISBN: 978-3-030-41083-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)