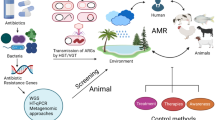
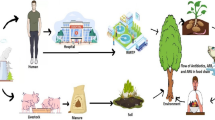
Abstract
Worldwide morbidity and mortality caused by infectious diseases is high, mandating high rates of antibiotic use among humans and animals. Antibiotics of anthropogenic origin often contaminate the environment. The arising ecological pressure results in alteration of bacterial “biomes,” high resistance rates in environmental microorganisms, and increase in the gene pool which contributes to antibiotic resistance. A number of such antibiotic resistance genes are carried on mobile genetic elements that can easily be exchanged between bacteria. The ecological net effect is an expanding population of resistant organisms contributing to spread of antibiotic resistance in both the clinical and the nonclinical environments. In nonclinical environments, antibiotics upset the natural symbiotic balance between microorganism and macroorganism communities. In clinical environments, while therapeutic antibiotic adverse effects are easily observed, the, impact of sub-inhibitory concentrations of antimicrobials on human health are less apparent and require investigations. In summary, impact of antimicrobial resistance is extensive, threatening not just health and food safety but also our environment. Actions are thus required to both safeguard efficacies of antimicrobial agents, and also to protect the environment from damage by them.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
The development of antimicrobials changed the course of medicine. With the availability of antimicrobials many infectious diseases previously associated with considerable morbidity and mortality could be overcome. Modern medicine progressed by leaps and bounds secure in the knowledge that infections would be managed by antimicrobial agents. However, this confidence was challenged by two things: slowing down of the pipeline for new antimicrobials (Koulenti et al. 2019; Durand et al. 2019) and the development of antimicrobial resistance.
Of the currently known antibiotics a number are synthetic compounds, but most are natural products of microorganisms. As such antimicrobials, and indeed resistance to them, are both part of normal microbial defense mechanisms (Durand et al. 2019). Antimicrobial resistance however, is not a new phenomenon. Phylogenetic of OXA genes (that encode beta-lactamases) have shown that much of the diversity in these genes is the result of ancient events, and that the OXA genes were mobilized to plasmids from chromosomes millions of years ago (Barlow and Hall 2002). Genes that confer antimicrobial resistance have been identified in samples of permafrost dating back thousands of years (D’Costa et al. 2011; Kashuba et al. 2017). Environmental sample of Lechuguilla cave in New Mexico, isolated for more than four million years, yielded 93 bacterial strains of which several species were resistant in vitro to three or more antibiotics classes (Bhullar et al. 2012). Antimicrobial resistant genes (ARGs) have been isolated from gut microbiomes of an ancient mummy (Lugli et al. 2017) and from antibiotic naïve glaciers in the Antarctic (Van Goethem et al. 2018). In recent times though, antimicrobial resistance has come to be viewed as one of the biggest health threats faced by mankind (O’Neill 2016).
12.1 Epidemiological Impact of Antibiotics and ARGs
12.1.1 Pharmacoepidemiology and Evolution of Antibiotic Use
The first antimicrobials to be made available commercially included arsphenamine, a chemical compound discovered by Paul Ehrlich marketed in 1911 as Salvarsan@ and Mapharsen@ for the treatment of syphilis (Gensini et al. 2007). This was followed by sulphanilamide marketed as Protonsil@ in 1935 (Lewis 2013). Penicillin though discovered by Fleming in 1928 was made commercially available in 1940 (Durand et al. 2019). Since then several synthetic or naturally derived molecules were explored and a number added to the antimicrobial armamentarium. The global expansion of pharmaceutical industry, reduced cost of production in particular following expiry of drug patents, together with increasing demands from health care providers and patients, allowed for an exponential increase in global availability of antimicrobials. In many parts of the world with limited access to health care antimicrobials came to be used as first-line agents for a number of conditions, and in countries with weak health systems, made available over the counter. Microorganisms develop antimicrobial resistance either by de novo mutations under clinical antibiotic selection or through acquisition of mobile genes from other bacteria that have evolved resistance following exposure to antimicrobials at some earlier point in time. The increasing and unregulated exposure to antimicrobials (which in many parts of the world included poor quality drugs) therefore selected for organisms that were resistant to them. Patients treated with antibiotics became colonized with resistant organisms carrying them as part of their microbiome. Inadequate infection control measures and poor access to hygiene allowed the spread of these resistant bacteria initially within health care facilities, but ultimately within the communities as well.
In parallel to human use, antimicrobials also came to be recognized as being valuable in protecting farmed animals against infections leading to increased productivity; antifungals were found to be useful in protecting crops seeds and bulbs safeguarding the farmers’ economic interest and ensuring affordable protein and food safety for many. To the extent that currently agricultural antibiotic use exceeds human consumption (Van Boeckel et al. 2015), excessive antimicrobial use in food production contributed not only to selection of antimicrobial resistance in the environment, but to its spread through the food chain (Kirchhelle 2018).
When exploring the spread of antimicrobial resistance another important link to be considered is the environment. Approximately 50–90% of antibiotics administered to humans and animals are reportedly excreted via urine and feces, as a mixture of parent drug and metabolite forms (Kümmerer 2009). Pharmaceutical industry too discharges waste (which includes waste containing antimicrobial agents) into sewage. The antimicrobial compounds thus discharged may associate with sewage sludge, or be released to rivers. Sludge-associated drugs will enter agricultural systems when the sludge is used as a fertilizer, or when wastewaters are used for irrigation (Kinney et al. 2006). Thus, significant levels of active drugs end up in the environment, where they may persist in soil and aquatic ecosystems (Kümmerer 2009; Wellington et al. 2013) contributing to generation of antimicrobial resistance among the environmental microbiota (including amongst microbiota of health care facilities), which in turn passes back to humans and to animals and into the food chain (Chamosa et al. 2017; Ekwanzala et al. 2018). Additionally, human/animal waste contains microbiome of those exposed to antimicrobials , antimicrobial resistant bacteria, and/or antimicrobial resistance genes, the resistome (Van Schaik 2015; Proia et al. 2018). Wastewater containing such resistomes and used for either agriculture or indeed for drinking too represents a significant risk for spreading bacteria and antimicrobial resistance among both humans and animals (Lamba et al. 2018; Bougnom et al. 2019).
12.1.2 Economic Epidemiology
The significance of and risk from AMR is enormous. It is estimated that by 2050, ten million lives a year and a cumulative 100 trillion USD of economic output are at risk due to the rise of drug-resistant infections (O’Neill 2016). A recent report from the Organization for Economic Co-operation and Development (OECD) predicts that 2.4 million people in Europe, North America, and Australia will die from infections with resistant microorganisms in the next 30 years and that such infections could cost up to US$3.5 billion per year (OECD 2018). Analysis of European Antimicrobial Resistance Surveillance Network (EARS-Net) 2015 data suggests that the burden of infections due to antibiotic resistance organisms has increased between 2007 and 2015 and was similar to the cumulative burden of influenza, tuberculosis, and HIV (Cassini et al. 2019). Yet antimicrobial usage continues to increase. A recent study analyzing the trends of antibiotic consumption in 76 countries from 2000 to 2015 reports that antibiotic consumption, expressed in defined daily doses (DDD), increased 65% (21.1–34.8 billion DDDs), and the antibiotic consumption rate increased 39% (11.3–15.7 DDDs per 1000 inhabitants per day). The study further found that the increase was in particular driven by low- and middle-income countries (LMICs), where rising consumption was correlated with gross domestic product per capita (GDPPC) growth (Klein et al. 2018).
In response to these concerns at its sixty-eight World Health Assembly in 2015 World Health Organization endorsed a global action plan to tackle antimicrobial resistance (World Health Organization 2015) One year later the United Nations General Assembly (UNGA) termed antimicrobial resistance as one of the biggest threats to global health endangering major priorities including human development (World Health Organization 2016). In response to these concerns countries were called upon to develop their national AMR action plans encompassing a One Health agenda. The challenge remains in prioritizing implementation and balancing cost benefit of each component. The Organisation for Economic Cooperation and Development (OECD) report recommends five simple measures to reduce AMR: hand and environmental hygiene, antibiotic stewardship, rapid testing to distinguish bacterial from viral infections, delayed antibiotic prescriptions, and mass media campaigns. The report further estimates that in OECD countries this could be achieved by investing as little as US$2 per person per year and could avoid 75% of deaths caused by infections with resistant microorganisms (OECD 2018; Hofer 2019). In contrast, a multivariate analysis of 2008–2014 data from 103 countries by Collignon et al. (2018) conclude that given the importance of resistant bacteria and of resistance genes in spreading AMR, reducing antibiotic consumption alone would not be sufficient for the problem and call for improved sanitation, clean water, good governance, increased investment in public health care, and the importance of regulating the private health sector (Collignon et al. 2018; Hofer 2019). Equally, curbing antimicrobial use in the animal and agricultural sector remains a challenge. A number of wealthy countries like Korea, Japan, the USA, and EU member states have managed to stall decades of increasing antibiotic use and establish surveillance systems. However, in the absence of long-term funding commitments and international controls, antibiotic stewardship remains patchy in middle- and low-income countries. As such regulating food supply chains and reducing antibiotic consumption in the farm and agricultural industry across the world will require global solutions that are subject to transparent evaluation (Kirchhelle 2018).
12.2 Ecological Impact of Antibiotics and ARGs
Ecosystems are complex structures with significant heterogeneity with respect to geography, time, and components of the environment and animate species involved. It follows therefore that ecological impact of antibiotics and ARGs should vary within different ecosystems. With what little knowledge and evidence is available however, it can be surmised that antibiotics have a significant impact on micro- and macroecosystems and should be treated as ecotoxins.
Despite its ubiquity, microbial ecology is understudied. Since much was unknown about microbial ecology and its predictors before the beginning of this decade, relatively few planned studies have evaluated the impacts of antibiotics, antibiotic residues, and ARGs on various ecosystems.
12.2.1 Impact on Natural Ecosystems
12.2.1.1 Importance of Microbial Diversity in Terrestrial and Aquatic Ecosystems
Microbial diversity in the environment affects biogeochemical cycles (Panizzon et al. 2015), which in turn are critical to the sustainability of macroecosystems. Although very little is known about diversity of microbes owing, in part, to the inherent difficulty of studying non-cultivable microbes, it is well accepted that microbial abundance and diversity in natural ecosystems is essential to their productivity (Prosser et al. 2007).
Antibiotics from anthropogenic (man-made) sources in the environment affect bacterial diversity by inducing a selective pressure, causing a shift in the natural competition toward those microbes that have the ability to withstand this selective pressure. This change in the structural and/or functional composition of microbial communities can affect nitrogen cycles, nutrient cycles, biodegradation in nature (through affecting the proportion and function of anaerobic digesters), and other natural processes that are crucial for optimal functioning of other ecosystems (Blaser et al. 2016). Any effect on microorganism communities therefore extends to indirect impact on larger ecosystems.
12.2.1.2 Impact on Agroecosystems
Ecological risk assessment of the impact of pesticides on plant communities, alluvial soil, and related aquatic environments is a prime example of the effect of toxins on these larger ecosystems (Khetan and Collins 2007). Since antibiotics affect bacterial communities which in turn affect plant rhizospheres etc., a similar conceptual framework applied to antibiotics as plant toxins has been favored by Brandt et al. (2015) and Grenni et al. (2018). Several studies have demonstrated the effect of small sub-inhibitory concentrations of antibiotics and antibiotic residues on bacterial community and rhizosphere structure which can potentially lead to low productivity of agroecosystems with significant agronomic impact (Revellin et al. 2018; Topp et al. 2017). Studies on plants have also identified measured environmental concentrations (MECs) and the predicted no-effect concentrations (PNECs)/non-observed effect concentrations (NOECs) of major antibiotics (Park and Choi 2008; Santos et al. 2010) as toxicological endpoints. Further research into more antibiotic classes and similar limits for different ecosystems is warranted.
ARGs present in soil and agroecosystems impact constituent microbial populations through transmission into different bacterial species. It has been proposed that ARGs in bovine manure can make way into the farm environment with subsequent risk of transmission to consumers via farm produce (Doyle et al. 2017).
12.2.1.3 Impact on Aquatic Ecosystems
Through direct and indirect means, anthropogenic (human and veterinary use) antibiotics enter the natural aquatic environment. Presence of antibiotics and their residues in these aquatic microcosms, wastewaters, as well as in natural marine and freshwaters impacts microbial diversity as well as aquatic animal species and their functions at different trophic levels (Kümmerer 2009).
Aquatic algal populations are affected by the presence of macrolides, tetracyclines, sulfonamides, and quinolones in the order of micrograms per liter (Santos et al. 2010). Both acute and chronic toxic effects have been observed. Macrolides and tetracyclines also impact photosynthesis in cyanobacteria and also disrupt the balance between these beneficial bacteria and toxic weeds, increasing populations of the latter (Pomati et al. 2004). Crustaceans also exhibit chronic toxicity as a result of macrolide and quinolone accumulation (Yamashita et al. 2006). Although studies show no observable effects on fish populations (Isidori et al. 2005), an effect on algal species is thought to eventually affect fish by impacting the aquatic food chain.
Water reuse is also impacted by the presence of antibiotics, antibiotic residues, and ARGs. Microbial water quality is dependent on the presence and quantitation of microbes and absence of pathogenic bacteria. As antibiotics directly affect bacterial compositions, their presence in water ultimately impacts water reusability (Liu et al. 2016).
Aquatic environments are ideal for horizontal gene transfer from one bacterial species to another, thus facilitating spread of antibiotic resistance. Presence of antibiotic concentrations favors survival of organisms carrying ARGs and also perhaps the presence, dispersal, and transmittance of mobile genetic elements (MGEs) (Martínez et al. 2015).
12.2.1.4 Impact on Human and Animal Microbiomes
Evidence on the role of the human microbial ecosystem—the human microbiome —in human health and disease is vast. Diversity and composition of the human microbiome are currently of great interest (Costello et al. 2012). Antibiotic use by humans, whether therapeutic or prophylactic, or resulting from contamination of food, water, or presence in other products, affect the microbiome through modification of this diversity and composition. Microbiomes are present on the human skin, epithelialized orifices, the respiratory tract, the gastrointestinal tract, and the genitourinary tract. Although normal gastrointestinal microbiota are sufficiently resilient to counter major ecological shifts in species, repeated courses of antibiotics are thought to lead to major and perhaps more persistent changes in an individual’s microbiome (Jakobsson et al. 2010). Changes thereof can potentially lead to a shift in the microbiome toward less resilient bacteria more easily overcome by pathogens, and also putatively a larger microbial resistome.
Understanding of the human-microbial symbiosis has now evolved to an extent that ecological paradigms have been applied to clinical situations. Transplantation of feces from a healthy donor (therefore with a “healthy” microbiome) into patients with Clostridium difficile colitis has not only revolutionized the management of this disease, but has also impacted the understanding of the role of microbiome in human health (Bakken et al. 2011).
Antibiotics are common stressors also of the animal microbiome, inducing a so-called “dysbiotic” state (Zaneveld et al. 2017) leading to possible animal health effects. Presence of therapeutic and subtherapeutic (through indirect exposure) concentrations of antibiotics in animal gut has been shown to affect quantities of ARGs (Field and Hershberg 2015).
The presence of resistance genes against trimethoprim, a synthetic antibiotic in human, mammalian, and farm soil microbiomes, alike suggests that some exchange interface exists between these ecosystems, suggesting a much wider impact (Fitzpatrick and Walsh 2016). As applications of metagenomics, metabolomics, and proteomics enable more accurate studies on composition and diversity of the human and animal microbiome, the ecological impact of antibiotics, possible toxicological endpoints, and the resulting overarching health effects will be better understood.
12.2.2 Impact on Artificial Ecosystems
Man-made environments have their own unique ecosystems, and individuals and surfaces within these built environments have their own microbial ecosystems. Metagenomic approaches to some of these ecosystems have been applied recently, and although antibiotic concentrations have not yet been studied widely, evidence from natural ecosystems suggests that antibiotics within these environments also affect microbial ecosystems and constituent ARGs. Among these ecosystems which affect human life significantly and have their own microbiomes are food production facilities (e.g., food manufacturing/packaging plants) (Doyle et al. 2017), aquaculture ecosystems, intensive urban farming, horticulture, and hospitals. The recently initiated Hospital Microbiome Project (Westwood et al. 2014) is likely to reveal further aspects of the built hospital environment and surfaces which can lead to improved understanding of microbial ecology therein. It has been postulated that the presence of antibiotic-impregnated surfaces in hospitals can lead to increase in resistance among resident microbial flora (Strachan et al. 1991; Caselli et al. 2016). Furthermore, patients within hospitals may be considered individual microbial ecosystems where antibiotics significantly impact the microbiome and ARGs (Lofgren et al. 2016).
12.3 Public Health Impact of Antibiotics and ARGs
Antibiotics have revolutionized the treatment of infectious diseases. While this has tremendously impacted the health of those suffering from disease, there are other consequences of using antibiotic that are fast becoming clear and relevant in recent years.
Consequences of antibiotic use may be direct, resulting from intentional therapeutic or prophylactic use, or indirect, due to the presence of subtherapeutic concentrations in the environment, food, and water. Both acute and chronic effects of antibiotic use have been observed at the individual and population level in humans. These result from three main mechanisms or drivers: an increasing resistance gene pool or “resistome,” alterations in microbiomes, and direct tissue toxicity (Fig. 12.1).
12.3.1 Consequences of Expanding Resistome
The hidden resistome in the animate and inanimate environment is a direct public health threat due to its causal association with clinical AMR. Spread of environmental antibiotic resistance genes has resulted in very high clinical resistance among pathogens, severely restricting treatment options for life-threatening infections. Attributable mortality resulting from infections due to resistant pathogens is high in critically ill patients (De Kraker et al. 2011), and also contributes to high health care costs due to prolonged hospital stays alone (De Kraker et al. 2011). By 2050, it is predicted that AMR will cause around 4 million deaths in Asia alone, and 10 million deaths per annum globally (O’Neill 2016).
AMR has also forced older antibiotics and some repurposed drugs as antibiotics into clinical usage. Repurposed drugs and older antibiotics such as polymyxins have uncertain dosages pharmacokinetics, and adverse effect profiles (Palomino and Martin 2012). Clinical use of these drugs therefore often results in toxicities that require further medical interventions, burdening the individual, health care facilities, and health systems.
Intensive management of drug-resistant infections also necessitates investment in diagnosis and treatment of AMR pathogens. This increases health expenditure, and competes with investments in preventive health programs especially in resource limited settings.
12.3.2 Consequences of an Altered Microbiome
As highlighted previously, environmental antibiotics impact microbiomes, which differ remarkably between healthy and diseased humans. Deviations in population structure of microorganisms constituting the “healthy microbiome” affect the health of individuals and populations. This functional impact of the microbiome has been observed in skin disease (Byrd et al. 2018), bowel disease (Heeney et al. 2018), and liver disease (Adolph et al. 2018), and it is expected that further research into this domain will uncover more systemic relationships. Microbiomes are also thought to have a bearing on the nervous system and behavior (Ghaisas et al. 2016).
Alteration of the microbiome in pregnancy is associated with gestational diabetes and adverse fetal outcomes such as preterm birth (Bassols et al. 2016; Prince et al. 2016). In utero exposure to antibiotics can potentially alter the developmental microbiome of the fetus, which modulates the infant immune system (Stiemsma and Michels 2018). Although causality has not been proven, exposure to antibiotics prenatally and in infancy is thought to lead to noncommunicable diseases such as asthma (Hoskin-Parr et al. 2013) or inflammatory bowel disease (Hviid et al. 2011) in later life. The recent obesity epidemic has also been associated with antibiotic exposure and microbiome modification in early childhood (Leong et al. 2018). Antibiotics induce microbial dysbiosis even at subinhibitory concentrations (Berendonk et al. 2015). The association of subinhibitory exposure with dysbiosis and disease needs to be determined through further research.
Gastrointestinal microbiome structure affects tumor formation, as suggested by murine studies (Zackular et al. 2013). In a recent study, long-term antibiotic use in adults was associated with colorectal adenomas; however, independent association with antibiotics remains to be elucidated (Cao et al. 2018). Breast cancer may result from a disturbance of the “estrobolome”—the collective enteric bacterial engine that can metabolize estrogens (Kwa et al. 2016).
12.3.3 Consequences of Antibiotic Toxicity
Data on direct toxic effects of subtherapeutic antibiotics on humans are limited. However, recognition of antimicrobials as toxins in food and water is increasing (Hanekamp and Bast 2015), and no-effect antibiotic concentrations in food have been proposed and revised (Barton 2000). Despite this, comprehensive information on risk assessment and Thresholds of Toxicological Concern (TTC) are lacking for antibiotic classes (Hanekamp and Bast 2015). TTC studies at a population level are also critical to understanding of the various types of anti-infective side effects potentially caused by subtherapeutic concentrations of antibiotics (Barton 2000). Three categories of antibiotic toxicities merit special mention. These are tumorigenesis, allergic and pseudoallergic reactions, and idiosyncratic immune reactions.
Antibiotics such as quinolones and tetracyclines are potentially genotoxic and cytotoxic to mammalian cells (Smart and Lynch 2011; Çelik and Eke 2011). While these antibiotics are monitored in food and biosolids, other mutagenic antibiotics such as furazolidones (Magee et al. 2018) are often overlooked and metabolites of which may contribute to side effects (Hoogenboom et al. 2002).
Allergenic potential of antibiotics is well known; however, the allergenic concentrations and potentials at subtherapeutic levels present in food and water are poorly understood. Veterinary antibiotics also have several toxicological effects observed at high concentrations, and occasional occupational exposure to these can lead to serious side effects (Joint FAO/WHO Expert Committee 2009). Penicillin residues in milk have been documented to trigger a Type I hypersensitivity reaction (Dewdney et al. 1991) rarely. However, reports of other antibiotics triggering allergic reactions are often unsubstantiated and thought to be insignificant (Dayan 1993).
The most well-known example of an immune-mediated reaction to small amounts of antibiotic contamination in food is that of chloramphenicol. This antibiotic is known to cause a dose-independent aplastic anemia (Settepani 1984) through exposure in food. Other reported instances of smaller concentrations of antibiotics causing adverse effects in humans are rare; however, this may be due to the inherent difficulty of monitoring for such effects at the population level. The overall health impact and spectrum of toxicities of contaminant antibiotics may be much wider and hitherto unrecognized.
Assessing public health impact of any intervention is central to influencing policy decisions. The adverse impact of clinical AMR on human health and economy is indisputable. The projected future impact on morbidity and mortality as predicted by the O‘Neill report makes it essential to revise current policies in light of emerging evidence to not only safeguard efficacy of antimicrobial agents, but also to protect the environment from damage by them.
The interdependence of environments, humans, animals, and microbiota is evident in the manner in which changes at epidemiological, ecological, and the public health level are impacted by the presence of antibiotics and spread of ARGs through the ecosystem. As these environments are interlinked, it is necessary that risk assessment and solutions to prevent antimicrobial resistance encompass all sectors where the antibiotics and ARGs interface with the animate world.
References
Adolph TE, Grander C, Moschen AR, Tilg H (2018) Liver–microbiome axis in health and disease. Trends Immunol 39(3):712–723
Bakken JS, Borody T, Brandt LJ, Brill JV, Demarco DC, Franzos MA, Kelly C, Khoruts A, Louie T, Martinelli LP, Moore TA (2011) Treating Clostridium difficile infection with fecal microbiota transplantation. Clin Gastroenterol Hepatol 9(12):1044–1049
Barlow M, Hall BG (2002) Phylogenetic analysis shows that the OXA b-lactamase genes have been on plasmids for millions of years. J Mol Evol 55(3):314–321
Barton MD (2000) Antibiotic use in animal feed and its impact on human healt. Nutr Res Rev 13(2):279–299
Bassols J, Serino M, Carreras-Badosa G, Burcelin R, Blasco-Baque V, Lopez-Bermejo A, Fernandez-Real JM (2016) Gestational diabetes is associated with changes in placental microbiota and microbiome. Pediatr Res 80(6):777
Berendonk TU, Manaia CM, Merlin C, Fatta-Kassinos D, Cytryn E, Walsh F, Bürgmann H, Sørum H, Norström M, Pons MN, Kreuzinger N (2015) Tackling antibiotic resistance: the environmental framework. Nat Rev Microbiol 13(5):310
Bhullar K, Waglechner N, Pawlowski A, Koteva K, Banks ED, Johnston MD, Barton HA, Wright GD (2012) Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS One 7(4):e34953
Blaser MJ, Cardon ZG, Cho MK, Dangl JL, Donohue TJ, Green JL, Knight R, Maxon ME, Northen TR, Pollard KS, Brodie EL (2016) Toward a predictive understanding of Earth’s microbiomes to address 21st century challenges. MBio 13;7(3):pii: e00714–16
Bougnom BP, Zongo C, McNally A, Ricci V, Etoa FX, Thiele-Bruhn S, Piddock LJ (2019) Wastewater used for urban agriculture in West Africa as a reservoir for antibacterial resistance dissemination. Environ Res 168:14–24
Brandt KK, Amézquita A, Backhaus T, Boxall A, Coors A, Heberer T, Lawrence JR, Lazorchak J, Schönfeld J, Snape JR, Zhu YG (2015) Ecotoxicological assessment of antibiotics: a call for improved consideration of microorganisms. Environ Int 85:189–205
Byrd AL, Belkaid Y, Segre JA (2018) The human skin microbiome. Nat Rev Microbiol 16(3):143
Cao Y, Wu K, Mehta R, Drew DA, Song M, Lochhead P, Nguyen LH, Izard J, Fuchs CS, Garrett WS, Huttenhower C (2018) Long-term use of antibiotics and risk of colorectal adenoma. Gut 67(4):672–678
Caselli E, D’Accolti M, Vandini A, Lanzoni L, Camerada MT, Coccagna M, Branchini A, Antonioli P, Balboni PG, Di Luca D, Mazzacane S (2016) Impact of a probiotic-based cleaning intervention on the microbiota ecosystem of the hospital surfaces: focus on the resistome remodulation. PLoS One 11(2):e0148857
Cassini A, Högberg LD, Plachouras D, Quattrocchi A, Hoxha A, Simonsen GS, Colomb-Cotinat M, Kretzschmar ME, Devleesschauwer B, Cecchini M, Ouakrim DA (2019) Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European economic area in 2015: a population-level modelling analysis. Lancet Infect Dis 19(1): 56–66
Çelik A, Eke D (2011) The assessment of cytotoxicity and genotoxicity of tetracycline antibiotic in human blood lymphocytes using CBMN and SCE analysis, in vitro. Int J Hum Genet 11(1):23–29
Chamosa LS, Álvarez VE, Nardelli M, Quiroga MP, Cassini MH, Centrón D (2017) Lateral antimicrobial resistance genetic transfer is active in the open environment. Sci Rep 7(1):513
Collignon P, Beggs JJ, Walsh TR, Gandra S, Laxminarayan R (2018) Anthropological and socioeconomic factors contributing to global antimicrobial resistance: a univariate and multivariable analysis. Lancet Planet Health 2(9):e398–e405
Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA (2012) The application of ecological theory toward an understanding of the human microbiome. Science 336(6086):1255–1262
D’Costa VM, King CE, Kalan L, Morar M, Sung WW, Schwarz C, Froese D, Zazula G, Calmels F, Debruyne R, Golding GB (2011) Antibiotic resistance is ancient. Nature 477(7365):457
Dayan AD (1993) Allergy to antimicrobial residues in food: assessment of the risk to man. Vet Microbiol 35(3-4):213–226
De Kraker ME, Davey PG, Grundmann H, BURDEN Study Group (2011) Mortality and hospital stay associated with resistant Staphylococcus aureus and Escherichia coli bacteremia: estimating the burden of antibiotic resistance in Europe. PLoS Med 8(10):e1001104
Dewdney JM, Maes L, Raynaud JP, Blanc F, Scheid JP, Jackson T, Lens S, Verschueren C (1991) Risk assessment of antibiotic residues of β-lactams and macrolides in food products with regard to their immuno-allergic potential. Food Chem Toxicol 29(7):477–483
Doyle CJ, O'toole PW, Cotter PD (2017) Metagenome-based surveillance and diagnostic approaches to studying the microbial ecology of food production and processing environments. Environ Microbiol 19(11):4382–4391
Durand GA, Raoult D, Dubourg G (2019) Antibiotic discovery: history, methods and perspectives. Int J Antimicrob Agents 53(4):371–382
Ekwanzala MD, Dewar JB, Kamika I, Momba MNB (2018) Systematic review in South Africa reveals antibiotic resistance genes shared between clinical and environmental settings. Infect Drug Resist 11:1907
Field W, Hershberg R (2015) Alarmingly high segregation frequencies of quinolone resistance alleles within human and animal microbiomes are not explained by direct clinical antibiotic exposure. Genome Biol Evol 7(6):1743–1757
Fitzpatrick D, Walsh F (2016) Antibiotic resistance genes across a wide variety of metagenomes. FEMS Microbiol Ecol 92(2):p.fiv168
Gensini GF, Conti AA, Lippi D (2007) The contributions of Paul Ehrlich to infectious disease. J Infect 54(3):221–224
Ghaisas S, Maher J, Kanthasamy A (2016) Gut microbiome in health and disease: linking the microbiome–gut–brain axis and environmental factors in the pathogenesis of systemic and neurodegenerative diseases. Pharmacol Ther 158:52–62
Grenni P, Ancona V, Caracciolo AB (2018) Ecological effects of antibiotics on natural ecosystems: a review. Microchem J 136:25–39
Hanekamp JC, Bast A (2015) Antibiotics exposure and health risks: chloramphenicol. Environ Toxicol Pharmacol 39(1):213–220
Heeney DD, Gareau MG, Marco ML (2018) Intestinal lactobacillus in health and disease, a driver or just along for the ride? Curr Opin Biotechnol 49:140–147
Hofer U (2019) The cost of antimicrobial resistance. Nat Rev Microbiol 17(1):3
Hoogenboom LA, van Bruchem GD, Sonne K, Enninga IC, van Rhijn JA, Heskamp H, Huveneers-Oorsprong MB, van der Hoeven JC, Kuiper HA (2002) Absorption of a mutagenic metabolite released from protein-bound residues of furazolidone. Environ Toxicol Pharmacol 11(3-4):273–287
Hoskin-Parr L, Teyhan A, Blocker A, Henderson AJW (2013) Antibiotic exposure in the first two years of life and development of asthma and other allergic diseases by 7.5 yr: a dose-dependent relationship. Pediatr Allergy Immunol 24(8):762–771
Hviid A, Svanström H, Frisch M (2011) Antibiotic use and inflammatory bowel diseases in childhood. Gut 60(1):49–54
Isidori M, Lavorgna M, Nardelli A, Pascarella L, Parrella A (2005) Toxic and geno- toxic evaluation of six antibiotics on non-target organisms. Sci Total Environ 346:87–98
Jakobsson HE, Jernberg C, Andersson AF, Sjölund-Karlsson M, Jansson JK, Engstrand L (2010) Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One 5(3):e9836
Joint FAO/WHO Expert Committee. on Food Additives Meeting and World Health Organization(2009) Toxicological evaluation of certain veterinary drug residues in food, vol 70. World Health Organization
Kashuba E, Dmitriev AA, Kamal SM, Melefors O, Griva G, Römling U, Ernberg I, Kashuba V, Brouchkov A (2017) Ancient permafrost staphylococci carry antibiotic resistance genes. Microb Ecol Health Dis 28(1):1345574
Khetan SK, Collins TJ (2007) Human pharmaceuticals in the aquatic environment: a challenge to green chemistry. Chem Rev 107(6):2319–2364
Kinney CA, Furlong ET, Zaugg SD, Burkhardt MR, Werner SL, Cahill JD, Jorgensen GR (2006) Survey of organic wastewater contaminants in biosolids destined for land application. Environ Sci Technol 40(23):7207–7215
Kirchhelle C (2018) Pharming animals: a global history of antibiotics in food production (1935–2017). Palgrave Communications. https://doi.org/10.1057/s41599-018-0152 https://www.nature.com/articles/s41599-018-0152-2
Klein EY, Van Boeckel TP, Martinez EM, Pant S, Gandra S, Levin SA, Goossens H, Laxminarayan R (2018) Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc Natl Acad Sci. 10;115(15):E3463–E3470
Koulenti D, Song A, Ellingboe A, Abdul-Aziz MH, Harris P, Gavey E, Lipman J (2019) Infections by multidrug-resistant gram-negative bacteria: what's new in our arsenal and what's in the pipeline? Int J Antimicrob Agents 53(3):211–224
Kümmerer K (2009) Antibiotics in the aquatic environment–a review–part I. Chemosphere 75(4):417–434
Kwa M, Plottel CS, Blaser MJ, Adams S (2016) The intestinal microbiome and estrogen receptor–positive female breast cancer. JNCI: J Natl Cancer Inst 108(8). https://doi.org/10.1093/jnci/djw029
Lamba M, Gupta S, Shukla R, Graham DW, Sreekrishnan TR, Ahammad SZ (2018) Carbapenem resistance exposures via wastewaters across New Delhi. Environ Int 119:302–308
Leong KS, Derraik JG, Hofman PL, Cutfield WS (2018) Antibiotics, gut microbiome and obesity. Clin Endocrinol 88(2):185–200
Lewis K (2013) Platforms for antibiotic discovery. Nat Rev Drug Discov 12(5):371
Liu W, Ma J, Shen C, Wen Y, Liu W (2016) A pH-responsive and magnetically separable dynamic system for efficient removal of highly dilute antibiotics in water. Water Res 90:24–33
Lofgren ET, Egizi AM, Fefferman NH (2016) Patients as patches: ecology and epidemiology in healthcare environments. Infect Control Hosp Epidemiol 37(12):1507–1512
Lugli GA, Milani C, Mancabelli L, Turroni F, Ferrario C, Duranti S, van Sinderen D, Ventura M (2017) Ancient bacteria of the Ötzi’s microbiome: a genomic tale from the copper age. Microbiome 5(1):5
Magee HY, Maurer MM, Cobos A, Pycke BF, Venkatesan AK, Magee D, Scotch M, Halden RU (2018) US nationwide reconnaissance of ten infrequently monitored antibiotics in municipal biosolids. Sci Total Environ 643:460–467
Martínez JL, Coque TM, Baquero F (2015) What is a resistance gene? Ranking risk in resistomes. Nat Rev Microbiol 13(2):116–123
O’Neill J (2016) Review on antimicrobial resistance: tackling drug-resistant infections globally—final report and recommendations (Wellcome Trust, UK Government, 2016)
OECD (2018) Stemming the superbug tide: just a few dollars more. Available at: oe.cd/amr-2018
Palomino JC, Martin A (2012) Is repositioning of drugs a viable alternative in the treatment of tuberculosis? J Antimicrob Chemother 68(2):275–283
Panizzon JP, Pilz Júnior HL, Knaak N, Ramos RC, Ziegler DR, Fiuza LM (2015) Microbial diversity: relevance and relationship between environmental conservation and human health. Braz Arch Biol Technol 58(1):137–145
Park S, Choi K (2008) Hazard assessment of commonly used agricultural antibiotics on aquatic ecosystems. Ecotoxicology 17(6):526–538
Pomati F, Netting AG, Calamari D, Neilan BA (2004) Effects of erythromycin, tetracycline and ibuprofen on the growth of Synechocystis sp. and Lemna minor. Aquat Toxicol 67(4):387–396
Prince AL, Ma J, Kannan PS, Alvarez M, Gisslen T, Harris RA, Sweeney EL, Knox CL, Lambers DS, Jobe AH, Chougnet CA (2016) The placental membrane microbiome is altered among subjects with spontaneous preterm birth with and without chorioamnionitis. Am J Obstet Gynecol 214(5):627–6e1
Proia L, Anzil A, Borrego C, Farrè M, Llorca M, Sanchis J, Bogaerts P, Balcázar JL, Servais P (2018) Occurrence and persistence of carbapenemases genes in hospital and wastewater treatment plants and propagation in the receiving river. J Hazard Mater 358:33–43
Prosser JI, Bohannan BJ, Curtis TP, Ellis RJ, Firestone MK, Freckleton RP, Green JL, Green LE, Killham K, Lennon JJ, Osborn AM (2007) The role of ecological theory in microbial ecology. Nat Rev Microbiol 5(5):384
Revellin C, Hartmann A, Solanas S, Topp E (2018) Long term exposure of agricultural soil to veterinary antibiotics changes the population structure of symbiotic nitrogen-fixing Rhizobacteria occupying nodules of soybeans (Glycine max). Appl Environ Microbiol 84:AEM-00109
Santos LH, Araújo AN, Fachini A, Pena A, Delerue-Matos C, Montenegro MCBSM (2010) Ecotoxicological aspects related to the presence of pharmaceuticals in the aquatic environment. J Hazard Mater 175(1-3):45–95
Settepani JA (1984) The hazard of using chloramphenicol in food animals. J Am Vet Med Assoc 184(8):930–931
Smart DJ, Lynch AM (2011) Evaluating the genotoxicity of topoisomerase-targeted antibiotics. Mutagenesis 27(3):359–365
Stiemsma LT, Michels KB (2018) The role of the microbiome in the developmental origins of health and disease. Pediatrics 141(4):e20172437
Strachan CJL, Newsom SWB, Ashton TR (1991) The clinical use of an antibiotic-bonded graft. Eur J Vasc Surg 5(6):627–632
Topp E, Larsson DJ, Miller DN, Van den Eede C, Virta MP (2017) Antimicrobial resistance and the environment: assessment of advances, gaps and recommendations for agriculture, aquaculture and pharmaceutical manufacturing. FEMS Microbiol Ecol 94(3):fix185
Van Boeckel TP, Brower C, Gilbert M, Grenfell BT, Levin SA, Robinson TP, Teillant A, Laxminarayan R (2015) Global trends in antimicrobial use in food animals. Proc Natl Acad Sci 112(18):5649–5654
Van Goethem MW, Pierneef R, Bezuidt OK, Van De Peer Y, Cowan DA, Makhalanyane TP (2018) A reservoir of ‘historical’antibiotic resistance genes in remote pristine Antarctic soils. Microbiome 6(1):40
Van Schaik W (2015) The human gut resistome. R Soc Publ 370:1–9
Wellington EM, Boxall AB, Cross P, Feil EJ, Gaze WH, Hawkey PM, Johnson-Rollings AS, Jones DL, Lee NM, Otten W, Thomas CM (2013) The role of the natural environment in the emergence of antibiotic resistance in gram-negative bacteria. Lancet Infect Dis 13(2):155–165
Westwood J, Burnett M, Spratt D, Ball M, Wilson DJ, Wellsteed S, Cleary D, Green A, Hutley E, Cichowska A and Hopkins S (2014) The hospital microbiome project: meeting report for the UK science and innovation network UK-USA workshop ‘beating the superbugs: hospital microbiome studies for tackling antimicrobial resistance’, October 14th 2013
World Health Organization (2015) Global action plan on antimicrobial resistance
World Health Organization (2016) United Nations High-level Meeting on Antimicrobial Resistance. In 2017-01-01]. http://www.who.int/antimicrobial-resistance/events/UNGA-meeting-amr-sept2016/en
Yamashita N, Yasojima M, Nakada N, Miyajima K, Komori K, Suzuki Y, Tanaka H (2006) Effects of antibacterial agents, levofloxacin and clarithromycin, on aquatic organisms. Water Sci Technol 53(11):65–72
Zackular JP, Baxter NT, Iverson KD, Sadler WD, Petrosino JF, Chen GY, Schloss PD (2013) The gut microbiome modulates colon tumorigenesis. MBio 4(6):e00692–e00613
Zaneveld JR, McMinds R, Thurber RV (2017) Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat Microbiol 2(9):17121
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Shakoor, S., Hasan, Z., Hasan, R. (2020). Epidemiological, Ecological, and Public Health Effects of Antibiotics and AMR/ARGs. In: Hashmi, M. (eds) Antibiotics and Antimicrobial Resistance Genes. Emerging Contaminants and Associated Treatment Technologies. Springer, Cham. https://doi.org/10.1007/978-3-030-40422-2_12
Download citation
DOI: https://doi.org/10.1007/978-3-030-40422-2_12
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-40421-5
Online ISBN: 978-3-030-40422-2
eBook Packages: Earth and Environmental ScienceEarth and Environmental Science (R0)