Abstract
In metastatic breast cancer the role of circulating tumor cells (CTCs) enumeration for predicting clinical outcome is supported by many studies, most of them dealing with strictly epithelial cells. However, it is becoming clear that CTCs are a heterogeneous cell population characterized by plasticity and including also cells which have lost the epithelial phenotype. Here we review literature data on CTC heterogeneity both at phenotype and at molecular level and discuss the possible contribute of single cell analyses in precision medicine. We conclude with some remarks about the steps still necessary to achieve clinical validity and utility when considering also CTC phenotypic and molecular heterogeneity beyond a simple enumeration.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Breast cancer
- Metastasis
- Circulating tumor cells (CTCs)
- CTC heterogeneity
- CTC molecular characterization
6.1 Introduction
Metastasis is the leading cause of death in cancer patients, but still the biology of tumor dissemination and metastases formation is poorly understood. Contrary to what initially thought, tumor cell dissemination leading to the formation of clinically overt metastases is a process that starts early [1, 2]. Cells giving origin to metastases improve their fitness for invasion and colonization by acquiring new characteristic either in parallel to the primary tumor or within the primary tumors as progression occurs giving raise to late dissemination. In particular, metastatic cells lose drug sensitivity respect to the primary tumor [3]. Moreover, in metastatic disease, the primary tumor itself is characterized by increasing heterogeneity due to its own evolution and to the reseeding of cancer cells among different sites [4].
Circulating Tumor Cells (CTCs) are the seeds of metastases and their study is instrumental for understanding the metastatic process and tumor complexity. In fact, CTCs originate from established tumor masses, either primary or metastatic foci, migrate into the bloodstream, and acquire the potential to change their fate by undergoing different phenomena driven by epigenetic events, but also by interaction with other cells in the blood such as platelets that enable them to seed metastases [5]. The capacity to lose the lineage commitment for acquiring different features and to direct cell fate by switching to another differentiated cell type is defined as “plasticity”. CTC plasticity includes different programs affecting invasion, survival and proliferation, and is to a certain extent mirrored by the typical heterogeneity of cancer [6, 7]. Moreover, CTCs reflect in part the spectrum of mutations present in either the primary and/or metastatic tumor [4].
Heterogeneity is a hallmark of cancer that is responsible for its complexity, tracks its development and is regarded as a main culprit for the failure of cancer therapies [3]. CTCs in particular are characterized by different types of heterogeneity: an extrinsic and an intrinsic one. The extrinsic heterogeneity of CTCs is linked to the tumor of origin, and is therefore due to the tumor type and to its specific tumor driver mutations. All these factors give rise to different tumor cell phenotype even within the same tissue. Intrinsic heterogeneity of CTCs instead deals with mechanisms of adaptation occurring during the metastatization process that are instrumental for tumor spread, and includes conversions between cellular phenotypes [6].
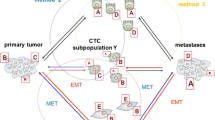
The main mechanisms by which CTCs develop intrinsic heterogeneity is the epithelial-to-mesenchymal transition (EMT) that is influenced by the type of tumor, the tissue of origin, the local microenvironment of cancer cells and by the treatment. Later, before the colonization step, the disseminated cancer cells undergo an inverted process called mesenchymal-to-epithelial transition (MET) that allows their settlement in metastatic foci. The EMT core network controls feedback loops between the two extreme fates (epithelial and mesenchymal phenotype). Those however, are not binary processes, accordingly cells retaining a hybrid epithelial/mesenchymal phenotype are often observed and can promote multicellular aggregate migrations thanks to their mixed traits. Thus, genes regulating the EMT phenotype are differently expressed in distinct CTC populations, promoting their intrinsic heterogeneity and influencing drug resistance and tumor dormancy. Importantly, intermediate states could also induce cells to exhibit stemness traits, although, the acquisition of mesenchymal and stemness traits can be uncoupled [4, 7, 8].
In such a scenario, the key question is how to better understand and possibly best classify CTC heterogeneity in order to obtain a biomarker or a set of biomarkers useful for clinical applications in metastatic patients. It is in fact important to establish which approaches are more suitable for defining CTC features that have a direct impact on prognosis, treatment response prediction and that are able to inform the clinical decision by also taking into account intra-patient heterogeneity, both in space and time.
In this chapter, the CTC heterogeneity issue will be reviewed limiting to Metastatic Breast Cancer (MBC).
6.2 CTC Phenotypic Heterogeneity
CTCs can be isolated from patients’ peripheral blood and are used to monitor tumor cell populations during disease progression and in response to therapies. Many CTC isolation technologies have been developed, but only the CellSearch® system was warranted FDA-approval. It is based on enrichment of CTCs by epithelial markers (EpCAM) and subsequent software-assisted manual enumeration of CTCs defined as nucleated cells expressing cytokeratins and not expressing the pan-leukocyte antigen CD45 [9].
The role of CTC detection by CellSearch® as a survival predictive biomarker in MBC has been extensively tested by Cristofanilli et al. [10]. In patients with measurable metastatic disease, the detection of ≥5 CTC/7.5 mL of blood before treatment was demonstrated to represent an independent predictor of both progression-free survival (PFS) and overall survival (OS). In a later study, Cristofanilli et al. followed CTCs enumeration in serial samples collected at different times, showing that their detection before first-line therapy and after 4 weeks of treatment was significantly predictive of PFS and OS. Persistence of CTCs at restaging time was also significantly associated with worst prognosis [11].
In a pooled analysis including 51 centers across Europe, Bidard et al. reported the clinical validity of CTCs changes during treatment and showed that addition of CTC enumeration to prognostic model including currently used clinicopathological variables improves prognostication by adding significant independent information [12]. In keeping with this, in a recent retrospective study pooled analysis including almost 2500 MBC patients, CTC enumeration proved to be able to stratify patients based on their disease aggressiveness (indolent vs aggressive) supporting the introduction of CTC to improve staging in MBC [13].
To improve CTC enumeration by the CellSearch® system, de Wit et al. studied the relevance of EpCAMlow-expressing cells, which are considered as EpCAM-negative cells by the CellSearch® system and discarded. EpCAMlow-CTCs were collected on microfilters from the blood fraction discarded after CellSearch® CTC-enrichment, were fluorescently labeled and scored for enumeration using the classic CellSearch® critera. The authors analyzed the presence of these cells in the blood of castration-resistant prostate cancer and MBC patients. In both tumor types the number of patients with positive detection of CTCs increased when both EpCAMhigh and EpCAMlow CTCs were considered. However, the presence of EpCAMlow CTCs was not associated with survival, and it deserves to be further investigated [14]. These cells may in fact be a different subpopulation of CTCs, however, although their epithelial origin is proven by the expression of cytokeratin (CK), no direct evidence exist for their malignant nature. Only genetic analysis could provide direct evidence and help clarifying specific features of such cells.
Despite the success of CellSearch® in predicting risk in MBC, concerns arise on the possible clinical role of CTC subpopulations that do not strictly meet CellSearch® criteria and that are missed by such method. Therefore, studies aiming at selecting and identifying all CTC subpopulations have been done using marker-independent approaches for CTC-enrichment such as magnetic beads selection (AdnaTest, Myilteni Biotec), filters (ScreenCell), gradient centrifugation (Oncoquick®), size and deformability selection (Parsortix™) and exploiting dielectrophoretic properties (DEPArray™ System) [15, 16].
The DETECT study run a direct head-to-head comparison between CellSearch® and the AdnaTest on a prospective series of 254 women with MBC. Fifty percent of patients were defined as CTC-positive by the CellSearch® system versus 40% by the AdnaTest, which employs magnetic beads functionalized with antibodies against EpCAM and MUC1 for CTC-enrichment and a multiplex PCR assessing EPCAM, MUC1 and HER2 expression for CTC detection. Overall the concordance rate between the two assays was 64%, but no data on the direction of discordances were reported. The association between CTC positivity by CellSearch® and shorter PFS and OS was confirmed, whereas no statistically significant associations were observed between AdnaTest CTC–positivity and clinical outcome [17].
To better investigate the impact of CTC detection methods, also Aaltonen et al. studied CTCs from MBC patients in parallel by CellSearch® and AdnaTest. They used a new developed kit for CTC capture, the EMT2, which adds antibodies against HER2 and EGFR to the CTC-enrichment antibody cocktail to improve the capture efficiency compared to the traditional AdnaTest (EMT1). Enriched samples from AdnaTest EMT1 and EMT2 were then analyzed by multiplex qPCR for 38 genes associated with cancer. Evaluating CTC-positivity, a number of patients was positive by both methods, whereas some patients resulted as positive only by CellSearch® or AdnaTest. In addition, some of the samples defined as CTC-negative by both the CellSearch® and the classic AdnaTest (based on transcripts for EPCAM, HER2, MUC1), did instead express KRT19 or ERBB2 questioning their negativity [18]. These results, although lacking strong clinical evidence, definitely highlight the potential of combining different markers to improve circulating cells classification and possibly their association with clinical outcome.
Using the AdnaTest only, Aktas et al. investigated expression of EMT markers (Akt-2, Twist1 and PI3Kα) and of ALDH1 (marker for stem cell) in different MBC patients undergoing different types of palliative therapy. Treatment response was evaluated according to RECIST criteria. Only 10% of responders were CTC-positive, respect to 71% among non-responder group. Interestingly, in 81% of CTC-positive patients, at least one of EMT markers, ALDH1 or both, were expressed. Conversely EMT and stem-cell markers were expressed in only 11% of CTC-negative samples. Such data suggest that beside the presence of CTCs themselves, also their specific transcriptomic program needs to be considered in order identify CTC subpopulations and understand clinical associations [19].
Overall, the conclusions of the above described studies lead us to the exploration of circulating cells exhibiting mesenchymal traits.
The presence of mesenchymal and intermediate epithelial/mesenchymal cells in blood samples enriched for CTCs in MBC patients was investigated in a landmark study by Yu et al. [20]. These authors investigated the presence of cells exhibiting mesenchymal traits both in primary tumor and in blood samples and reported only rare mixed epithelial/mesenchymal cells within the primary tumors whereas mixed phenotype cells were frequently present among CTCs. Blood samples were enriched for CTC with the microfluidic herringbone-chip using epithelial and tumor-specific antibodies (EpCAM, EGFR, HER2) and studied at single cell level with in situ approaches. Based on the results of RNA-ISH analysis that evaluated a series of epithelial (KRT5, 7, 8, 18, 19; EpCAM, CDH1) and mesenchymal (FN1, CDH2, SERPINE/PAI1) markers five categories of CTCs were defined: exclusively epithelial cells (E), 3 categories of intermediate cells (E > M, E = M, E < M), and exclusively mesenchymal cells (M). Using a cutoff of 5 CTCs/3 ml 41% of MBC, at various treatment stages, scored positive for CTCs, and EMT features were different between lobular and ductal histotypes. Also when comparing pre- and post-treatment blood samples (n = 10), different CTCs features were found. In post-treatment samples from patients who responded to therapy (n = 5), the absolute CTC numbers decreased and/or the proportion of M-CTCs decreased. Conversely, in patients who experienced progression while on therapy (n = 5), the number of M-CTCs increased in the post-treatment samples supporting a role of EMT in treatment sensitivity [20].
To understand the prognostic relevance of single CTCs with specific phenotypes, Papadaki et al. detected and characterized CTCs pre- and post-treatment in patients with MBC. They identified four different CTCs subpopulations by performing triple immunofluorescence on cytospin preparations of peripheral blood mononuclear cell (PBMC) with antibodies against CK8, CK18 and CK19 (epithelial markers), ALDH1 and TWIST1. In patients not responding to the treatment, the number of samples with CTCs showing stem and partial EMT features increased. Conversely, positivity percentages slightly decreased after treatment for the other types of CTCs (i.e., CTC showing stem but lacking partial EMT features, and CTCs lacking stem features and/or positive or negative for EMT features). In keeping with this, only the presence of CTCs with stem and partial EMT features was associated to shorter PFS and OS. This finding was interpreted by the authors as a suggestion that partial EMT increases the chance of the cells to subsequently undergo mesenchymal-epithelial transition (MET), a step necessary to allow colonization at the metastatic site [21].
The importance of such mixed-phenotype CTCs is also supported by other studies. Using CD45 MicroBeads for depletion of leukocytes and the DEPArray™ system, Bulfoni et al. identified four different CTC subtypes: epithelial CTCs (E-CTC) expressing only epithelial markers (EpCAM, E-cadherin), CTCs undergoing EMT (EM CTC) co-expressing epithelial and mesenchymal markers (CD44, CD146, N-cadherin), putative mesenchymal cells (MES) expressing only mesenchymal markers, and negative cells (NEG) not expressing the tested markers. Some associations were highlighted between CTC subpopulations and breast cancer molecular subtype, proliferative rates and metastatic localization, however only the EM-CTCs were significantly associated with shorter PFS and OS [22].
The identification of the so far described CTC-subpopulations is strongly influenced by the type of CTC-enrichment. In such a context, to avoid underestimation of CTC subpopulations, the Parsortix™ system that selects CTCs in an epitope-independent way, exploiting size and deformability as selection criteria appears to be particularly promising [23]. In our laboratory we combined the Parsortix™ with the DEPArray™ system. Thanks to the presence of a fluorescent microscope equipped with a camera, and to a microfluidic chip exploiting dielectrophoresis to entrap single cells, the DEPAarray™ allows visualization of cells labeled for epithelial, leukocyte, mesenchymal or other type of markers, coupled with the selection and recovery by the operator of the cells of interest [24]. Using this system, we were able to observe the presence of specific CTCs subpopulations in all blood samples (n = 14) from women who underwent mastectomy for early triple negative breast cancer (TNBC), collected at the time of imaging-proven distant site relapse. Our data support the concept that CTCs identification cannot rely on a single CTC phenotype, but should rather broaden the phenotypic criteria used for selection and include direct molecular evidence for the malignant nature of the selected cells. In fact, using antibodies against epithelial (EpCAM, panCK, EGFR) and leukocyte (CD45, CD14, CD16) markers, we succeeded in distinguishing two different CTCs subpopulations: epithelial CTCs (eCTCs), and non-conventional “putative” CTCs (ncCTCs), i.e. cells lacking both leukocyte and epithelial (tumoral) but with malignant genotype. We therefore suggest that besides mixed CTC (epithelial and mesenchymal phenotype) a third CTC subpopulation characterized by the lack of expression of epithelial and leukocyte markers, but with confirmed aberrant genotypes is detectable in the blood collected at the time of imaging-documented relapse, of women who underwent mastectomy for early TNBC (manuscript under preparation).
Whereas many studies have addressed both technical aspects involved in isolation of CTC subpopulations and the clinical role of CTC subpopulations, few studies are instead available on the mechanisms involved in induction of mesenchymal traits in CTCs. Interaction with platelets and secretion of TGFβ have been described as a possible mechanism for induction of EMT [5] and some heterogeneity in methylation of genes involved in EMT has been reported for single CTCs [25] thus suggesting an epigenetic control.
Recently a different mechanism has been suggested as possibly causally involved in promoting CTC heterogeneity and in the generation of specific CTC subpopulations, i.e. heterotypic cell fusion between epithelial cells and blood cells, in particular with macrophages [26]. Such a mechanism, so far experimentally investigated in preclinical models but poorly validated in clinical samples, deserves further attention as it may open the way to the identification of even more CTC subpopulations and possibly also to new pharmaceutical targets for interfering with tumor dissemination.
All the above reported data emphasize that studies evaluating the role of circulating cells in cancer evolution should not be limited to circulating cells expressing epithelial and lacking leukocyte markers, but must be broadened to include other phenotypes. Unfortunately, at the moment clinical data available on CTC subpopulations are not impressive and often limited to small studies lacking statistical power: nonetheless the field appears as very promising. However, we must underline the lack of both clearly defined criteria for selection and of a proof of the malignant nature of the various CTC subpopulations.
6.3 CTC Molecular Characterization
Discrepancies between different methods for CTC-identification by phenotypic features suggest the need for more accurate criteria for CTCs classification. In fact, as described above the phenotype alone is not sufficient to classify a single cell as a bona-fide CTC and thus in the case of ncCTCs (i.e. CTCs lacking the conventional identification markers) only a characterization of the genotype can definitely ascertain the actual malignant nature.
A possible approach to test for the malignant nature is performing an analysis of copy number alterations (CNA) at a single cell level. By running a low-coverage whole genome sequencing it is in fact possible to obtain CNA profiles for each cell, indicating if the genome is diploid and thus presents a flat CNA profile as expected for normal cells or if, as expected for a bona-fide CTC, it contains regions characterized by genomic gains and losses. This approach, which leads to a molecular proof of tumor origin for each single-cell, is particularly important for cells where the phenotype does not allow a clear distinction.
Molecular characterization however, does offer much more than simply ascertaining the malignant nature, as it contains the information on the clonal origin of each analyzed cell.
In current practice tissue biopsies are used to test tumors for actionable genomic abnormalities despite well-known limitations dealing with spatial and temporal heterogeneity. In the metastatic setting, limitations affecting tissue profiling are even greater, since the tumor cells are homed in different anatomical sites and might have evolved in distinct ways. In this context, the analysis of CTCs deriving from both primary tumor and metastatic lesions would instead provide a comprehensive molecular portrait of the entire tumor burden just by a single blood test. In principle, obtaining a molecular profile of CTCs could therefore facilitate individual patient treatment management thus helping to reach the ambitious aim of achieving a true precision medicine. However, although this approach appears promising from a theoretical point of view, it still has many limitations. Accordingly, revision of the literature data mostly shows results obtained on very few patients and by different technical approaches that limit comparability. Still some general messages can be derived.
Among the first in assessing the feasibility of mutational analysis on single CTCs isolated with the DEPArray™, Mu et al. investigated mutations in CTCs isolated from one woman with inflammatory MBC. CTCs were enriched by an unbiased method based on size selection, using ScreenCell filters, and were thereafter analyzed with the DEPArray™ to select and isolate single CTCs. After whole genome amplification (WGA) using Ampli1™ WGA kit, mutational analysis on amplified DNA from 7 CTCs was performed by Sanger sequencing, in order to investigate the presence of a specific TP53 mutation which had been previously identified in the primary tumor: the TP53 exon 6 p.R248W missense point mutation. The same mutation was investigated both in single and pooled CTCs. Heterozygous TP53 mutation was found in 1 single cell and 1 pool of 3 CTCs, and homozygous TP53 mutation was instead detected in 1 single CTCs and 1 pool of 2 CTCs. The mutation was not present in one WBC analyzed as control [27]. These results show the feasibility of a molecular approach to investigate relevant mutations in CTCs, and suggest that mutations identified in the tissue can also be traced in single or pooled CTCs, which might therefore be regarded as representative of the tissue of origin. No information is however provided on the possible presence of CTC-private mutations that may have resulted from tumor clonal evolution, as has instead been reported in other clinical settings [28].
CTC private mutations, if identified with reliable methods, are very interesting since they provide a genomic/clonal tracking of the disease evolution. In the case of MBC, searching for CTC private mutations in the ESR1 is particularly promising, since those mutations (some of them directly involved in endocrine treatment resistance) are rarely detected prior to treatment start and only appear with the onset of treatment resistance [29, 30]. This represents therefore a typical clinical scenario where CTCs molecular analysis would be useful to dissect time-related heterogeneity. Moreover, CTCs represent the ideal biological sample for tracking in real-time the onset of endocrine resistance, since they potentially allow evaluating at the same time both mutations as well as splicing variants. Nonetheless, a recent study planned to evaluate ESR1 mutations and splice variants in CellSearch-enriched bulk CTCs before start of endocrine therapy and at the time of progression, failed to detect an enrichment for ESR1 mutations at progression [31]. Such mutations were instead found to be enriched in ctDNA. These results, which apparently rule out a role for CTCs in monitoring the onset of endocrine resistance, as suggested by the authors themselves, can be instead interpreted as an indication of the need to perform CTC molecular analyses on single isolated cells rather than on bulk samples, albeit enriched for CTCs. Indeed, only by considering the genotype of each single CTC we may be able to capture their message on heterogeneity and evolution of the disease. This poses technical problems that can be overcome as reported above, but also challenges in the interpretation of results.
In a study to assess the possibility of detecting mutations in CTCs, Paolillo et al. investigated ESR1 mutations by Sanger sequencing in single CTCs from MBC patients. They analyzed 40 CTCs recovered combining CellSearch® and DEPArray™ platforms from 3 ER-positive MBC patients. Their protocol was technically robust since 12 white blood cells (WBCs) analyzed as controls were all correctly classified as wild-type for ESR1. The first investigated patient presented 5 CTCs, all wild-type for ESR1. The second patient carried a single ESR1 activating mutation (Y537S) in exon 8 in heterozygosis in 3 CTCs, and the same mutation in homozygosis in 1 CTC, the last CTC was instead wild-type for the same mutation. In this patient, CTCs’ molecular heterogeneity and the detection of activating mutations were in keeping with the observed treatment failure. For the third patient, serial samples collected during treatment at different time points were available. In the first sample, the authors could study 12 CTCs, all wild-type for ESR1 mutations, thus suggesting that the patient was still endocrine sensitive. In the second blood sample, the authors detected high heterogeneity: 8 CTCs were negative for estrogen receptor (ER) expression and wild-type for ESR1 mutation, 4 CTCs were positive for ER expression and wild-type for ESR1 mutation, 3 CTCs were positive for ER expression and carried the mutation Y537s in heterozygosis, and 1 CTCs was positive for ER expression and carried 2 different mutation in exon 8 of ESR1 in homozygosis [32]. In this latter patient too, the appearance of ESR1 mutations in CTCs was mirrored by failure to respond to conventional endocrine treatment. Overall, these results, although still anecdotal, show that CTC characterization might be more informative than tissue, giving new hints on resistance mechanisms to endocrine therapy in ER-positive MBC patients.
In the clinical management of MBC the most frequently used treatment-predictive biomarkers, such as ER and HER2, are evaluated at protein level on the primary tumor. Thus, besides molecular characterization at genomic level, also transcriptomic analysis of single CTC should be useful to guide treatment and to inform on possible changes of the molecular phenotype with respect to the primary tumor. Such studies would indeed provide hints on the heterogeneity of typical treatment targets.
In a recent study, the status of the therapeutic biomarkers ER and HER2 was examined in CTCs isolated from 105 women with MBC. Immune enrichments for EpCAM and FACS analysis were used for isolation of single cells prior to performing genome wide CNA by aCGH and transcription analysis of 64 genes by low-density array qPCR. Combined transcriptional and genomic profiling showed presence of different CTCs subpopulations at different frequencies (26% ESR1 − ERBB2−, 47% ESR1 + ERBB2−, and 27% ERBB2+). Serial testing of longitudinally collected samples showed that ERBB2 status was more stable over time compared to ESR1 status. Moreover, discordance in ESR1/ER (27%) and ERBB2/HER2 (23%) status between CTCs and matched primary tumors emerged by comparative analysis [33]. Based on the results it was concluded that CTC molecular analysis has the potential to help treatment decision in a clinical setting of a progressing disease, but the clinical utility of CTC-biomarkers is far from being demonstrated.
All studies listed above are concordant about the concept that CTC molecular analysis could improve clinical practice, although so far they only demonstrate the technical feasibility. The reported results underline a high level of intra-patient heterogeneity indirectly suggesting that currently used criteria for patients’ stratification could in some cases not represent the patients’ real tumor “status”. Patients classified as similar at tissue level could instead display differences at molecular level, therefore molecular approaches represent an opportunity for more accurate patients’ profiling for personalized medicine.
In the meantime a new scenario is slowly appearing, where liquid biopsy can improve understanding of metastatization processes bringing out different characteristics between the primary tumor and the circulating tumor elements and elucidating the extrinsic heterogeneity of the tumor. Single-cell analysis may also help to rebuild the origin of detected variants, understanding if they coexist in a single cell or derive from distinct clones.
Using the CellSearch® system for CTC enrichment, the DEPArray™ for single cell isolation and next generation sequencing (NGS), De Luca et al. investigated mutations in single CTCs in patient with MBC. In this study, the authors could compare CTCs and primary tissue (limiting to the variants found in the single CTCs) in 3 patients, and they found correspondence only for a benign PDGFRA variant (1 patient) and a deleterious somatic mutation in TP53. For all the other variants discovered in CTCs, there was no correspondence in primary tissue [34]. After analyzing 14 CTCs derived from 4 patients, for 51 sequence variants in 25 genes, it was observed that almost all mutations were present in only one single CTC. This represents an interesting result, which might highlight the importance of single cell analyses for studying heterogeneity, but it also poses technical questions. The mandatory step in single-cell analyses of performing a WGA might have been responsible for the introduction of technical artifacts and, despite the fact that high coverage increases the confidence of variants called in each single cell, DNA polymerase fidelity still represents a concern. However, also given the technical reliability of the data, can such result be considered robust enough for a clinical decision? This is at the moment the main issue, questioning the application of single cell NGS in the clinics. Nonetheless, this is also possibly its strength, due to the ability to potentially suggest new biologically/clinically relevant variants as it is illustrated by the example reported below.
Paoletti et al. investigated the possibility of obtaining paired information from CTCs and tissue metastases by NGS in 12 patients. For this purpose the authors processed whole blood from MBC patients using the CellSearch® system, isolated single cells with the DEPArray™ system and applied Ampli1™ protocol for DNA isolation and analysis. Targeted NGS was performed using the Oncomine Comprehensive Assay, and selected mutations were confirmed by Sanger Sequencing. In parallel, frozen tissue from primary tumor and fresh tissue from metastatic biopsies were used to generate exome libraries to be processed for Whole Exome Sequencing (WES). High concordance (85%) between CTCs and fresh metastatic tissue was found, in term of genomic alterations. However, private alterations were detected both in tissue and in CTCs, though at low frequencies [35]. Since potentially clinically informative/actionable mutations may either be exclusively present in metastatic tissue or in CTCs, the authors suggest performing genomic profiling of both, since results may be considered as complementary in order to achieve a true clinical impact.
Within the same study, in one single patient with endocrine treatment refractory disease, 32 individual CTCs and CTC-pool samples were recovered and analyzed by comprehensive NGS revealing the presence of a well-known ESR1 mutation associated with endocrine resistance, the ESR1p.Y537S. However, in one single CTC such mutation was not detected, and a new heterozygous ESR1 mutation (ESR1p.A569S) was instead detected and confirmed by ddPCR (droplet digital PCR). To assess its role, Paoletti et al. stably overexpressed the ESR1p.A569S mutation by lentivirus-mediated infection in the endocrine sensitive MCF7 cells showing increased estradiol and tamoxifen-induced growth, thus validating its role in conferring a modest treatment resistance. In this case, a specific somatic variant detected in a single CTC allowed increasing our knowledge on endocrine resistance mechanisms.
With the exception of specific cases as the one reported by Paoletti et al. detection of a somatic mutations in one single CTC is still of questionable biological or clinical utility. In a study run on 112 women with MBC, CTCs were enumerated by CellSearch®, and 5 patients with CTC counts ≥100 were chosen for performing targeted NGS (Custom Cancer Hot Spot panel V2), globally analyzing 40 CTCs. Mutational heterogeneity was observed among CTCs, and it was reflected by ctDNA analysis run in matched samples. Minor subclonal mutations, likely acquired during tumor progression, were observed only in liquid biopsy and were undetectable at tissue level. These data suggest that ctDNA mutational profile can reflect the CTC heterogeneity in patients with high CTC counts, however no data are provided on patients with low CTC counts. It may be concluded that, despite the good correlation with ctDNA, still the occurrence of a new somatic variant in one single CTC remains difficult to interpret [36].
6.4 Conclusions and Future Perspectives
Since the original observation of the presence of tumoral cell in the blood of patients with solid tumors, enormous progresses have been made. In last two decades, thanks to efforts in technical standardization, enumeration of epithelial-CTCs has become a widely used tool for prognostication and treatment monitoring across many tumor types. Indeed, in MBC CTC enumeration with the CellSearch® is technically and clinically valid [12] and has been proposed as useful for clinical staging of MBC [13].
Now, thanks to new technical refinements both in CTC enrichment and in CTC molecular characterization, the possibility of using CTCs for a real-time assessment of the disease and of its evolution at molecular level is becoming reality. However, there are still many aspects that need to be better investigated. Among those, although we know that CTC are present as phenotypically distinct subpopulations, and we have achieved at least a technical validation of the methods used for detection of CTC subpopulations, we still ignore the role of each subpopulation and are thus far from having obtained a clinical validation for each CTC phenotype.
In addition, molecular characterization of single CTCs is technically feasible both at genotype and at transcriptional level. So far results are definitely suggestive of a possible role of CTC as tissue surrogate, and as a means to capture and to overcome tumor heterogeneity, but again we are far from having reached a clinical validation and still confused about the possible future clinical utility. We are however confident on the fact that further advances in methods that not only facilitate CTC isolation and characterization, but also allow their functional characterization, will bring us closer to achieve clinical utility (Fig. 6.1).
Schematic representation of the steps in the validation of CTC as a biomarker. Vertical bars represent the consecutive steps in the path towards the development of a biomarker: technical validity, clinical validity and clinical utility. Arrows represent the current achievements and dots represent future steps necessary for considering CTC-enumeration, CTC-phenotypic characterization, CTC-molecular characterization and CTC-functional characterization as a biomarker
References
Klein CA. Parallel progression of primary tumours and metastases. Nat Rev Cancer. 2009;9(4):302–12. https://doi.org/10.1038/nrc2627.
Gray JW. Evidence emerges for early metastasis and parallel evolution of primary and metastatic tumors. Cancer Cell. 2003;4(1):4–6.
Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell. 2017;168(4):670–91. https://doi.org/10.1016/j.cell.2016.11.037.
Brown HK, Tellez-Gabriel M, Cartron PF, Vallette FM, Heymann MF, Heymann D. Characterization of circulating tumor cells as a reflection of the tumor heterogeneity: myth or reality? Drug Discov Today. 2019;24(3):763–72. https://doi.org/10.1016/j.drudis.2018.11.017.
Labelle M, Begum S, Hynes RO. Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer Cell. 2011;20(5):576–90. https://doi.org/10.1016/j.ccr.2011.09.009.
Gupta PB, Pastushenko I, Skibinski A, Blanpain C, Kuperwasser C. Phenotypic plasticity: driver of cancer initiation, progression, and therapy resistance. Cell Stem Cell. 2019;24(1):65–78. https://doi.org/10.1016/j.stem.2018.11.011.
Nieto MA, Huang RY, Jackson RA, Thiery JP. Emt: 2016. Cell. 2016;166(1):21–45. https://doi.org/10.1016/j.cell.2016.06.028.
Jolly MK, Boareto M, Huang B, Jia D, Lu M, Ben-Jacob E, et al. Implications of the hybrid epithelial/mesenchymal phenotype in metastasis. Front Oncol. 2015;5:155. https://doi.org/10.3389/fonc.2015.00155.
Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10(20):6897–904. https://doi.org/10.1158/1078-0432.CCR-04-0378.
Cristofanilli M, Reuben JM, Budd GT, Ellis MJ, Stopeck A, Matera J et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. 2004.
Cristofanilli M, Hayes DF, Budd GT, Ellis MJ, Stopeck A, Reuben JM, et al. Circulating tumor cells: a novel prognostic factor for newly diagnosed metastatic breast cancer. J Clin Oncol. 2005;23(7):1420–30. https://doi.org/10.1200/JCO.2005.08.140.
Bidard FC, Peeters DJ, Fehm T, Nole F, Gisbert-Criado R, Mavroudis D, et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. Lancet Oncol. 2014;15(4):406–14. https://doi.org/10.1016/S1470-2045(14)70069-5.
Cristofanilli M, Pierga JY, Reuben J, Rademaker A, Davis AA, Peeters DJ, et al. The clinical use of circulating tumor cells (CTCs) enumeration for staging of metastatic breast cancer (MBC): international expert consensus paper. Crit Rev Oncol Hematol. 2019;134:39–45. https://doi.org/10.1016/j.critrevonc.2018.12.004.
de Wit S, Manicone M, Rossi E, Lampignano R, Yang L, Zill B, et al. EpCAM(high) and EpCAM(low) circulating tumor cells in metastatic prostate and breast cancer patients. Oncotarget. 2018;9(86):35705–16. https://doi.org/10.18632/oncotarget.26298.
Ferreira MM, Ramani VC, Jeffrey SS. Circulating tumor cell technologies. Mol Oncol. 2016;10(3):374–94. https://doi.org/10.1016/j.molonc.2016.01.007.
Alix-Panabieres C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer. 2014;14(9):623–31. https://doi.org/10.1038/nrc3820.
Muller V, Riethdorf S, Rack B, Janni W, Fasching PA, Solomayer E, et al. Prognostic impact of circulating tumor cells assessed with the CellSearch System and AdnaTest Breast in metastatic breast cancer patients: the DETECT study. Breast Cancer Res. 2012;14(4):R118. https://doi.org/10.1186/bcr3243.
Aaltonen KE, Novosadova V, Bendahl PO, Graffman C, Larsson AM, Ryden L. Molecular characterization of circulating tumor cells from patients with metastatic breast cancer reflects evolutionary changes in gene expression under the pressure of systemic therapy. Oncotarget. 2017;8(28):45544–65. https://doi.org/10.18632/oncotarget.17271.
Aktas B, Tewes M, Fehm T, Hauch S, Kimmig R, Kasimir-Bauer S. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 2009;11(4):R46. https://doi.org/10.1186/bcr2333.
Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339(6119):580–4. https://doi.org/10.1126/science.1228522.
Papadaki MA, Stoupis G, Theodoropoulos PA, Mavroudis D, Georgoulias V, Agelaki S. Circulating tumor cells with stemness and epithelial-to-mesenchymal transition features are chemoresistant and predictive of poor outcome in metastatic breast cancer. Mol Cancer Ther. 2019;18(2):437–47. https://doi.org/10.1158/1535-7163.MCT-18-0584.
Bulfoni M, Gerratana L, Del Ben F, Marzinotto S, Sorrentino M, Turetta M, et al. In patients with metastatic breast cancer the identification of circulating tumor cells in epithelial-to-mesenchymal transition is associated with a poor prognosis. Breast Cancer Res. 2016;18(1):30. https://doi.org/10.1186/s13058-016-0687-3.
Xu L, Mao X, Imrali A, Syed F, Mutsvangwa K, Berney D, et al. Optimization and evaluation of a novel size based circulating tumor cell isolation system. PLoS One. 2015;10(9):e0138032. https://doi.org/10.1371/journal.pone.0138032.
Reduzzi C, Motta R, Bertolini G, Miodini P, Martinetti A, Sottotetti E, et al. Development of a protocol for single-cell analysis of circulating tumor cells in patients with solid tumors. Adv Exp Med Biol. 2017;994:83–103. https://doi.org/10.1007/978-3-319-55947-6_4.
Pixberg CF, Raba K, Muller F, Behrens B, Honisch E, Niederacher D, et al. Analysis of DNA methylation in single circulating tumor cells. Oncogene. 2017;36(23):3223–31. https://doi.org/10.1038/onc.2016.480.
Gast CE, Silk AD, Zarour L, Riegler L, Burkhart JG, Gustafson KT, et al. Cell fusion potentiates tumor heterogeneity and reveals circulating hybrid cells that correlate with stage and survival. Sci Adv. 2018;4(9):eaat7828. https://doi.org/10.1126/sciadv.aat7828.
Mu Z, Benali-Furet N, Uzan G, Znaty A, Ye Z, Paolillo C, et al. Detection and characterization of circulating tumor associated cells in metastatic breast cancer. Int J Mol Sci. 2016;17(10) https://doi.org/10.3390/ijms17101665.
Heitzer E, Auer M, Gasch C, Pichler M, Ulz P, Hoffmann EM, et al. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res. 2013;73(10):2965–75. https://doi.org/10.1158/0008-5472.CAN-12-4140.
Jeselsohn R, Yelensky R, Buchwalter G, Frampton G, Meric-Bernstam F, Gonzalez-Angulo AM, et al. Emergence of constitutively active estrogen receptor-alpha mutations in pretreated advanced estrogen receptor-positive breast cancer. Clin Cancer Res. 2014;20(7):1757–67. https://doi.org/10.1158/1078-0432.CCR-13-2332.
Toy W, Shen Y, Won H, Green B, Sakr RA, Will M, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013;45(12):1439–45. https://doi.org/10.1038/ng.2822.
Beije N, Sieuwerts AM, Kraan J, Van NM, Onstenk W, Vitale SR, et al. Estrogen receptor mutations and splice variants determined in liquid biopsies from metastatic breast cancer patients. Mol Oncol. 2018;12(1):48–57. https://doi.org/10.1002/1878-0261.12147.
Paolillo C, Mu Z, Rossi G, Schiewer MJ, Nguyen T, Austin L, et al. Detection of activating estrogen receptor gene (ESR1) mutations in single circulating tumor cells. Clin Cancer Res. 2017;23(20):6086–93. https://doi.org/10.1158/1078-0432.CCR-17-1173.
Magbanua MJM, Rugo HS, Wolf DM, Hauranieh L, Roy R, Pendyala P, et al. Expanded genomic profiling of circulating tumor cells in metastatic breast cancer patients to assess biomarker status and biology over time (CALGB 40502 and CALGB 40503, Alliance). Clin Cancer Res. 2018;24(6):1486–99. https://doi.org/10.1158/1078-0432.CCR-17-2312.
De Luca F, Rotunno G, Salvianti F, Galardi F, Pestrin M, Gabellini S, et al. Mutational analysis of single circulating tumor cells by next generation sequencing in metastatic breast cancer. Oncotarget. 2016;7(18):26107–19. https://doi.org/10.18632/oncotarget.8431.
Paoletti C, Cani AK, Larios JM, Hovelson DH, Aung K, Darga EP, et al. Comprehensive mutation and copy number profiling in archived circulating breast cancer tumor cells documents heterogeneous resistance mechanisms. Cancer Res. 2018;78(4):1110–22. https://doi.org/10.1158/0008-5472.CAN-17-2686.
Shaw JA, Guttery DS, Hills A, Fernandez-Garcia D, Page K, Rosales BM, et al. Mutation analysis of cell-free DNA and single circulating tumor cells in metastatic breast cancer patients with high circulating tumor cell counts. Clin Cancer Res. 2017;23(1):88–96. https://doi.org/10.1158/1078-0432.CCR-16-0825.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Vismara, M., Reduzzi, C., Daidone, M.G., Cappelletti, V. (2020). Circulating Tumor Cells (CTCs) Heterogeneity in Metastatic Breast Cancer: Different Approaches for Different Needs. In: Piñeiro, R. (eds) Circulating Tumor Cells in Breast Cancer Metastatic Disease. Advances in Experimental Medicine and Biology, vol 1220. Springer, Cham. https://doi.org/10.1007/978-3-030-35805-1_6
Download citation
DOI: https://doi.org/10.1007/978-3-030-35805-1_6
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-35804-4
Online ISBN: 978-3-030-35805-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)