Abstract
Automated instrument localization during cardiac interventions is essential to accurately and efficiently interpret a 3D ultrasound (US) image. In this paper, we propose a method to automatically localize the cardiac intervention instrument (RF-ablation catheter or guidewire) in a 3D US volume. We propose a Pyramid-UNet, which exploits the multi-scale information for better segmentation performance. Furthermore, a hybrid loss function is introduced, which consists of contextual loss and class-balanced focal loss, to enhance the performance of the network in cardiac US images. We have collected a challenging ex-vivo dataset to validate our method, which achieves a Dice score of 69.6% being 18.8% higher than the state-of-the-art methods. Moreover, with the pre-trained model on the ex-vivo dataset, our method can be easily adapted to the in-vivo dataset with several iterations and then achieves a Dice score of 65.8% for a different instrument. With segmentation, instruments can be localized with an average error less than 3 voxels in both datasets. To the best of our knowledge, this is the first work to validate the image-based method on in-vivo cardiac datasets.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Cardiac intervention therapies, such as cardiac electrophysiology (EP) and transcatheter aortic valve implantation (TAVI), have been broadly applied to achieve lower risk and shorter recovery time for patients. To guide the instruments inside the heart during intervention, fluoroscopy imaging is typically considered using a contrast agent to visualize the vessel and tissue. However, radiation dose, harmful agents, invisible soft tissue and lack of 3D spatial information in X-ray imaging complicate the interpretation of the instrument during the interventions. To address this, 3D ultrasound (US) is considered as an alternative solution for intervention guidance, which has a richer spatial information and no radiation exposure. Nevertheless, the low-resolution and low-contrast imaging of 3D US lead to difficulty for a sonographer to timely localize the instrument during the surgery. Therefore, automatic instrument segmentation and localization methods are highly demanded for clinical practice. As a promising approach, 3D US image-based instrument localization has been studied in recent years [1, 5, 6, 8, 9]. Conventional machine learning approaches with handcrafted features were applied to localize the catheter in a phantom heart or an ex-vivo dataset [5, 9]. However, the limited discriminating capacity of handcrafted features cannot always handle the complex anatomical structures in 3D US images. More recently, deep learning methods, such as convolutional neural networks (CNNs), have achieved a significant performance improvement in medical applications. For instruments detection or localization in 3D US using deep learning, two main approaches have been studied: voxel-based classification by a CNN [6, 8] and slice-based semantic segmentation [6]. Although they achieved better results than the approaches with handcrafted features, these deep learning methods still have limitations. Particularly for the slice-based semantic segmentation method, the authors [6] employ a 2D convolution method on the decomposed 2D slices. However, the 3D contextual information in 3D US cannot be fully exploited.
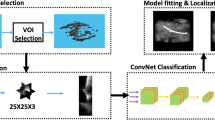
To better exploit 3D contextual information, we propose as the first contribution a 3D CNN for instrument localization in 3D US, which is shown in Fig. 1. More specifically, we propose a compact UNet with pyramid structure (Pyramid-UNet), which is able to keep both high-level and low-level features simultaneously at different image scales, while reducing the complexity of the standard UNet [2, 10]. From our experiments, our proposed Pyramid-UNet improves the segmentation performances when compared to a standard UNet structure. Moreover, as the second contribution, we design a hybrid loss function, which consists of contextual loss and class-balanced focal loss, to learn a better discriminating representation. The contextual loss controls the CNN towards high-level contextual information encoding for the prediction domain. The class-balanced focal loss enables the network to balance and focus more on challenging voxels of difficult structures. To validate our method, we first performed an experiment on the collected ex-vivo dataset for RF-ablation catheter localization (for EP operation), which successfully segmented the instrument with Dice score 69.6%. Furthermore, we conducted an experiment on an in-vivo dataset for guidewire localization (for TAVI operation). With limited images of the in-vivo dataset, we performed fine-tuning on this dataset by using the pre-trained model from the ex-vivo dataset, which achieved Dice score of 65.8%. Based on the successful segmentation result, the instrument’s tip can be localized with an average error less than 3 voxels on both datasets. To the best of our knowledge and as the third contribution, this paper is the first one to validate the image-based cardiac instrument localization in an in-vivo dataset.
2 Methods
The block diagram of our proposed method is shown in Fig. 1, which is based on three stages: (1) the input 3D image is decomposed into smaller patches; (2) each patch is segmented by our proposed network and the output patches are combined back; (3) the instrument axis and its tip are extracted after the segmentation, which can then be visualized for clinical experts.
2.1 Pyramid-UNet
We adopt the popular segmentation net 3D UNet [2, 10] as our backbone architecture, but we introduce the following modifications for our application, as shown in Fig. 1(b). Because of the limited amount of images in the dataset, we experimentally reduce the number of multi-scale levels of UNet and convolutional channels at each level, which leads to less trainable parameters and avoids over-fitting. When compared to a standard UNet (19.4M parameters), our re-designed compact UNet is more compact and efficient (4.6M parameters). Typically as the network goes deeper, the discriminating information at low level can vanish or be omitted. Although UNet [2] employs skipping connections to preserve low-level information, it still cannot fully preserve the information at different levels. To address this, we design a Pyramid-UNet, which is shown in Fig. 1(b). We consider the multi-scale inputs at different UNet levels to preserve more low-level information within the encoding stage. The proposed image pyramid scaling at the input is attractive, since it potentially compensates the information loss during the feature pyramid of UNet [4]. Furthermore, to better supervise and synchronize the features at different scales, we employ deep supervision at the decoding stage [3], but introduce an extra convolutional block for a better stability. Specifically, we apply the deconvolution operation at each decoding level to generate the prediction with original patch size, which avoids further artifacts in the ground truth and preserves the accuracy. By combining the pyramid inputs and outputs, the proposed network potentially preserves more information at different feature scales than the standard UNet for US images.
2.2 Hybrid Loss Function
To better supervise the Pyramid-UNet and to enforce learning more contextual information rather than a conventional voxel-based loss function, such as cross-entropy and Dice loss, we propose a hybrid loss function. It consists of a contextual loss and a class-balanced focal loss. As shown in Fig. 1(b), the three outputs of the 3D Pyramid-UNet are denoted as \(\hat{Y}_1\), \(\hat{Y}_2\) and \(\hat{Y}_3\). The hybrid loss function is defined as
where Y is the ground truth of the input patch, \(Loss_{FL}\) denotes the class-balanced focal loss and \(Loss_{CL}\) is the contextual loss.
Typically, networks are learned by employing a voxel-wise loss function, such as cross-entropy or Dice loss, which are ignoring the high-level difference between prediction and ground truth. To enforce the network to learn a better contextual representation, we introduce a novel contextual loss, which formulates the contextual difference in a latent space. The prediction and ground truth are encoded by a contextual encoder, which is depicted in Fig. 1(c), to generate a high-level representation in latent space, denoted as \(S_{{\hat{Y}}}\) and \(S_{Y}\), respectively. As a consequence, the contextual loss \(Loss_{CL}\) is characterized by
where \(||\cdot ||_2\) is the norm-2 distance and \(CE(\cdot )\) is the context encoder in Fig. 1(c).
The loss function, such as Dice or cross-entropy, is typically applied for segmentation tasks in medical imaging. However, it is not optimized when segmented objects have large size variations and imbalanced class distribution in the ground truth [7]. Moreover, when the instrument has a small size in 3D space and hard/challenging classified boundary voxels are more important than easy classified voxels at the center part of the instrument, the commonly used loss functions might not be optimized. Therefore, to focus more on challenging voxels and concerning the imbalanced classes of the previous focal loss [7], we adopt them into the class-balanced hybrid focal loss function, which is defined as
where \(y_{ci}\) denotes an instrument voxel from the ground truth, \(\hat{y}_{ci}\) represents the voxel’s prediction probability for the instrument class, while \(y_{ni}\) and \(\hat{y}_{ni}\) are a non-instrument voxel and its corresponding prediction probability, respectively. Parameters \(\beta \) and \(\omega \) are controlling the weight between different classes, which are calculated as the square root of the inverse of the classes ratio. Parameters \(\gamma \) and \(\sigma \) are controlling the slope of the loss curve, which are empirically selected as \(\gamma =0.3\) and \(\sigma =2\), respectively. Parameter \(\eta \) is the weight between two different focal losses, which is empirically chosen as \(\eta =0.8\).
2.3 Training Stage: Dense Sampling
The common training strategy for patch-wise segmentation is based on a random patch cropping from the full volumes [10]. However, this approach fails to train the network for instrument segmentation in 3D US, since the instrument occupies relatively small space in the volume and random cropping leads to an extremely imbalanced information distribution. To address this, we propose a dense sampling approach on catheter voxels: for each instrument voxel in the training volume, a 3D patch with size \(48^3\) voxels is generated that surrounds the voxel being at the center. As a result, the training patches are focusing on a sub-space surrounding the instruments rather than sampling irrelevant information. The network is trained by minimizing the joint loss function of Eq. (1) using the Adam optimizer with initial learning rate equal to 0.001. Empirically, we empirically select loss weights in Eq. (1) as \(\alpha _1=1\), \(\alpha _2=0.6\) and \(\alpha _3=0.4\), respectively. The learning is terminated after convergence. To generalize the network, data augmentations are applied on-the-fly, like random mirroring, flipping, contrast transformation, etc. The dropout rate is 0.5 during the training.
2.4 Instrument Localization
The full volume of 3D US is decomposed into patches to generate the segmentation results, which are combined back into a volume as the segmentation output, as shown in Fig. 1(a). A typical instrument localization method is using a pre-defined model to fit the instrument in 3D space [9], which could be complex and time-consuming. In our method, with the high segmentation performance of the proposed network, we directly extract the largest connected group as the instrument after the segmentation. As a result, our method avoids a complex post-processing stage. With the selected group, the instrument axis is extracted and the instrument’s tip is localized as the point closest to the image center.
(a) Our ex-vivo dataset collection setup with RF-ablation catheter; (b) Porcine heart placed in the water tank, the US probe is placed under the heart while the catheter is going through the vein; (c) (d) Example slices of ex-vivo image with RF-ablation catheter; (e) (f) Example slices of in-vivo image with guidewire.
3 Experiments
3.1 Experiment on ex-vivo dataset
Materials: We have first validated our method on an ex-vivo dataset, examples of data collection setup and corresponding US images are shown in Fig. 2. The ex-vivo dataset consists of 92 3D cardiac US images from porcine hearts. During the recording, the hearts were placed in water tanks with an RF-ablation catheter for EP (diameter range from 2.3 mm to 3.3 mm) inside the left ventricle or right atrium. The US probes were placed next to the heart to capture the images containing the catheter. The dataset includes the volumes of size range \(120\times 69\times 92\) to \(294\times 283\times 202\) voxels, in which the voxel size was isotropically resampled to the range of 0.4–0.7 mm. The datasets were manually annotated by clinical experts to generate the binary segmentation mask as the ground truth. The ex-vivo dataset was randomly divided into 62/30 volumes for training/testing. The evaluation metrics are Dice Score (DSC) and Hausdorff Distance (HD).
Segmentation in ex-vivo: We have extensively compared our method with state-of-the-art medical instrument segmentation approaches on the ex-vivo data-set, including handcrafted feature methods using Gabor features (GF-SVM) [5], Multi-scale and multi-definition features (MF-AdaB) [9], LateCNN for voxel-based catheter classification (LateCNN) [8], and ShareFCN using a cross-section approach to decompose 3D information for needle segmentation (ShareFCN) [6]. Moreover, we also compared a standard 3D UNet for 3D US in another task (3D-UNet) [10]. The results are compared with our method and shown in Table 1. Ablation studies are also performed to validate our proposed compact UNet with standard Dice loss (Compact-UNet), Pyramid-UNet with standard Dice loss and our Pyramid-UNet with hybrid loss (denoted as Proposed in the Table). From the results in Table 1, the Compact-UNet has better performance than 3D-UNet because of using less parameters and avoiding of over-fitting. Moreover, it also has better performance than other medical instrument segmentation approaches. Our proposed Pyramid-UNet with hybrid loss is able to further boost the performance by exploiting more semantic information.
(a) Learning curves for testing patches under two different scenarios at first 2k iterations in ex-vivo, with corresponding Dice score on testing volumes. (b) Boxplots of instrument tip error in different segmentation methods. (c) 3D volume with ground truth (green), segmentation (red), and enlarged visualization. (d) 2D slices of 3D volume, which is tuned to have the best view. (Color figure online)
3.2 Experiment on in-vivo dataset
Materials: The collected in-vivo dataset includes 18 volumes from TAVI operations. During the recording, the sonographer recorded images from different locations of the chamber without any influence on the procedure. The volumes were recorded with a mean volume size of \(201\times 202\times 302\), where the volume voxel size was resampled to 0.6 mm. The applied instrument in the in-vivo dataset is a guidewire (0.889 mm). Threefold cross-validation was performed on the in-vivo dataset with fine-tuning, based on the pre-trained ex-vivo model for the RF-ablation catheter. All ethical guidelines for human studies were followed.
Segmentation in in-vivo: When comparing our challenging datasets, ex-vivo possess more information of 3D cardiac images because of a larger training dataset, which could be beneficial to the in-vivo dataset using the concept of fine-tuning (from RF-ablation catheter to guidewire). As a consequence, we trained the model on ex-vivo data from scratch and fine-tuned it on the in-vivo data. Example curves of testing Dice score are shown in Fig. 3(a), which are obtained by random samples from testing images. These results come from two different scenarios with respect to training iterations: train from scratch (TFS) and fine-tuning. As we can observe, even the trained model is used for different instrument types, the pre-trained model promises a fast convergence less than 10 iterations. Moreover, it provides a better segmentation performance when compared to training from scratch. The fine-tuned model has 4% higher Dice score with 2,000 iterations than training from scratch with 20,000 iterations. Corresponding segmentation results are shown in Table 1.
3.3 Instrument Localization
With a robust segmentation performance, instruments are directly localized by selecting the largest connected component. The accuracy of instrument localization is evaluated in terms of instrument tip error, defined as the point-plane distance between the tip on the ground truth to the cross-section plane containing the instrument. The statistical results of errors in two different datasets are shown in the boxplots in Fig. 3(b). From the results, our proposed method achieves the best localization error less than 3 voxels.
4 Conclusion
In this paper, we have proposed a novel automatic instrument localization method for US-guided cardiac intervention therapy. In the proposed method, we design a network to provide segmentation of the instruments. With the aid of hybrid loss, the performance of the network achieved a Dice score of 69.6% and 65.8% in challenging ex-vivo and in-vivo datasets, respectively. Based on the proposed networks, the experiments show that our method obtains an instrument localization error that is less than 3 voxels without complex post-processing, which reduces the localization complexity and provides an accurate localization result.
References
Arif, M., Moelker, A., van Walsum, T.: Automatic needle detection and real-time bi-planar needle visualization during 3D ultrasound scanning of the liver. Med. Image Anal. 53, 104–110 (2019)
Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T., Ronneberger, O.: 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 424–432. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_49
Dou, Q., Chen, H., Jin, Y., Yu, L., Qin, J., Heng, P.-A.: 3D deeply supervised network for automatic liver segmentation from CT volumes. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 149–157. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_18
Lin, T.Y., Dollár, P., Girshick, R., He, K., Hariharan, B., Belongie, S.: Feature pyramid networks for object detection. In: IEEE CVPR, pp. 2117–2125 (2017)
Pourtaherian, A., et al.: Medical instrument detection in 3-dimensional ultrasound data volumes. IEEE Trans. Med. Imaging 36(8), 1664–1675 (2017)
Pourtaherian, A., Zanjani, F.G., Zinger, S., Mihajlovic, N., Ng, G.C., Korsten, H.H., et al.: Robust and semantic needle detection in 3D ultrasound using orthogonal-plane convolutional neural networks. IJCARS 13(9), 1321–1333 (2018)
Wong, K.C.L., Moradi, M., Tang, H., Syeda-Mahmood, T.: 3D segmentation with exponential logarithmic loss for highly unbalanced object sizes. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11072, pp. 612–619. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00931-1_70
Yang, H., Shan, C., Kolen, A.F., de With, P.H.: Catheter detection in 3D ultrasound using triplanar-based convolutional neural networks. In: IEEE ICIP, pp. 371–375. IEEE (2018)
Yang, H., Shan, C., Pourtaherian, A., Kolen, A.F., et al.: Catheter segmentation in three-dimensional ultrasound images by feature fusion and model fitting. J. Med. Imaging 6(1), 015001 (2019)
Yang, X., et al.: Towards automatic semantic segmentation in volumetric ultrasound. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10433, pp. 711–719. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66182-7_81
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Yang, H., Shan, C., Tan, T., Kolen, A.F., de With, P.H.N. (2019). Transferring from ex-vivo to in-vivo: Instrument Localization in 3D Cardiac Ultrasound Using Pyramid-UNet with Hybrid Loss. In: Shen, D., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2019. MICCAI 2019. Lecture Notes in Computer Science(), vol 11768. Springer, Cham. https://doi.org/10.1007/978-3-030-32254-0_30
Download citation
DOI: https://doi.org/10.1007/978-3-030-32254-0_30
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-32253-3
Online ISBN: 978-3-030-32254-0
eBook Packages: Computer ScienceComputer Science (R0)