Abstract
The early diagnosis of colorectal cancer (CRC) traditionally leverages upon the microscopic examination of histological slides by experienced pathologists, which is very time-consuming and rises many issues about the reliability of the results. In this paper we propose using Convolutional Neural Networks (CNNs), a class of deep networks that are successfully used in many contexts of pattern recognition, to automatically distinguish the cancerous tissues from either healthy or benign lesions. For this purpose, we designed and compared different CNN-based classification frameworks, involving either training CNNs from scratch on three classes of colorectal images, or transfer learning from a different classification problem. While a CNN trained from scratch obtained very good (about 90%) classification accuracy in our tests, the same CNN model pre-trained on the ImageNet dataset obtained even better accuracy (around 96%) on the same testing samples, requiring much lesser computational resources.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Colorectal cancer
- Histological image analysis
- Convolutional Neural Networks
- Deep learning
- Transfer learning
- Pattern recognition
1 Introduction
Colorectal carcinoma (CRC) is one of the most diffused cancers worldwide and one of the leading causes of cancer-related death. Based on most recent epidemiological studies, this type of cancer is particularly frequent in the highly-developed countries, especially Europe, and it is associated with very high mortality rates compared to other tumors [9]. Hence, the early diagnosis and differentiation of CRC is crucial for the survival and well-being of a large number of patients.
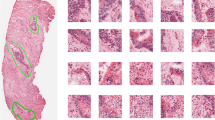
The primary diagnosis of CRC is traditionally performed by means of colonoscopy, that is the endoscopic examination of the large and the distal part of the small bowel. During this procedure, the surgeon will typically perform a biopsy on the suspicious colorectal lesions, which implies the resection of a thin sample of tissue for histopathological evaluation (see Fig. 1).
The samples, fixed and stained by means of Hematoxylin and Eosin (H&E), are then visually examined by a pathologist, either directly under the microscope, or on a computer monitor. In the latter case, the physical slide is first digitalised by a scanner in the form of a so-called virtual slide or whole-slide-image (WSI), a very large multi-resolution zoomable image file, and then visualised on a screen by means of specific viewing software.
The presence and level of malignancy is assessed by observing the organisational changes in the tissues, which are highlighted by the two stains. As shown in Fig. 2, normal colon tissues have a well-defined organisation, with the epithelial cells forming glandular structures and the non-epithelial cells (i.e. stroma) lying in between these glands. The main benign precursor of CRC, adenoma, is characterised by enlarged, hyper-chromatic and elongated nuclei arranged in a typically stratified configuration, characterised by either tubular or villous (finger-like) tissue architecture. Adenocarcinomas, on the other hand, produce abnormal glands that infiltrate into the surrounding tissues.
Traditional visual examination has two major drawbacks, that are widely pointed out by literature. First, it is time-consuming, especially for large image datasets. Second, it is highly subjective, which translates into large variability, both inter and intra observer [2]. To solve these drawbacks, there are growing efforts towards the automatisation of the analysis flow and the development of computer-aided diagnostic techniques. The major directions of the research efforts were mainly two in the last few years: (i) automated segmentation, aimed at partitioning the heterogeneous colorectal samples into regions of interest that are homogeneous in terms of tissue architecture. (ii) automated classification, aimed at partitioning the homogeneous tissue regions into a number of histological categories, either normal or malignant, leveraging quantitative features extracted from the image. In both the tasks, the large intra-class image variability is the main challenge to be tackled. This work focuses on the automated classification task, targeting the three histological categories that are most relevant for CRC diagnosis: (i) healthy tissue, (ii) adenocarcinoma, (iii) tubulovillous adenoma (see Fig. 2).
Recent literature on automated classification of histological images has been extensive not only on colon but also on brain, breast, prostate and lungs cancer applications. These works generally propose solutions based on automated texture analysis, where a limited set of local descriptors (for example, statistical features based on grey level co-occurrence matrix, GLCM, local binary patterns, LBP, Gabor and wavelet transforms, etc.) are computed from the input images and then fed into a classifier. The texture descriptors, eventually encoded into a compact dictionary of visual words, are used as input of machine learning techniques such as Support Vector Machines (SVM), Random Forests or Logistic Regression classifiers [4]. In spite of the good level of accuracy obtained by some of these works, the dependence on a fixed set of handcrafted features is a major limitation to the robustness of the classical texture analysis approaches. First, because it leverages upon a priori knowledge about the image characteristics that are best suited for classification, which is not obvious for all types of cancers. Second, because it puts severe constraints to the generalisation and transfer capabilities of the proposed classifiers, especially in the presence of inter-dataset variability.
As an answer to such limitations, deep learning (DL) architectures, and more specifically Convolutional Neural Networks (CNNs), have now become a major trend [6, 7]. In CNNs a number of convolutional and pooling layers learn by backpropagation a set of features that are best for classification, thus avoiding the extraction of handcrafted texture descriptors. Nonetheless, the need of computational resources and the necessity of extensively training the networks with a huge number of independent samples are open issues in histopathology, and put limits to the usability of this approach in the everyday clinical setting. Transfer learning techniques (i.e applying CNNs pre-trained on a different type of images, for which large datasets are available) might be a promising solution to this problem [13], which deserves better investigations.
In this work, we apply a CNN-based approach to automatically differentiate healthy tissues and tubulovillous adenomas from cancerous samples, which is a challenging task in histological image analysis. For this purpose, we fully train a CNN on a large set of colorectal samples, and assess its accuracy on a completely independent test set. This technique is experimentally compared with two different transfer learning approaches, both leveraging upon a CNN pre-trained on a completely different classification problem. The first approach uses the pre-trained CNN to extract a set of discriminative features that will be fed into a separate SVM classifier. The second approach fine-tunes on CRC histological images only the last stages of the pre-trained CNN. By doing so, we investigate and discuss the transfer learning capabilities of CNNs in the domain of CRC classification.
This paper revises and extends [10]. In this version we provide a better introduction to histopathological image analysis, ameliorated pictorial representations of the proposed methodologies, as well as a new section on the visualisation and exploitation of the CNN outcome for CRC tissue classification.
2 Materials and Methods
2.1 Image Dataset
The dataset used in this study was extracted from a public repository of H&E stained whole-slide images (WSIs) of colorectal tissues, that can be freely downloaded from [1], together with their anonymised clinical information.
In order to obtain a statistically significant dataset in terms of inter-subjects and inter-class variability, we selected 27 WSIs, obtained from univocal subjects (i.e. one WSI per patient). As a WSI is typically very wide, it may contain different types of tissues (e.g. healthy and cancerous portions). Hence, the original WSI cannot be given a unique histological label. With the supervision of a pathologist, we identified on each WSI large regions of interest (ROIs) that are homogeneous in terms of tissue architecture (see example of Fig. 3). Hence, each ROI can be univocally associated to one out of the three tissue subtypes: (i) adenocarcinoma (AC); (ii) tubuvillous adenoma (TV) and (iii) healthy tissue (H). Then, the obtained ROIs were cropped into a total number of 13500 \(1089 \times 1089\) patches (500 per patient), at a 40x magnification level, without applying any data augmentation.
CRC classification by means of CNN: schematic representation. Homogeneous ROIs are cropped into small non-overlapping patches, that are fed into the CNN after normalisation by mean and standard deviation on the training set. The output of the CNN is a probability map of the input patch into one three tissue classes: healthy, adenocarcinoma or tubulovillous adenoma.
The original image cohort was randomly split into two disjoint subsets, one for training and one for testing purposes, respectively containing 9000 and 4500 patches. In order to ensure a complete independence of the two sets, the training and testing patches belong to different subjects. More specifically, 18 patients were used to generate the training patches, and 9 for the testing patches.
The random sampling was stratified, so that both the training and the testing set are balanced among the three classes of interest (H, AC and TV). Hence, the accuracy assessment was not affected by class prevalence.
Before being fed into the CNN, each patch was down sampled by a factor five, that was empirically set as a trade-off between computational burden of the processing and architectural detail of the images. To compensate for possible color inconsistencies, all the patches were normalised by mean and standard deviation, computed over the whole training dataset. A pictorial representation of the patches preparation and classification process is shown in Fig. 4.
2.2 Convolutional Neural Network: Architecture and Training Paradigm
Convolutional Neural Networks (CNNs) consist of a sequence of multiple locally connected trainable stages, aimed at learning the image representation at a progressively increasing level of abstraction, and of two or more fully-connected layers as the last step, aimed at learning the class partitioning task like a traditional multi-layer perceptron.
The locally connected part of a CNN implements two main types of building blocks:
-
1.
Convolutional (CONV) blocks basically act like kernel filters with trainable parameters, performing a 2D convolution operation on the input image. Based on the value of these parameters, a filter is able to detect different types of local patterns. After convolution, typically the stage applies a non-linear transfer function, such as Rectified Linear Unit (ReLU).
-
2.
Pooling (POOL) blocks perform a non-linear down-sampling of the input, typically applying a max function. Down-sampling has the two-fold effect of reducing the number of parameters of the network that need to be learned (and hence of controlling overfitting), as well as of introducing space invariance into the image representation.
The higher the number of CONV and POOL layers in a CNN, the higher the depth of the network, and the higher the level of detail that can be achieved by the hierarchical representation of the image. Hence, deeper networks are usually able to achieve much better classification performance that their shallow counterparts. Nonetheless, a higher depth also translates into a higher number of parameters that need to be learned, and hence into a much higher computational cost of the training process.
The training paradigm of the CNN is usually a classic backpropagation scheme: that is, an iterative process involving multiple passes of the whole input dataset until convergence of the optimisation algorithm. At each training step, the whole dataset flows from the first to the last layer in order to compute a classification error, quantified by a loss function. Such error flows backward through the net, and at each training step the model parameters (i.e. the weights of the network) are tuned in the direction that minimises the classification error on the training data.
In our work we used a VGG16 CNN model [12], that ensured the best compromise between representation capabilities (and hence, depth) and computational costs of the training. The model is schematically reported in Fig. 5.
VGG16 model was successfully applied to a large number of computer vision tasks. In spite of its large depth (16 layers, including convolutional and fully-connected stages), its architecture is very simple and repetitive. More specifically, the model consists of a linear sequence of 13 \(3 \times 3\) CONV layers, that can be conceptually grouped into 5 macro-blocks, each ending with a \(2 \times 2\) POOL, and of 3 fully-connected (FC) layers as final classification stage. All the non-linearities are ReLU, except for the last fully-connected layer (FC3), that is a softmax activation function. The convolution stride and the padding are fixed to 1 pixel and the max pooling stride to 2. Differently from the original VGG16 model, we modified the architecture by implementing a final FC3 stage of 3 units, matching the number of categories targeted by our research problem. The output values of this final stage can be interpreted as the probability of the input patch belonging respectively to the healthy, the adenocarcinoma or the tubulovillous adenoma class.
The net was developed within Keras framework [3] and trained with a classic backpropagation paradigm. More specifically, we applied a stochastic gradient descent (SGD), implemented with a momentum update approach [11] as iterative optimisation algorithm to minimise the categorical cross-entropy function between the three classes of interest (H, AC and TV).
The monitoring of the training process and the optimisation of the hyper-parameters of the net leverage upon 10% of the training set, that were appointed as independent validation data and excluded from the training per se. This validation set was solely used to compute the validation accuracy metric upon which the training process is optimised. Based on validation, we set the learning rate (LR) to 0.0001, the momentum (M) to 0.9 and the batch size (BS) to 32.
To reduce overfitting, the learning process implemented a early stopping strategy (i.e., the training is stopped when validation accuracy does not improve for 10 subsequent epochs), as well as a progressive reduction of LR each time the validation accuracy does not improve for 5 consecutive epochs [14].
The CNN was trained for 30 epochs on the training set, which lasted 8 h on Linux Infiniband-QDR MIMD Distributed Shared-Memory Cluster provided with single GPU (NVIDIA Tesla K40 - 12 GB - 2880 CUDA cores).
As shown by the trend of loss and accuracy curves on both the training and validation datasets (see Fig. 6(a) and (b), respectively), the model converged quite quickly. Indeed, while training accuracy was still increasing, the value of validation accuracy saturated within 15 epochs. The decay speed of the validation loss curve indicates that the learning rate was appropriate. On top of that, validation and training accuracy were fairly similar. This reasonably rules out overfitting.

Figure from [10].
Training vs validation loss per epoch (a) and training vs validation accuracy per epoch (b).
2.3 Transfer Learning from Pre-trained CNN
CNNs are cascades of trainable filter banks, where the first blocks of filters are devoted to the detection of low-level features (i.e. edges or simple shapes), and the following ones are activated by high-level semantic aggregations of the previous patterns, that are more problem-specific. Hence, while the top-most blocks are generally tailored to a specific classification task, the lower-level features can be ideally generalised to a large number of applications. This concept is at the basis of transfer learning techniques, that leverage CNNs pre-trained on a very large set of examples, with significant variability of image characteristics, to solve a different classification problem.
In our work, we used a pre-trained CNN model with the same architecture and topology of the one used for full training on colorectal cancer images (VGG16, shown in Fig. 5). The model was pre-trained on the ImageNet dataset, from the Large Scale Visual Recognition Challenge 2012 (ILSVRC-2012). This dataset contains 1.2 million photographs depicting 1000 different categories of natural objects. Hence, the training images are completely different from our specific target, in terms of imaging technology, image content as well as of number of categories of the classification problem.
Figure 7 shows two different transfer learning strategies that we implemented and compared:
-
(a)
CNN as a fixed feature generator. The CRC patches are given as input to the pre-trained CNN only for inference. The output of the convolutional blocks are fed into a separate machine learning framework, consisting of a feature reduction stage and a supervised classifier.
-
(b)
Fine-tuning the CNN. The CNN model is re-trained on our training set of histological images, keeping all the parameters of the low-level blocks fixed to their initial value. Hence, only the weights of the top-most layers are fine-tuned for colorectal cancer classification.

Figure from [10].
Transfer learning approaches. (a) Pre-trained CNN as a fixed feature generator. (b) Fine tuning of pre-trained CNN.
As a preliminary step to both the two approaches, we analysed the discriminative capabilities of the features generated by all the major blocks of the pre-trained CNN, as follows. We randomly selected a small subset of the training images (500 per class) and fed them into the pre-trained CNN for inference. The output of each successive macro-block of the CNN was then analysed, to assess the degree of separation of samples belonging to the three different classes. As a trade-off between thoroughness and computational burden of the investigation, we analysed the intermediate output of the CNN only at the end of the pooling layers (i.e. POOL1 to 5, as represented Fig. 8). As POOL layers apply a dimensionality reduction operation, their output are expected to have lower redundancy compared to CONV layers.
The degree of class separation was assessed by means of t-Distributed Stochastic Neighbour Embedding (t-SNE) [8], a non-linear dimensionality reduction algorithm that is used for the visualisation of high-dimensional datasets in a reduced 3-dimensional space. More specifically, t-SNE represents each high-dimensional object (in our case, the feature vector obtained at the output of a POOL layer) by means of a three-dimensional point in a Cartesian space, so that similar feature vectors are represented by nearby points and dissimilar vectors by distant points. The distance between different categories of points in the Cartesian space drawn by t-SNE provides a qualitative measure of class separability in the original feature space. Hence, by repeating the analysis for all the five feature vectors, we were able to establish which of the five POOL blocks ensures the best image representation for our specific classification problem (see the examples at the bottom of Fig. 8). In our experiments, POOL3 outperformed all the other blocks.
Using the pre-trained CNN to generate discriminative features. The output of the five macro-blocks of the CNN are analysed by means of t-SNE, to assess the class separability obtained by each of the five feature vectors in a reduced 3-dimensional feature space. For example, features extracted from POOL3 (bottom-right) ensure much better class separation than features extracted from POOL2 (bottom-left).

Figure from [10].
Sequential forward procedure to select the optimal number of principal components for PCA.
Pre-trained CNN as a Fixed Feature Generator
As a first transfer learning methodology, the output of the most discriminative POOL layer of the pre-trained CNN (POOL3, in our case) was used to generate a feature vector for colorectal tissue classification. The feature vector was fed into the machine learning framework represented by Fig. 7(a), that consists of a feature reduction followed by a classification step.
-
(a)
Feature Reduction. To reduce further the dimensionality and redundancy of the data and prevent overfitting we applied Principal Component Analysis (PCA). PCA applies an orthogonal transformation of the original features into a reduced number of so-called principal components, that are linear combinations of the original characteristics. As PCA works towards the minimisation of the correlation between the features, the new descriptors are expected to be the most representative for the classes of interest. In our work, we empirically set the optimal number of principal component by implementing a sequential forward procedure. More specifically, we computed the mean classification accuracy on the training set at increasing number of principal components (step of 50) and selected the minimum number of principal components after which the classification accuracy had started decreasing. As shown in the graph of Fig. 9, this value was found to be 250).
-
(b)
Classification. The final classification into the three categories of interest (H, AC, TV) was performed by a Support Vector Machine (SVM) with Gaussian radial basis function kernel. The hyper-parameters of the kernel were set by means of a Bayesian Optimisation (BO) algorithm [5], implementing a 10-fold cross-validation on the training images. In our preliminary experiments, BO was found to provide much better and faster results compared to classic methods based on grid search or heuristic techniques.
Fine-Tuning of Pre-trained CNN
As a second transfer learning methodology, we adapted the pre-trained VGG16 net to our specific classification task, using it as a standalone feature extractor and classifier. For this purpose, we first initialised all the weights of the network to the values learned on the ImageNet dataset, as represented in Fig. 7(b). Then, we started a backpropagation algorithm on our CRC dataset, keeping the weights of the first blocks of the net frozen. More specifically, we froze all the weights up-to the most discriminative pooling layer (POOL3), as determined by t-SNE. The rationale of this strategy is trying to maintain the low-level features, that are expected to describe the most generic and generalisable details (e.g. edges and simple shapes), as they were learned from the ImageNet. Hence, all the computational efforts can be focused on the top-most layers, which are expected to learn high-level task-specific features for colorectal image classification.
3 Results and Discussion
The classification accuracy was assessed on the colorectal dataset described in Sect. 2.1. As already pointed out, the test dataset is completely independent from the one used for training the network and optimising the classification parameters and it is balanced among the three categories of interest. The accuracy of the system was assessed at two different levels of abstraction (per patch and per patient, respectively). For this purpose, in [10] we introduced the following performance metrics.
-
(a)
Patch score: (\(S_{P}\)), defined as the fraction of patches of the test set that were correctly classified:
$$S_{P} = \frac{N_C}{N},$$where \(N_C\) is the number of patches correctly classified and N the total number of patches in the test set.
-
(b)
Patient score: (\(S_{Pt}\)), defined as the fraction of patches of a single patient that were correctly classified (i.e. per-patient patch score), averaged over all the patients in the test set:
$$S_{Pt} = \frac{\sum _i S_P(i)}{N_P},$$where \(S_P(i)\) is the patch score of the \(i-th\) patient and \(N_P\) the total number of patients in the test set.
The patch and patient scores obtained for the three classification frameworks described in Sect. 2 are reported in Table 1. More specifically, in the first row (full-train-CNN) we report the values of the CNN fully trained on CRC samples. In the second row (CNN+SVM) we refer to the SVM, with pre-trained CNN used as fixed feature generator. Finally, in the last row (fine-tune-CNN) we quantify the accuracy of the pre-trained CNN with fine-tuning of the stages after POOL3. The first column of the table reports the patch score \(S_{P}\), that is a value in [0, 1] range, and the second column the patient score \(S_{Pt}\) as mean ± standard deviation.
From the values in Table 1 we can observe that all the proposed classification frameworks obtained very good accuracy (above 90%), both in terms of patch and patient scores. Hence, our experiments confirm the promising results obtained by CNNs in other contexts. The patch-wise accuracy (\(S_P\)) was very similar to the patient-wise accuracy (\(S_{Pt}\)), with a very small standard deviation of the latter value, suggesting a good robustness of the classification frameworks. Hence, the CNN-based classifiers cope well with inter-patient variability, that is a typical challenge of histopathological image analysis.
The same conclusions hold if we analyse the per-class accuracy values, that are reported in the form of \(3 \times 3\) confusion matrices in Fig. 10. From such results we can easily gather that the performance of the classification frameworks was fairly homogeneous for the three classes of interest (H, AC and TV).

Figure from [10].
Patch-wise confusion matrices for (a) CNN fully trained on CRC samples, (b) SVM with pre-trained CNN as fixed features generator, (c) pre-trained CNN with fine-tuning of the stages after POOL3 block.
The most interesting point arising from our results is that both the transfer learning methodologies overcome the accuracy obtained by the CNN fully trained on colorectal samples by almost 7%. More specifically, the pre-trained CNN with fine-tuning of the blocks following POOL3 obtained the best accuracy values among all the tested methodologies. This is quite surprising, given that the dataset used for the training (i.e. ImageNet) was extremely different from the one used for testing the network.
From this result we can draw the following considerations.

Figure from [10].
Mean accuracy in relation to the first block till back-propagation is continued.
Most probably, even though the full training seemed to converge well and without overfitting on the training images (see the graph in Fig. 6), our CRC training sample is not large enough to burst the generalisation capability of the CNN. The full training methodology works best only with a very large cohort of training examples, especially in the presence of very high inter-class variability. On the other hand, obtaining much larger training dataset is not always viable, especially in a clinical context.
In spite of the fact that the pre-training was performed on a completely different dataset (i.e. the ImageNet, which contains photographs of every-day objects and natural scenes, and not histological samples), the low-level features learned by the first stages of a CNN can be successfully generalised to the context of CRC image classification. Hence, as a matter of fact the CNNs are capable of extracting useful semantic knowledge from totally different domains. This partially avoids the computational problems and overfitting risks associated with full-training. Indeed, fine-tuning a pre-trained CNN took only two hours against the eight taken by full-training on the same hardware.
To investigate further on these findings, we run few additional experiments on fine-tuned CNNs by changing the starting block for the backpropagation algorithm. The results of this experiment are reported in Fig. 11, where we show the patch score obtained on the test set at different configurations of the fine-tuning. More specifically, POOL-i in the x-axis means that only the weights after the i-th POOL block were learned on the CRC training set, while all the rest of the parameters were frozen to the values learned on the ImageNet. Likewise, FC means that only the fully-connected stage of the network was re-trained.
The trend of the patch score values suggests that the maximum accuracy is reached when the CNN is fine-tuned after POOL3, which confirms the qualitative results of t-SNE. On top of that, we can observe that fully-training the network obtains more or less the same results than training only the last fully-connected stage. This further confirms that colorectal tissue classification can be performed successfully using CNNs, transferring features that were learned from the ImageNet.
4 Exploitation and Visualisation
The outcome of a CNN can be exploited in many ways in histopathology, that go beyond simply labeling a colorectal image patch into a certain class or another. In this section, we show how the analysis and visualisation of the network response during the inference process can benefit the histological assessment of colorectal images.
More specifically, we describe and exemplify two different types of visualisations, namely attention heat-maps and activation maps. Both the maps were built on top of the CNN that provided the best classification results in our experiments (i.e. a VGG16 model pre-trained on ImageNet and fine-tuned on colorectal samples).
As the name attention heat-map suggests, this map serves the specific purpose of driving the observer’s attention towards the areas of the slide that mostly deserve it, i.e. the potentially cancerous ones. As we discussed is Sect. 2, the output layer of our CNN implements a three-dimensional softmax. Hence, when a histological image is given as input to the CNN, the value returned by each of the three output units can be interpreted as the probability of this image being a healthy tissue, adenocarcinoma, or tubulovillous adenoma, respectively. Taking this into consideration, we implemented a visualisation framework that exploits the value of the output unit associated to the adenocarcinoma class to generate the attention heat-map for CRC assessment, as follows. First, the input WSI is cropped into patches, and given as input to the CNN for inference. Then, by aggregating the CNN outcome of each patch, a heat-map of the same size of the input WSI is built, where the saturation of the red color in each pixel is proportional to the likelihood of cancer in that pixel, as predicted by the CNN. Hence, the most intensely red areas are the ones that should catch the pathologist’s attention during the slide assessment. In Fig. 12 we show an example of such heat-map, where the input WSI is the same that was shown in Fig. 3.
Besides the output stage of the CNN, even the hidden layers can be a very useful source of information. As discussed in Sect. 2, the hidden layers of the CNN are filter banks that are triggered by local patterns in the image. Hence, visually representing the response of such filters into so-called activation maps can provide interesting insights into the architectural characteristics of a tissue that are mostly responsible for the image being classified into one histological class or the other. For example, in Fig. 13 we show the activation maps of the POOL3 layer (i.e. the most class-discriminative of our CNN model, as demonstrated in Sect. 2), for input patches belonging respectively to adenocarcinoma (a) and healthy (b) class. More specifically, the color of each pixel in the activation maps is proportional to the level of activation of the POOL3 layer during the inference, with blue color associated to the lowest activation and red to the highest.
As we can easily gather from Fig. 13, POOL3 activation maps capture very well the dysplasia of the colonic glands in the adenocarcinomas (a). Very interestingly, the highest activations are actually localised where the dysplasya is more pronounced. On the other hand, the activation maps of the healthy slides (b) highlight the regularity of normal colonic glands.
5 Conclusions
This work leverages Convolutional Neural Networks, a powerful class of deep learning architectures, to automatically classify colorectal histological slides. Our specific target is the primary diagnosis of colorectal cancer. Hence, we address three main histological classes: healthy tissue, adenocarcinoma or tubulovillous adenoma.
To seek a solution to our problem, we investigated both training a CNN from scratch on a large dataset of pre-annotated images of colorectal samples, as well as transfer learning methodologies leveraging upon CNNs pre-trained on the ImageNet.
According to our experiments, full training obtained satisfactory results (i.e. accuracy in the order of 90%). Nonetheless, this solution was costly both in terms of computational resources as well of number of annotated samples required for the training. Quite surprisingly, transfer learning largely outperformed the full training approach, obtaining classification accuracy above 96% with much lesser training time. This proves that the low-level features learned by the CNNs can be successfully generalised to very different classification problems, such as colorectal image classification, and offers a promising solution to cases with limited availability of training samples per class. In the future, we plan to investigate more thoroughly on this aspect.
As we show in our work, besides classifying an input patch into a certain class or another, the outcome of a CNN can be exploited in many ways in clinics. The analysis and visualisation of the filters’ response at different depth of the network, as well as of the probability map provided by the final softmax layer, provide useful insights into the patterns that most triggered the classification outcome, possibly guiding the pathologists towards local architectural alterations otherwise difficult to spot.
References
Virtual pathology at the university of leeds (2018). http://www.virtualpathology.leeds.ac.uk/
Young, A., Hobbs, R., Kerr, D.: ABC of Colorectal Cancer, 2nd edn. Wiley, Hoboken (2011)
Chollet, F., et al.: Keras (2015). https://github.com/fchollet/keras
Di Cataldo, S., Ficarra, E.: Mining textural knowledge in biological images: applications, methods and trends. Comput. Struct. Biotechnol. J. 15, 56–67 (2017). https://doi.org/10.1016/j.csbj.2016.11.002
Hastie, T., Tibshirani, R., Friedman, J.: Overview of supervised learning. In: Hastie, T., Tibshirani, R., Friedman, J. (eds.) The Elements of Statistical Learning. SSS, pp. 9–41. Springer, New York (2009). https://doi.org/10.1007/978-0-387-84858-7_2
Janowczyk, A., Madabhushi, A.: Deep learning for digital pathology image analysis: a comprehensive tutorial with selected use cases. J. Pathol. Inform. 7(1), 29 (2016). https://doi.org/10.4103/2153-3539.186902
Korbar, B., et al.: Deep learning for classification of colorectal polyps on whole-slide images. J. Pathol. Inform. 8, 30 (2017). https://doi.org/10.4103/jpi.jpi_34_17
Van der Maaten, L., Hinton, G.: Visualizing data using t-SNE. J. Mach. Learn. Res. 9(Nov), 2579–2605 (2008)
Marley, A.R., Nan, H.: Epidemiology of colorectal cancer. Int. J. Mol. Epidemiol. Genet. 7(3), 105–114 (2016)
Ponzio, F., Macii, E., Ficarra, E., Di Cataldo, S.: Colorectal cancer classification using deep convolutional networks - an experimental study. In: Proceedings of the 11th International Joint Conference on Biomedical Engineering Systems and Technologies, BIOIMAGING, vol. 2, pp. 58–66. INSTICC, SciTePress (2018). https://doi.org/10.5220/0006643100580066
Qian, N.: On the momentum term in gradient descent learning algorithms. Neural Netw. 12(1), 145–151 (1999)
Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556 (2014)
Weiss, K., Khoshgoftaar, T.M., Wang, D.: A survey of transfer learning. J. Big Data 3(1), 9 (2016). https://doi.org/10.1186/s40537-016-0043-6
Yao, Y., Rosasco, L., Caponnetto, A.: On early stopping in gradient descent learning. Constr. Approx. 26(2), 289–315 (2007)
Acknowledgements
We thank the anonymous reviewers for their helpful suggestions.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Ponzio, F., Macii, E., Ficarra, E., Di Cataldo, S. (2019). Going Deeper into Colorectal Cancer Histopathology. In: Cliquet Jr., A., et al. Biomedical Engineering Systems and Technologies. BIOSTEC 2018. Communications in Computer and Information Science, vol 1024. Springer, Cham. https://doi.org/10.1007/978-3-030-29196-9_7
Download citation
DOI: https://doi.org/10.1007/978-3-030-29196-9_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-29195-2
Online ISBN: 978-3-030-29196-9
eBook Packages: Computer ScienceComputer Science (R0)