Abstract
The supra- and subgingival plaque biofilm communities of plaque are composed of hundreds of different microbes. These communities are spatially and temporally structured, largely due to cell–cell communications that coordinate synergistic interactions, and intracellular signaling systems to sense changes in the surrounding environment. Homeostasis is maintained through metabolic communication, mutualistic cross-feeding, and cross-respiration. These nutritional symbioses can reciprocally influence the local microenvironments by altering the pH and by detoxifying oxidative compounds. Signal transduction mechanisms include two-component systems, tyrosine phosphorelays, quorum sensing systems, and cyclic nucleotide secondary messengers. Signaling converges on transcriptional programs and can result in synergistic or antagonistic interbacterial interactions that sculpt community development. The sum of all these interactions can be a well-organized polymicrobial community that remains in homeostasis with the host, or a dysbiotic community that provokes pathogenic responses in the host.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
Introduction
The human mouth supports several hundred species of bacteria, viruses, archaea, and fungi that reside in complex, polymicrobial communities on every hard and soft surface. Typically, this oral microbiome exists in homeostasis with the host. The close proximity of oral bacteria in supra- and subgingival plaque allows for constant communication both at an intracellular level as the bacteria are able to sense and respond to their environment, but also at the interspecies level with a wide variety of complex synergistic and antagonistic relationships (Table 1). The environment oral bacteria find themselves in is constantly changing, with local fluctuations in pH, oxygen availability, and temperature gradients, a constant flow of saliva and gingival crevicular fluid, as well as changes in immune effectors and nutrient availability. Indeed, the composition of the oral community dynamically responds to environmental changes, and through the acquisition of new species can mold the environment to its preferences rather than adapting to its surroundings. These microbiomes can remain in mutualistic balance with the host or can become dysbiotic, or destructive to the host, resulting in increased risk of dental caries and gingivitis along with other more severe periodontal diseases (Lamont et al. 2018). Our goal in this chapter is to provide a broad understanding of the strategies employed by oral bacteria to sense and respond to their environment with special emphasis on those signaling systems participating in cell–cell communication that may coordinate the polymicrobial community and promote dysbiosis.
Two-Component Signaling in Oral Bacteria
Oral bacteria located in both the supra- and subgingival environments must be able to respond to changes in local pH, osmotic shifts, nutrient availability, as well as chemical and immunological stresses. Bacteria have widely conserved signal transduction systems that sense and respond to these environmental conditions in order to survive and thrive in the dynamic oral environment. There are several broadly conserved classes of signal transduction systems of various complexities that operate through phosphorelay signaling. The best characterized of the signaling mechanisms in bacteria are the two-component systems (TCSs), which are primarily involved in adaptation to external stimuli, but are often involved in interspecies interactions (Stock et al. 2000).
Archetypically, each TCS is composed of two proteins, a transmembrane stimulus-sensing protein and a cytosolic response regulator (RR), which carries out some cellular change in response to the stimulus (Gao and Stock 2009). The sensor protein (normally a histidine kinase; HK) functions as a periplasmic sensing protein composed of a sensing domain and a cytoplasmic autophosphorylation domain. The N-terminal sensing domain typically comprises a signal receiver flanked by one or more transmembrane domains. While this architecture is generally conserved among HKs, the sensing domain is highly variable, allowing for broad specificity of signals to be detected. The highly conserved C-terminus is composed of the autophosphorylation domain followed by the ATPase-like catalytic domain. Almost all HKs function as homodimers with dimerization occurring within the autophosphorylation domain. Autophosphorylation of HKs was previously thought to only occur in trans, but some recent reports indicate HKs can autophosphorylate in cis (Sankhe et al. 2018). The RR is also a modular protein composed of a receiver domain and an output domain. Recognition of the extracellular signal by the HK stimulates ATP-dependent phosphorylation of a histidine residue followed by subsequent transfer of the phosphate to an aspartate residue in the receiver domain of the RR. Phosphorylation of the RR promotes conformation changes and, in some cases dimerization of the RR, that activate the RR output domain to carry-out various functions (discussed further below). Most of the TCSs are encoded as an operon with the HK, RR, and some accessory genes being co-transcribed. In some cases, termed “orphan” systems, the HK and RR are not associated and the genes encoded the two components are spatially separated.
TCSs of P. gingivalis
The P. gingivalis genome contains relatively few complete TCSs with some strain variability on the number of complete and orphan TCS encoding genes (Table 2). The best characterized TCS in P. gingivalis is FimSR (33277: PGN_0904-0903; W83: PG_1432-1431). The FimSR system regulates the production of the long FimA-component fimbriae that are associated with binding to gingival epithelial cells, host matrix proteins, other bacteria, and with biofilm development. Functional analysis demonstrated that FimR positively regulates the expression of FimA and that a fimS-deficient strain does not express fimA or produce the FimA fimbriae (Nishikawa et al. 2004). Most of the work to characterize the regulation of FimSR has been done in the 33277 strain as the W83 strain contains a mutation in the HK kinase domain, preventing FimS from binding ATP and autophosphorylation. As expected, W83 lacks production of the FimA fimbriae (Nishikawa and Duncan 2010). The regulation of fimA gene expression is an indirect function of FimR as it does not directly bind the fimA promoter. The exact mechanism by which FimR regulates FimA expression remains unclear, with conflicting studies involving the genes immediately upstream of the fimA operon. Deletion of the HK FimS also abolishes FimA fimbrial production, confirming that a fully functional FimSR TCS is required for fimbrial gene transcription (Nishikawa et al. 2004). There is also evidence that FimR can regulate the Mfa1-component minor fimbriae, and that this regulation is mediated by direct binding of FimR to the mfa1 promoter (Wu et al. 2007). The FimSR system appears to be constitutively active during in vitro growth. The FimS sensor domain contains multiple tetratricopeptide (TPR) domains involved in protein-binding interactions, suggesting that FimSR may sense and respond to protein or peptide levels, regulating costly fimbrial production in nutrient-deplete conditions for the asaccharolytic P. gingivalis.
P. gingivalis possesses multiple systems to acquire and transport heme as an essential source of iron (Smalley and Olczak 2017). The HaeSR system (33277: PGN_0752-0753; W83: PG_0719-0720) was identified and named for its regulation of genes involved in heme acquisition by P. gingivalis (Scott et al. 2013). HaeR is a transcriptional regulator, and based on chromatin immunoprecipitation and high-throughput sequencing (ChIP-seq) data, can recognize two promoter architectures allowing it to function as either an activator or a repressor. Utilizing ChIP-seq and electromobility shift assays (EMSA), Scott et al. showed that HaeR can bind to the 5′ UTR directly upstream of genes encoding known iron transport system proteins ihtA (PGN_0704; PG_0668), htrA (PGN_0687; PG_0648), and hmuS (PGN_0556; PG_1553) and, moreover, positively regulates their expression in response to heme (Scott et al. 2013). The same studies also identified a number of ABC transporters and TonB-dependent transporters negatively regulated by HaeR in response to heme concentrations. In addition to heme transport, HaeR is also known to directly control the expression of the Kgp and RgpA gingipains in response to heme-deplete conditions. The gingipains play a role in iron acquisition through the binding and degradation of hemoglobin, and other heme-containing proteins, to release heme. Gingipains work in concert with HmuY to capture the released heme for transport into the cytoplasm (Smalley et al. 2011). Additionally, gingipains are instrumental in the conversion of heme into μ-oxo-bisheme, which P. gingivalis accumulates on the surface, causing the normal black-pigmented appearance of colonies on solid media and providing protection against hydrogen peroxide (Smalley and Olczak 2017).
The PorXY TCSs are orphan proteins due to the fact they are not located in a genetic locus but they have been shown to function as a bona fide TCS. PorY (33277: PGN_2001; W83: PG_0052) is a HK that autophosphorylates and is able to transfer the phosphate to PorX (33277: PGN_1019; W83: PG_0928) (Kadowaki et al. 2016). PorXY have been shown to regulate expression of genes associated with the type IX secretion system (T9SS) of P. gingivalis that is responsible for transporting the gingipains and other important effector proteins across the outer membrane of the cell (Lasica et al. 2017). Mutations of either porX or porY have shown reduced secretion of T9SS proteins, reduced extracellular gingipain activity, and non-pigmentation of colonies (Sato et al. 2010). However, PorX is an atypical RR in that it does not contain a DNA-binding domain but rather is annotated to contain an alkaline phosphatase domain. Indeed, direct examination of PorX function using EMSAs found that PorX did not directly bind to the promoter regions associated with T9SS expression (Vincent et al. 2016). Rather, PorX acts indirectly by binding to the extracytoplasmic sigma factor, SigP, which has been shown to directly interact with the promoter of T9SS-encoding genes (Kadowaki et al. 2016). The exact role PorX plays in regulating SigP activity remains unclear, whether PorX can dephosphorylate SigP or potentially displace its anti-sigma factor to promote activity remains an open question for future study. Another study found that the PorX was able to bind to the T9SS component protein PorL, but again the biological significance of this interaction remains unstudied (Vincent et al. 2016).
P. gingivalis also possess a hybrid TCS, GppX (PGN_1768; PG_1797), in which the sensor and response regulator domains are contained in the same protein (Hasegawa et al. 2003). GppX regulates the post-translational maturation and localization of gingipains (Hasegawa et al. 2003) and also negatively regulates expression of the luxS gene encoding quorum sensing molecules (see below) (James et al. 2006). RNA-Seq of a gppX mutant strain revealed that in addition to luxS, GppX controls expression of proteins important for both monospecies and heterotypic biofilm formation (Hirano et al. 2013).
An orphan RR, RprY (33277: PGN_1186; W83: PG_1089), has also been well characterized in P. gingivalis. There is no protein encoded by P. gingivalis with homology to the cognate HK, RprX. Activation of RprY responds to sodium depletion as a RprY-LacZ fusion protein in E. coli was specifically induced in sodium-deplete media, and an rprY-deficient strain of P. gingivalis was unable to grow under sodium depletion (Krishnan and Duncan 2013). EMSA and ChIP-on-chip assays determined the RprY regulon to include oxidative stress response genes such as clpB, dnaK, groES, and alkyl hydroperoxide reductase C (Duran-Pinedo et al. 2007). In the absence of RprX, P. gingivalis activation of RprY may involve more complex regulation of transcriptional regulatory activities. Where most TCS proteins are co-transcribed, RprY is co-transcribed with a protein acetyltransferase (pat; PGN_1185), which has been shown to acetylate RprY in vitro (Li et al. 2018). Acetylation of RprY is reversed by the deacetylase CobB (PGN_0004). Acetylation of RprY was shown to reduce binding to target promoters while phosphorylation increased binding to promoter DNA, suggesting multiple layers of RprY regulation. RprY was determined to be acetylated in vivo; however, the exact role of in vivo acetylation of RprY and what protein(s) are responsible for phosphorylation of RprY remain uncharacterized (Li et al. 2018).
TCS of T. denticola
T. denticola ATCC 35405 encodes 5 complete TCSs along with 9 orphan TCS proteins (4 HK, 4 RR and 1 HK-RR hybrid protein) (Table 2). Only two of the TCSs of T. denticola have been characterized and will be discussed here. The first system is AtcSR (TDE0032-0033) and the second is Hpk2/Rrp2 (TDE1969-1970). Both of these TCSs are expressed throughout growth but are up-regulated during late-log and stationary phases (Frederick et al. 2011). Hpk2/Rrp2 is common in spirochetes and other oral bacteria, whereas AtcSR is unique to T. denticola, suggesting a possible role in niche-specific signaling. AtcS is a typical HK although it lacks a consensus sensing domain. It does, however, possess a conserved autoinducer (AI)-2 transport domain and may sense and respond to the presence of AI-2 (AI-2–dependent signaling will be discussed later in the chapter). AtcS has been shown to transfer phosphate to its cognate RR, AtcR (Frederick et al. 2008). AtcR contains a LytRT domain, the only RR in oral spirochetes to possess this DNA-binding domain, and indeed of the over 81,000 predicted bacterial RRs, only ~4% possess a LytTR domain (according to the prokaryotic two-component system database as of January 2019; http://www.p2cs.org/) (Ortet et al. 2015). A recent analysis in T. denticola identified 25 LytTR-binding sites composed of 3 distinct promoter architectures that regulate a possible 50 genes (Miller et al. 2014). These studies used bioinformatics approaches to identify the LytTR-binding sites in the ATCC 35405 genome and confirmed function by EMSA studies. AtcR was shown to directly bind to promoters upstream of TDE2770 (the first gene in the flagellar assembly operon), TDE1514 (an operon encoding an ABC transport system), TDE2496 (a methyl-accepting chemotaxis protein), and TDE1971 (first gene in the operon encoding Hpk2/Rrp2). Binding to TDE2770 and TDE2496 strongly suggests that AtcSR plays a role in regulating the motility and chemotaxis of T. denticola. Both motility and chemotaxis of T. denticola are required for biofilm development in mono- and dual-species biofilms (Yamada et al. 2005). Additionally, the ability to regulate the expression of another TCS (Hpk2/Rrp2) suggests that AtcSR sits atop a complex signaling cascade with global regulatory potential.
The other characterized treponemal TCS is Hpk2/Rrp2, so named for its homology with a TCS well characterized in the Lyme disease spirochete Borrelia burgdorferi (Sarkar et al. 2010). Hpk2 contains an N-terminal Per-Arnt-Sim (PAS) domain, which is predicted to sense redox potential, oxygen, and light. Preliminary characterization of Hpk2 revealed increased autophosphorylation under anoxic conditions compared to an aerobic environment. Deletion of the PAS domain from Hpk2 abolished the difference in phosphorylation under different environmental conditions, suggesting that the PAS domain of Hpk2 acts as an oxygen sensor (Sarkar et al. 2010). More recent structural and functional studies of Hpk2 reveal that binding of heme to the PAS domain promotes homodimerization of Hpk2 and enhances autophosphorylation (Sarkar et al. 2018). As T. denticola generally resides at the leading edge of the subgingival oral biofilm, it may be exposed to constant changes in oxygen tension, and Hpk2 through the PAS domain may sense and respond to these environmental cues. Rrp2 is a σ-54–dependent transcriptional regulator, named on the basis of homology with the borrelial Rrp2 RR. Yet, the Rrp2 in Borrelia activates the RpoN-RpoS regulatory pathway that controls the tick-to-host cycle, whereas T. denticola lacks both the enzootic lifecycle and any homolog for RpoN (Frederick et al. 2011). So while the regulon of Rrp2 in T. denticola remains unclear, it is sure to regulate genes in response to environmental cues as Hpk2 activity is oxygen dependent.
Serine/Threonine and Tyrosine Phosphorylation Systems in Oral Bacteria
Nearly 40 years ago, the first bacterial serine/threonine (Ser/Thr) phosphorylation systems were identified, and after decades of research, we now know that Ser/Thr phosphosystems play an integral role in bacterial pathogenicity regulating important cellular processes including metabolism, stress responses, and cell division. While our understanding of Ser/Thr phosphorylation in bacteria has developed over many years, only recently have we understood that phosphorylation of tyrosine residues also occurs in bacteria, previously thought to be an eukaryotic-exclusive post-translational modification (PTM). Emerging research into this relatively new class of PTM in bacteria also shows that Tyr phosphorylation can influence protein function and cellular localization to regulate key systems including capsule production, stress responses, motility, secondary metabolism, and virulence (Whitmore and Lamont 2012).
Ser/Thr phosphorylation in bacteria is primarily carried out by Hanks-type kinases, and these kinases are historically referred to as “eukaryotic-like” due to their homology with previously well-defined systems, although there is no evidence to suggest that these genes were horizontally transferred to bacteria from eukaryotic sources (Cozzone 2005). Recent phylostratigraphic analysis suggests that bacterial, archaeal, and eukaryotic Hanks-type Ser/Thr kinases share an evolutionary ancestor and did not arise from convergent evolution or a horizontal transfer event (Stancik et al. 2018). Due to this shared lineage, the structural domains and function of all Hanks-type kinases from all organisms are very similar. This is in contrast to the bacterial tyrosine kinases (BY-kinase), which are structurally and mechanistically distinct from those found in eukaryotic systems.
Ser/Thr/Tyr phosphorylation networks do not have specific DNA-binding elements that directly regulate transcription; they rather can phosphorylate multiple transcription factors to activate or repress gene transcription. In this way, Hanks-type kinases and BY-kinases are distinct from TCS in that they phosphorylate many substrates and often influence diverse cellular processes, while HK are highly restricted to a single RR substrate. An ever expanding number of microbiome studies and detailed molecular work in Bacillus subtilis and Mycobacteria tuberculosis clearly demonstrate that Ser/Thr/Tyr phosphorylation can all occur on the same substrates and often involves phosphorylation of other kinases (Shi et al. 2014; Prisic et al. 2010). There is mounting evidence that both BY-kinases and Hanks-type kinases serve as complex signaling hubs, and their roles in regulating the physiology of oral bacteria are an emerging topic.
Streptococcus mutans, the major etiological agent in dental caries, resides in the supragingival oral biofilm. S. mutans can efficiently metabolize carbohydrates to produce lactic acid, resulting in a pH reduction that causes demineralization of the enamel and dentin. Major virulence determinants of S. mutans, including adherence to hydroxyapatite surfaces and development of biofilms with acidic microenvironments, are all associated with the Hanks-type kinase PknB and a Ser/Thr protein phosphatase PppL. S. mutans contains only a single Ser/Thr protein kinase and phosphatase, which makes it an ideal model organism for studying the pleiotropic effects of Ser/Thr phosphorylation networks (Hussain et al. 2006). PppL is located immediately upstream of PknB, and the two genes are co-expressed. Genetic deletion of PknB attenuated biofilm development by reducing biofilm thickness and maturation of nutrient channels compared to the parental strain but without a significant change in long-term growth of S. mutans, suggesting a specific role in biofilm development (Hussain et al. 2006). Additional biofilm studies using the pknB-deficient strain as well as strains with deletion of pppL and a pknB/pppL double mutation revealed that the kinase- and phosphatase-deficient strains had comparable reduced biofilm thickness, while the double mutant had an increased reduction in biofilm thickness (Banu et al. 2010). Further study of the S. mutans deletion strains showed reduced resistance to acidic growth conditions and diminished ability to survive oxidative and osmotic stresses. The impact of PknB and PppL on S. mutans was also demonstrated in vivo using a rat dental caries model (Banu et al. 2010). All three mutants showed reduced advanced caries compared to the parental strain. These data suggest that Ser/Thr phosphorylation plays an important role in the pathogenicity of S. mutans and the development of dental caries.
S. mutans competes with other oral streptococci in the oral cavity through the production of bacteriocins, antimicrobial peptides that target closely related bacteria. One particular antagonistic relationship is with S. sanguinis, with which S. mutans primarily competes for nutrients, and which is reciprocally antagonistic to S. mutans through the production of H2O2 (Kreth et al. 2008). Ser/Thr signaling is instrumental to the fitness of S. mutans in this competitive oral ecosystem. In one study, examination of a pknB-deficient strain demonstrated that PknB is involved in the resistance to oxidative stress from H2O2, and this strain was thus more sensitive to competition with S. sanguinis than the wild type (Zhu and Kreth 2010). The exact role of PknB in oxidative stress remains unclear; however, this study confirms the results previously discussed. Transcriptomic analysis of S. mutans suggests that there is signaling cross talk between PknB/PppL and the VicKR and ComDE TCSs (Banu et al. 2010). Deletion of PknB reduced the expression of two bacteriocins and genetic competence genes, which were also down-regulated in a vicK mutant. The overlap of the PknB/PppL regulon and that of VicKR may also include resistance to oxidative stress. While no direct evidence has been shown that PknB can either activate the HK or phosphorylate the RR of either system, the phosphorylation of the RR CovR by the Streptococcus agalactiae Ser/Thr kinase was found to occur within the signal receiver domain and this prevented binding of CovR to DNA (Lin et al. 2009).
P. gingivalis produces a haloacid dehydrogenase family phosphoserine phosphatase, SerB, which is located on the outer membrane of the cell and is secreted upon contact with the gingival epithelium. SerB is transported into epithelial cells and regulates cytoskeletal architecture involving tubulin microtubules and actin microfilaments (Hasegawa et al. 2008; Tribble et al. 2006; Zhang et al. 2005). Most importantly for the oral community as a whole, SerB is able to down-regulate IL-8 secretion from epithelial cells, hence, reducing PMN recruitment (Hasegawa et al. 2008; Takeuchi et al. 2013). The localized dysregulation of IL-8 will induce a transient and localized state of chemokine paralysis and disrupt the local mucosal defense. In this function, P. gingivalis may exert immune regulatory functions that can impact the whole subgingival community, consistent with its role as a keystone pathogen (Darveau et al. 2012; Hajishengallis et al. 2012). While SerB has extensively been characterized, a Ser/Thr kinase has yet to be characterized for this species. There is one gene annotated as a Ser/Thr kinase in the P. gingivalis genome (33277: PGN_0762; W83: PG_0731) due to a conserved PASTA (penicillin binding and Ser/Thr kinase associated) domain; however, this protein is yet to be tested for kinase activity.
P. gingivalis is the only oral species with a characterized Tyr phosphorelay system, consisting of a single BY-kinase (33277: PGN_1524; W83: PG_0436) named Ptk1 and a low-molecular weight protein tyrosine phosphatase named Ltp1 (33277: PGN_0491; PG_1641). The first set of studies sought to identify P. gingivalis genes that are differentially regulated in dual-species communities with S. gordonii and determined that Ltp1 was significantly induced upon incubation with S. gordonii (Simionato et al. 2006). An ltp1 mutant strain showed increased homotypic and heterotypic biofilm formation with S. gordonii, suggesting that it functions to constrain community development (Maeda et al. 2008; Simionato et al. 2006). The regulation of P. gingivalis biofilm development may be twofold (at least) as Ltp1 negatively regulates the expression of luxS (enzyme responsible for AI-2 synthesis and quorum sensing) and EPS production. Additional studies suggested that Ltp1 operates through the transcriptional regulator CdhR (Chawla et al. 2010). CdhR was determined to negatively regulate expression of mfa1 and luxS while positively regulating the expression of ltp1 and the hmu hemin acquisition operon (Chawla et al. 2010; Wu et al. 2009). Molecular and functional characterization of the BY-kinase Ptk1 demonstrated that it autophosphorylates up to seven Tyr residues within a C-terminal cluster and that kinase activity is essential for EPS production and biofilm development with S. gordonii (Liu et al. 2017; Wright et al. 2014). Substrate phosphorylation studies also determined that Ptk1 can phosphorylate EPS-associated genes PGN_0224 and PGN_0613 as well as CdhR (Liu et al. 2017; Wright et al. 2014). The pleiotropic regulation of the Tyr phosphorelay system suggests that it may act as a global regulator of community development and mediate other polymicrobial interactions. While P. gingivalis is the only oral bacteria with a characterized Tyr phosphorylation system, there are several other oral bacteria including Prevotella spp., oral streptococci, and Neisseria, as well as lactobacilli with proteins homologous to known BY-kinases.
Secondary Messengers and Metabolic Signaling
Quorum Sensing in Oral Bacteria
The oral biofilm is composed of complex community derived from multiple species that transition from assemblages of individual organisms to stable communities. These stable communities in fact undergo fluctuations in bacterial composition in response to environmental and microbial factors. Community development and maturation is a complex process that is the sum of various synergistic and antagonistic interactions all occurring in oral community but the net effect is a stable polymicrobial environment that is more beneficial to the bacteria for nutrient acquisition and processing, protection from immune responses, and resistance to environmental stresses. The bacteria within the oral community are able to sense and respond to each other by the production and detection of small chemical signals, secondary messengers, and metabolites. One method oral bacteria utilize to coordinate activities in the subgingival biofilm is quorum sensing. Bacteria can indirectly sense bacterial cell density through detection of increasing concentrations of quorum sensing molecules, termed autoinducers (Waters and Bassler 2005; Whiteley et al. 2017). There are two, chemically distinct autoinducers recognized in bacteria. The first, autoinducer-1 (AI-1), is an acyl-homoserine lactone, and this molecule functions in intraspecies cell–cell signaling. Quorum sensing mediated by AI-1 has not been found in periodontal pathogens and has been suggested to be absent in oral bacteria (Frias et al. 2001). The second cell–cell signaling system is composed of several interconvertible molecules that derive from 4,5-dihydroxy-2,3-pentanedione (DPD), which is a product of the LuxS enzyme. DPD spontaneously rearranges into a series of interconvertible molecules, collectively referred to as autoinducer-2 (AI-2). Interestingly, environmental conditions can influence the forms of AI-2 present at equilibrium, including pH or whether AI-2 can be boronated (Semmelhack et al. 2005). Additionally, two structurally distinct AI-2 receptors have been identified that allow specific detection of the multiple forms of AI-2. A number of luxS homologs have been identified in oral bacteria, suggesting that the ability for cell–cell communication via AI-2 is common.
Studies involving AI-2–based signaling among oral bacteria have predominantly focused on interactions with oral streptococci. Using an in vitro biofilm model, S. gordonii and P. gingivalis form AI-2–dependent dual-species biofilms (McNab et al. 2003). Deletion of luxS, and thus reduced AI-2, from both species attenuated biofilm development. These were some of the first studies to link AI-2 signaling to bacterial communication in dental plaque communities. AI-2 has also been shown to be essential for interactions between S. gordonii and Veillonella atypica, P. gingivalis and Filifactor alocis, and monospecies biofilm development in Eikenella corrodens and S. anginosus (Azakami et al. 2006; Wang et al. 2013; Mashima and Nakazawa 2014). Interesting work by Rickard et al. found that S. oralis and Actinomyces naeslundii dual-species biofilm formation is dependent upon AI-2 (Rickard et al. 2006). Unlike other studies, the interaction between S. oralis and A. naeslundii is extremely sensitive to the concentration of AI-2. Genetic deletion of luxS from S. oralis abolished biofilm development and the mutualistic interaction could only be restored using picromolar concentrations of AI-2. If the concentration was below or exceeded picomolar levels, biofilm development was delayed. Context for the significance of these findings is provided by studies showing that supernatant from late-colonizing bacteria (P. gingivalis, Fusobacterium nucleatum, and Prevotella intermedia) stimulated AI-2–dependent responses in a Veillonella harveyi reporter strain up to 100-fold more than early colonizers such as S. mutans, S. oralis, or S. mitis (Frias et al. 2001). The ability of pathogenic oral bacteria to induce significantly more quorum sensing responses suggests that they may have a disproportionate role in shaping oral community development and activities. This is most strongly supported by studies involving P. gingivalis, which has been shown to form dual-species biofilms with Aggregatibacter actinomycetemcomitans, T. denticola, and S. gordonii, which are all aborted in a P. gingivalis luxS mutant strain (McNab et al. 2003; Yamada et al. 2005).
In gram-positive bacteria, quorum sensing is mediated by competence stimulating peptides (CSPs), also termed autoinducing peptides (AIPs), that bind to a membrane HK and the response is transduced through a TCS. In most of these cases, the CSP is produced from the comC gene, which is proteolytically cleaved at a Gly-Gly site and the mature CSP is transported through the ComAB ABC transport system. These genes are also commonly found in an operon with the CSP-binding HK (ComD) and the cognate RR (ComE) (Havarstein et al. 1997). CSP-mediated signaling is typically species specific and has been shown to regulate bacteriocin production, genetic competence, and biofilm development. CSPs have been identified in several oral streptococci including S. mitis, S. gordonii, S. constellatus, S. mutans, and S. intermedius (Havarstein et al. 1997). In S. mutans, the ComC-dependent CSP produces a 18 amino acid peptide that is recognized by ComDE, and ComE has been shown to directly regulate genes involved in CipB bacteriocin biosynthesis (Hossain and Biswas 2012; Perry et al. 2009). With regard to competence, transcription of roughly 30 competence-associated genes is regulated by the sigma factor ComX (also known as SigX) (Khan et al. 2016). ComX is induced by both the ComC-CSP as well as a 7-amino acid CSP derived from the comS gene (also known as comX-inducing peptide or XIP) (Khan et al. 2012). While CSP signaling is species specific, a number of oral bacteria have been shown to antagonize S. mutans CSP-mediated signaling. Wang et al. reported that P. gingivalis and T. denticola reduced S. mutans transformation efficiency and bacteriocin production by degrading CSP, while S. gordonii, S. mitis, S. oralis, and S. sanguinis also degraded CSP leading to reduced S. mutans biofilm development (Wang et al. 2011a, b). Oral streptococci can also interact and form biofilms with the pathogenic fungus Candida albicans. Interestingly, CSP production by S. gordonii was shown to constrain extracellular DNA and dual-species biofilm development with C. albicans (Jack et al. 2015). While the complete roles of CSP signaling in the mouth remains unclear, these studies demonstrate the existence of complex, cross-species, and cross-kingdom signaling in the oral biofilm.
Cyclic Nucleotides as Secondary Messaging Molecules
Another important mechanism for oral bacteria to respond to changes in their environment is through secondary messaging molecules. One of the best characterized is cyclic dimeric guanosine 3′,5′-monophosphate (c-di-GMP), a molecule that has been extensively studied for its role in regulating biofilm development (Hengge et al. 2016). Levels of c-di-GMP in bacterial cells are regulated by the activities of diguanylate cyclases for synthesis, and phosphodiesterases (PDE) that degrade c-di-GMP. Diguanylate cyclases possess a conserved GG(D/E)EF domain that catalyzes the synthesis of c-di-GMP and two molecules of pyrophosphate from two molecules of GTP. The degradation of c-di-GMP is carried out by PDE enzymes that contain either an EAL or HD-GYP domain. Intracellular c-di-GMP is sensed by proteins that carry a PilZ-domain and by c-di-GMP-dependent riboswitches that regulate transcription of genes responsive to the concentrations of the cyclic nucleotide. A number of cellular processes including motility and chemotaxis, cell–cell communication, and exopolysaccharide production have all been shown to be controlled in response to c-di-GMP. Another very recently identified, similar messenger is c-di-AMP, which has also been shown to regulate bacterial biofilm development, fatty acid synthesis, and maintenance of the cell wall (Opoku-Temeng et al. 2016). C-di-AMP is synthesized by diadenylate cyclases and again degraded by the actions of PDEs. Signaling in oral bacteria by cyclic nucleotides has been severely understudied considering the significant impact it may play in promoting synergistic interactions and polymicrobial lifestyles.
T. denticola encodes seven proteins with a GGDEF domain, two proteins that contain both a GGDEF and an EAL domain, which are both inner membrane and cytosolic, and two PilZ-containing c-di-GMP binding proteins (Frederick et al. 2011). These systems were determined to be functional, as intracellular c-di-GMP was detected using high-performance liquid chromatography (Kostick et al. 2011). Genetic deletion of TDE0214, a PilZ-domain containing protein, reduced T. denticola motility and chemotaxis, biofilm development, and virulence in a murine infection model (Bian et al. 2013).
P. gingivalis was originally described as lacking a c-di-GMP signaling network; however, recent work identified intracellular c-di-GMP, thus suggesting that it can be synthesized by P. gingivalis. Chaudhuri et al. provided additional insight through the deletion of PGN_1932, revealing this protein to function as a diguanylate cyclase with a 60% reduction in intracellular c-di-GMP compared the parental strain (Chaudhuri et al. 2014). Deletion of PGN_1932 significantly altered FimA fimbriae production with a subsequent reduction in attachment to solid surfaces and HeLa cells. Bioinformatics analysis of the ATCC 33277 genome suggests that there are two genes (PGN_0239 and PGN_0282) annotated as PDE proteins, but no c-di-GMP binding proteins were identified. While clearly a large amount of remaining research into cyclic nucleotide signaling in P. gingivalis is required, these preliminary reports suggest that it’s a topic worthy of investigation.
In S. mutans, there is evidence that both c-di-GMP and c-di-AMP regulate EPS and biofilm development. Prior genome analysis of S. mutans strain UA159 did not identify any diguanylate cyclases genes; however, a study by Yan et al. described a UA159 protein (AAN59731) that when cloned into E. coli acted as a diguanylate cyclase (Yan et al. 2010). While AAN59731 does not contain a GG(D/E)EF domain and biochemical or structural characterization of its activity has not been described, an AAN59731 knockout does have a defective biofilm phenotype. Collectively, there is circumstantial evidence that S. mutans utilizes c-di-GMP, but there are several questions that remain as to whether the system is truly active and what the biological implication of such a system would be. There is, however, very strong evidence that c-di-AMP plays an important role in S. mutans signaling. S. mutans, like most gram-positive bacteria, encodes only a single diadenylate cyclase, CdaA. There are also two PDEs that function in S. mutans. The first is PdeA (also reported as GdpP for its homology to the first reported PDE in Bacillus subtilis), which degrades c-di-AMP to pApA the substrate for the second PDE, DhhP, finally producing two molecules of AMP. DhhP activity is specific for pApA, and c-di-AMP cannot be utilized as a direct substrate (Konno et al. 2018). There are also two c-di-AMP binding proteins, CabPA and CabPB. In initial studies, deletion of the cdaA gene resulted in reduced intracellular c-di-AMP levels, decreased resistance to H2O2, and increased EPS production (Cheng et al. 2016). However, a second study released in the same year found reduced EPS production in the cdaA-deficient strain compared to the parental strain (Peng et al. 2016a). More clarity on the role of c-di-AMP in S. mutans was gained from deletion of pdeA (Peng et al. 2016b). In these studies, c-di-AMP levels were elevated and biofilm development was enhanced. Increased biofilm development was dependent upon the up-regulation of a major glucan-producing protein, GtfB, as deletion of gtfB in the PdeA-knockout abolished the enhanced biofilm development. Further investigation convincingly showed that CabPA, but not CabPB, binds to the response regulator VicR, a known transcriptional regulator of gtfB (Peng et al. 2016b). Deletion of the second PDE, DhhP, and a double PDE mutant also showed increased biofilm development compared to wild type. Collectively, these results nicely show that c-di-AMP homeostasis in S. mutans is critical to the regulation of biofilm development (Konno et al. 2018).
Hence, although initial bioinformatic analysis of oral bacterial genomes determined that many bacteria did not have the essential enzymatic machinery for functional cyclic nucleotide signaling pathways (Romling et al. 2013), recent studies in P. gingivalis, F. nucleatum, and S. mutans dispute those initial annotations and suggest that there may be plenty more to be learned from studying these signaling systems in the future (Chaudhuri et al. 2014; Gursoy et al. 2017; Yan et al. 2010).
Metabolic Signaling
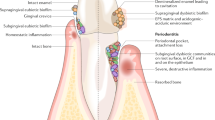
Within the subgingival biofilm, many bacteria are intimately associated with one or more physiologically compatible species. This close proximity facilitates the action of many of the signaling systems described in this chapter but also allows cooperative metabolism (Fig. 1). Metabolic communication can result from one of two processes; either metabolic cross-feeding where one species secretes small, simple metabolites, which may be utilized to benefit neighboring species or secondly metabolic syntrophy, the sequential metabolism of complex nutrients for community benefit. Multiple studies have demonstrated metabolic communication in dual-species experiments. In one case of synergistic growth, F. nucleatum enabled the growth of P. gingivalis in oxygenated environments, suggesting that the more aerotolerant F. nucleatum can consume oxygen, providing a microenvironment with differential redox potential in oral biofilms to benefit the growth of more oxygen-sensitive organisms (Diaz et al. 2002). In another study, A. naeslundii was incubated in a medium that did not support its growth, and then with the addition of S. oralis, both species displayed robust growth and co-aggregation, suggesting that close association promotes metabolic cross-feeding and enhanced growth (Palmer et al. 2001). Similarly, A. naeslundii enhances the growth of S. gordonii by increasing expression of arginine biosynthesis genes and detoxifying hydrogen peroxide (Jakubovics et al. 2008a, b). Metabolism may also have indirect benefits to other community members; the lactic acid utilizing Veillonella atypica and A. actinomycetemcomitans could prevent the acidification of the oral biofilm, thus protecting the acid-sensitive organisms such as P. gingivalis. A study by Egland et al. demonstrated that Veillonella atypica induces the expression of the amyB gene encoding an α-amylase of S. gordonii (Egland et al. 2004). Increased utilization of carbohydrates by S. gordonii results in increased production of lactic acid, the preferred energy source for V. atypica, and hence, this is a mutually synergistic nutritional interaction.
Overview of interactions in the oral biofilm. Abbreviations: A. actinomycetemcomitans (Aa), Veillonella atypica (Va), Actinomyces naeslundii (An), Streptococcus gordonii (Sg), Streptococcus mutans (Sm), Porphyromonas gingivalis (Pg), and Treponema denticola (Td). Synergistic interactions are shown with green arrows, and antagonistic interactions are shown with red arrows, with the direction of the arrows indicating the signaling information flow
S. gordonii is a participant in several instances of mutualistic growth. S. gordonii catabolizes carbohydrates and secretes H2O2 and l-lactate, a major carbon source for A. actinomycetemcomitans, and can promote A. actinomycetemcomitans growth by cross-feeding (Brown and Whiteley 2007). Interestingly, the relationship between A. actinomycetemcomitans and S. gordonii is a balance of synergistic cross-feeding and antagonistic interactions. Stacy et al. nicely demonstrated that A. actinomycetemcomitans spatially positions itself close enough to S. gordonii to benefit from the secretion of l-lactate, but far enough away for it to detoxify H2O2 through the production of catalase (Stacy et al. 2014). A. actinomycetemcomitans up-regulates dspB, a gene encoding the enzyme dispersin B that hydrolyzes polysaccharides and promotes A. actinomycetemcomitans biofilm dispersal, in response to oxygen and H2O2 (Kaplan et al. 2004). Many other oral streptococci produce H2O2 in the oral environment and similar homeostatic mechanisms to maximize energy gains from metabolic cross-feeding while minimizing oxidative stress may be present in shaping the spatial organization of dental plaque. An additional nuance in the interaction between S. gordonii and A. actinomycetemcomitans was revealed using an A. actinomycetemcomitans transposon-insertion pool followed by high-throughput sequencing (TN-Seq) (Stacy et al. 2016). S. gordonii increased the availability of oxygen to A. actinomycetemcomitans to use as a terminal electron acceptor, shifting the metabolism of A. actinomycetemcomitans from fermentation to oxidative respiration. This was termed cross-respiration and enhanced the growth and fitness of A. actinomycetemcomitans.
The presence of A. actinomycetemcomitans in an in vitro, multispecies biofilm is able to regulate the proteomic profile of the entire complex community, suggesting A. actinomycetemcomitans possesses the capability of global regulation of community development (Bao et al. 2015). Additional studies to understand this community regulator identified the histone-like family of nucleoid structuring (H-NS) protein of A. actinomycetemcomitans. H-NS proteins act as translational silencers in many gram-negative bacteria with global regulatory potential. In E. coli, H-NS regulates roughly 5% of the transcriptome and deletion of the hns gene attenuates biofilm development (Hommais et al. 2001). Characterization of an hns deletion strain of A. actinomycetemcomitans determined that H-NS promotes monospecies biofilm development as well as adhesin production (Bao et al. 2018). Analysis of the protein expression of the in vitro polymicrobial communities with the hns deletion and parental strains of A. actinomycetemcomitans demonstrated that H-NS regulates the community metabolic capability, specifically those processes involved in peptide, carbohydrate, and malate metabolism (Bao et al. 2018).
S. gordonii is also an essential partner with P. gingivalis promoting increased biofilm development and synergistic pathogenicity in rodent models of periodontitis (Daep et al. 2011; Periasamy and Kolenbrander 2009). In addition to the P. gingivalis tyrosine phosphorylation system discussed previously, S. gordonii metabolism can also regulate dual-species biofilm and virulence of P. gingivalis. In one study to examine the role of S. gordonii-associated genes that control biofilm development with P. gingivalis, spxB and cbe were determined to be instrumental in promoting biofilm development (Kuboniwa et al. 2006). SpxB is responsible for the production of H2O2 in aerobic conditions, but its role in promoting biofilm development with P. gingivalis remains unclear. Cbe (chorismate-binding enzyme) enzymes synthesize para-aminobenzoic acid (pABA) for secretion and folate biosynthesis. Interestingly, pABA was found to promote survival and colonization of P. gingivalis in vivo using a mouse oral model (Kuboniwa et al. 2017). Proteomic and metabolic analyses of P. gingivalis utilization of pABA showed that P. gingivalis can scavenge streptococcal-derived pABA for folate biosynthesis, and, moreover, pABA reduced the overall stress of P. gingivalis while promoting production of both FimA and Mfa1 fimbriae. Most surprisingly, pABA reduced the production of EPS and dampened the virulence of P. gingivalis in both mouse oral and abscess models. The regulation of EPS was determined to be through the pABA-induced regulation of Ltp1 previously discussed. These interactions will depend on density and spatial configuration, again supporting the developing narrative that metabolic communication promotes structure and polymicrobial synergy within the oral biofilm.
Both the interactions of S. gordonii with A. actinomycetemcomitans and P. gingivalis highlight the synergistic interactions between early colonizing commensal bacteria and more pathogenic late colonizers. One example of synergism involving two periodontal pathogens is the metabolic cross talk between T. denticola and P. gingivalis. When cultured together, P. gingivalis and T. denticola co-aggregate and display enhanced growth (Grenier 1992; Tan et al. 2014). Gas–liquid chromatography of co-grown culture supernatants demonstrated that P. gingivalis produces isobutyric acid that stimulates the growth of T. denticola, while T. denticola secretes succinic acid that is utilized by P. gingivalis (Grenier 1992). Additional transcriptome and metabolic analyses demonstrated that T. denticola induces the production and secretion of glycine by P. gingivalis to promote its own growth in mixed culture (Tan et al. 2014). Transcriptional evidence also suggests that P. gingivalis produces thiamine pyrophosphate, an essential nutrient for T. denticola; however, direct cross-feeding of this metabolite has not been established. Recent in vivo metatranscriptomic studies suggest that metabolic cross-feeding takes place during periodontal disease and may be a significant contributor to synergistic pathogenicity (Deng et al. 2018; Nowicki et al. 2018).
There are decades of studies demonstrating enhanced growth among oral bacteria in addition to the few examples detailed here. Mutualistic metabolism and communication promote stability of the oral biofilm and reduce direct competition for nutrients among biofilm constituents. Spatial and temporal arrangement of organisms in the complex polymicrobial community may also be a function of shared metabolism. A bioinformatics analysis of metabolic pathways of 11 oral bacteria found a large metabolic redundancy in the community, and metabolic capabilities varied among organisms associated with specific layers of the biofilm (Mazumdar et al. 2013). A recent metatranscriptomic analysis of periodontally diseased sites compared to patient-matched healthy sites revealed that metabolic capability was more stable during periodontitis than was species level diversity, suggesting the overall metabolic potential of the community may be more correlative with pathogenicity than species composition (Jorth et al. 2014). This overall metabolic stability is likely due to shared environmental conditions and stresses; however, true biofilm homeostasis is a balance between this metabolic redundancy and metabolic cross-feeding. If the metabolic capabilities and necessities of the community were too similar, individual species would be in constant competition for nutrients. However, the conservation of core metabolic functions and mutualistic metabolism between integral species reduces the antagonistic interactions and promotes community homeostasis.
Future studies will provide more insight into the complex web of cell–cell communication and signaling cascades utilized by oral bacteria to promote synergistic interactions. The oral biofilm is substantially more than the sum of its parts, with a community-dependent metabolic potential and a structure stabilized by interactions that promote survival of the community as a whole. Current and future technologies will allow for a better understanding of these community-level interactions. Demystifying the languages of bacterial communication may ultimately lead to more fruitful development of preventative and therapeutic interventions for periodontal diseases.
References
Azakami, H., Teramura, I., Matsunaga, T., Akimichi, H., Noiri, Y., Ebisu, S., & Kato, A. (2006). Characterization of autoinducer 2 signal in Eikenella corrodens and its role in biofilm formation. Journal of Bioscience and Bioengineering, 102, 110–117.
Banu, L. D., Conrads, G., Rehrauer, H., Hussain, H., Allan, E., & Van Der Ploeg, J. R. (2010). The Streptococcus mutans serine/threonine kinase, PknB, regulates competence development, bacteriocin production, and cell wall metabolism. Infection and Immunity, 78, 2209–2220.
Bao, K., Bostanci, N., Selevsek, N., Thurnheer, T., & Belibasakis, G. N. (2015). Quantitative proteomics reveal distinct protein regulations caused by Aggregatibacter actinomycetemcomitans within subgingival biofilms. PLoS One, 10, e0119222.
Bao, K., Bostanci, N., Thurnheer, T., Grossmann, J., Wolski, W. E., Thay, B., Belibasakis, G. N., & Oscarsson, J. (2018). Aggregatibacter actinomycetemcomitans H-NS promotes biofilm formation and alters protein dynamics of other species within a polymicrobial oral biofilm. NPJ Biofilms Microbiomes, 4, 12.
Bian, J., Liu, X., Cheng, Y. Q., & Li, C. (2013). Inactivation of cyclic Di-GMP binding protein TDE0214 affects the motility, biofilm formation, and virulence of Treponema denticola. Journal of Bacteriology, 195, 3897–3905.
Brown, S. A., & Whiteley, M. (2007). A novel exclusion mechanism for carbon resource partitioning in Aggregatibacter actinomycetemcomitans. Journal of Bacteriology, 189, 6407–6414.
Chaudhuri, S., Pratap, S., Paromov, V., Li, Z., Mantri, C. K., & Xie, H. (2014). Identification of a diguanylate cyclase and its role in Porphyromonas gingivalis virulence. Infection and Immunity, 82, 2728–2735.
Chawla, A., Hirano, T., Bainbridge, B. W., Demuth, D. R., Xie, H., & Lamont, R. J. (2010). Community signalling between Streptococcus gordonii and Porphyromonas gingivalis is controlled by the transcriptional regulator CdhR. Molecular Microbiology, 78, 1510–1522.
Cheng, X., Zheng, X., Zhou, X., Zeng, J., Ren, Z., Xu, X., Cheng, L., Li, M., Li, J., & Li, Y. (2016). Regulation of oxidative response and extracellular polysaccharide synthesis by a diadenylate cyclase in Streptococcus mutans. Environmental Microbiology, 18, 904–922.
Cozzone, A. J. (2005). Role of protein phosphorylation on serine/threonine and tyrosine in the virulence of bacterial pathogens. Journal of Molecular Microbiology and Biotechnology, 9, 198–213.
Daep, C. A., Novak, E. A., Lamont, R. J., & Demuth, D. R. (2011). Structural dissection and in vivo effectiveness of a peptide inhibitor of Porphyromonas gingivalis adherence to Streptococcus gordonii. Infection and Immunity, 79, 67–74.
Darveau, R. P., Hajishengallis, G., & Curtis, M. A. (2012). Porphyromonas gingivalis as a potential community activist for disease. Journal of Dental Research, 91, 816–820.
Deng, Z. L., Sztajer, H., Jarek, M., Bhuju, S., & Wagner-Dobler, I. (2018). Worlds apart—Transcriptome profiles of key oral microbes in the periodontal pocket compared to single laboratory culture reflect synergistic interactions. Frontiers in Microbiology, 9, 124.
Diaz, P. I., Zilm, P. S., & Rogers, A. H. (2002). Fusobacterium nucleatum supports the growth of Porphyromonas gingivalis in oxygenated and carbon-dioxide-depleted environments. Microbiology, 148, 467–472.
Duran-Pinedo, A. E., Nishikawa, K., & Duncan, M. J. (2007). The RprY response regulator of Porphyromonas gingivalis. Molecular Microbiology, 64, 1061–1074.
Egland, P. G., Palmer, R. J., Jr., & Kolenbrander, P. E. (2004). Interspecies communication in Streptococcus gordonii-Veillonella atypica biofilms: Signaling in flow conditions requires juxtaposition. Proceedings of the National Academy of Sciences of the United States of America, 101, 16917–16922.
Frederick, J. R., Rogers, E. A., & Marconi, R. T. (2008). Analysis of a growth-phase-regulated two-component regulatory system in the periodontal pathogen Treponema denticola. Journal of Bacteriology, 190, 6162–6169.
Frederick, J. R., Sarkar, J., McDowell, J. V., & Marconi, R. T. (2011). Molecular signaling mechanisms of the periopathogen, Treponema denticola. Journal of Dental Research, 90, 1155–1163.
Frias, J., Olle, E., & Alsina, M. (2001). Periodontal pathogens produce quorum sensing signal molecules. Infection and Immunity, 69, 3431–3434.
Gao, R., & Stock, A. M. (2009). Biological insights from structures of two-component proteins. Annual Review of Microbiology, 63, 133–154.
Grenier, D. (1992). Nutritional interactions between two suspected periodontopathogens, Treponema denticola and Porphyromonas gingivalis. Infection and Immunity, 60, 5298–5301.
Gursoy, U. K., Gursoy, M., Kononen, E., & Sintim, H. O. (2017). Cyclic dinucleotides in oral bacteria and in oral biofilms. Frontiers in Cellular and Infection Microbiology, 7, 273.
Hajishengallis, G., Darveau, R. P., & Curtis, M. A. (2012). The keystone-pathogen hypothesis. Nature Reviews. Microbiology, 10, 717–725.
Hasegawa, Y., Nishiyama, S., Nishikawa, K., Kadowaki, T., Yamamoto, K., Noguchi, T., & Yoshimura, F. (2003). A novel type of two-component regulatory system affecting gingipains in Porphyromonas gingivalis. Microbiology and Immunology, 47, 849–858.
Hasegawa, Y., Tribble, G. D., Baker, H. V., Mans, J. J., Handfield, M., & Lamont, R. J. (2008). Role of Porphyromonas gingivalis SerB in gingival epithelial cell cytoskeletal remodeling and cytokine production. Infection and Immunity, 76, 2420–2427.
Havarstein, L. S., Hakenbeck, R., & Gaustad, P. (1997). Natural competence in the genus Streptococcus: Evidence that streptococci can change phenotype by interspecies recombinational exchanges. Journal of Bacteriology, 179, 6589–6594.
Hengge, R., Grundling, A., Jenal, U., Ryan, R., & Yildiz, F. (2016). Bacterial signal transduction by cyclic Di-GMP and other nucleotide second messengers. Journal of Bacteriology, 198, 15–26.
Hirano, T., Beck, D. A., Wright, C. J., Demuth, D. R., Hackett, M., & Lamont, R. J. (2013). Regulon controlled by the GppX hybrid two component system in Porphyromonas gingivalis. Molecular Oral Microbiology, 28, 70–81.
Hommais, F., Krin, E., Laurent-Winter, C., Soutourina, O., Malpertuy, A., Le Caer, J. P., Danchin, A., & Bertin, P. (2001). Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Molecular Microbiology, 40, 20–36.
Hossain, M. S., & Biswas, I. (2012). An extracelluar protease, SepM, generates functional competence-stimulating peptide in Streptococcus mutans UA159. Journal of Bacteriology, 194, 5886–5896.
Hussain, H., Branny, P., & Allan, E. (2006). A eukaryotic-type serine/threonine protein kinase is required for biofilm formation, genetic competence, and acid resistance in Streptococcus mutans. Journal of Bacteriology, 188, 1628–1632.
Jack, A. A., Daniels, D. E., Jepson, M. A., Vickerman, M. M., Lamont, R. J., Jenkinson, H. F., & Nobbs, A. H. (2015). Streptococcus gordonii comCDE (competence) operon modulates biofilm formation with Candida albicans. Microbiology, 161, 411–421.
Jakubovics, N. S., Gill, S. R., Iobst, S. E., Vickerman, M. M., & Kolenbrander, P. E. (2008a). Regulation of gene expression in a mixed-genus community: Stabilized arginine biosynthesis in Streptococcus gordonii by coaggregation with Actinomyces naeslundii. Journal of Bacteriology, 190, 3646–3657.
Jakubovics, N. S., Gill, S. R., Vickerman, M. M., & Kolenbrander, P. E. (2008b). Role of hydrogen peroxide in competition and cooperation between Streptococcus gordonii and Actinomyces naeslundii. FEMS Microbiology Ecology, 66, 637–644.
James, C. E., Hasegawa, Y., Park, Y., Yeung, V., Tribble, G. D., Kuboniwa, M., Demuth, D. R., & Lamont, R. J. (2006). LuxS involvement in the regulation of genes coding for hemin and iron acquisition systems in Porphyromonas gingivalis. Infection and Immunity, 74, 3834–3844.
Jorth, P., Turner, K. H., Gumus, P., Nizam, N., Buduneli, N., & Whiteley, M. (2014). Metatranscriptomics of the human oral microbiome during health and disease. MBio, 5, e01012–e01014.
Kadowaki, T., Yukitake, H., Naito, M., Sato, K., Kikuchi, Y., Kondo, Y., Shoji, M., & Nakayama, K. (2016). A two-component system regulates gene expression of the type IX secretion component proteins via an ECF sigma factor. Scientific Reports, 6, 23288.
Kaplan, J. B., Velliyagounder, K., Ragunath, C., Rohde, H., Mack, D., Knobloch, J. K., & Ramasubbu, N. (2004). Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. Journal of Bacteriology, 186, 8213–8220.
Khan, R., Rukke, H. V., Ricomini Filho, A. P., Fimland, G., Arntzen, M. O., Thiede, B., & Petersen, F. C. (2012). Extracellular identification of a processed type II ComR/ComS pheromone of Streptococcus mutans. Journal of Bacteriology, 194, 3781–3788.
Khan, R., Rukke, H. V., Hovik, H., Amdal, H. A., Chen, T., Morrison, D. A. & Petersen, F. C. (2016). Comprehensive transcriptome profiles of Streptococcus mutans UA159 map core streptococcal competence genes. mSystems, 1.
Konno, H., Yoshida, Y., Nagano, K., Takebe, J., & Hasegawa, Y. (2018). Biological and biochemical roles of two distinct cyclic dimeric adenosine 3′,5’-monophosphate-associated phosphodiesterases in Streptococcus mutans. Frontiers in Microbiology, 9, 2347.
Kostick, J. L., Szkotnicki, L. T., Rogers, E. A., Bocci, P., Raffaelli, N., & Marconi, R. T. (2011). The diguanylate cyclase, Rrp1, regulates critical steps in the enzootic cycle of the Lyme disease spirochetes. Molecular Microbiology, 81, 219–231.
Kreth, J., Zhang, Y., & Herzberg, M. C. (2008). Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. Journal of Bacteriology, 190, 4632–4640.
Krishnan, K., & Duncan, M. J. (2013). Role of sodium in the RprY-dependent stress response in Porphyromonas gingivalis. PLoS One, 8, e63180.
Kuboniwa, M., Tribble, G. D., James, C. E., Kilic, A. O., Tao, L., Herzberg, M. C., Shizukuishi, S., & Lamont, R. J. (2006). Streptococcus gordonii utilizes several distinct gene functions to recruit Porphyromonas gingivalis into a mixed community. Molecular Microbiology, 60, 121–139.
Kuboniwa, M., Houser, J. R., Hendrickson, E. L., Wang, Q., Alghamdi, S. A., Sakanaka, A., Miller, D. P., Hutcherson, J. A., Wang, T., Beck, D. A. C., Whiteley, M., Amano, A., Wang, H., Marcotte, E. M., Hackett, M., & Lamont, R. J. (2017). Metabolic crosstalk regulates Porphyromonas gingivalis colonization and virulence during oral polymicrobial infection. Nature Microbiology, 2, 1493–1499.
Lamont, R. J., Koo, H., & Hajishengallis, G. (2018). The oral microbiota: Dynamic communities and host interactions. Nature Reviews. Microbiology, 16, 745–759.
Lasica, A. M., Ksiazek, M., Madej, M., & Potempa, J. (2017). The type IX secretion system (T9SS): Highlights and recent insights into its structure and function. Frontiers in Cellular and Infection Microbiology, 7, 215.
Li, Y., Krishnan, K., & Duncan, M. J. (2018). Post-translational regulation of a Porphyromonas gingivalis regulator. Journal of Oral Microbiology, 10, 1487743.
Lin, W. J., Walthers, D., Connelly, J. E., Burnside, K., Jewell, K. A., Kenney, L. J., & Rajagopal, L. (2009). Threonine phosphorylation prevents promoter DNA binding of the group B Streptococcus response regulator CovR. Molecular Microbiology, 71, 1477–1495.
Liu, C., Miller, D. P., Wang, Y., Merchant, M., & Lamont, R. J. (2017). Structure-function aspects of the Porphyromonas gingivalis tyrosine kinase Ptk1. Molecular Oral Microbiology, 32, 314–323.
Maeda, K., Tribble, G. D., Tucker, C. M., Anaya, C., Shizukuishi, S., Lewis, J. P., Demuth, D. R., & Lamont, R. J. (2008). A Porphyromonas gingivalis tyrosine phosphatase is a multifunctional regulator of virulence attributes. Molecular Microbiology, 69, 1153–1164.
Mashima, I., & Nakazawa, F. (2014). The influence of oral Veillonella species on biofilms formed by Streptococcus species. Anaerobe, 28, 54–61.
Mazumdar, V., Amar, S., & Segre, D. (2013). Metabolic proximity in the order of colonization of a microbial community. PLoS One, 8, e77617.
McNab, R., Ford, S. K., El-Sabaeny, A., Barbieri, B., Cook, G. S., & Lamont, R. J. (2003). LuxS-based signaling in Streptococcus gordonii: Autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. Journal of Bacteriology, 185, 274–284.
Miller, D. P., Frederick, J. R., Sarkar, J., & Marconi, R. T. (2014). The Treponema denticola AtcR LytTR domain-containing response regulator interacts with three architecturally distinct promoter elements: Implications for understanding the molecular signaling mechanisms that drive the progression of periodontal disease. Molecular Oral Microbiology, 29, 219–232.
Nishikawa, K., & Duncan, M. J. (2010). Histidine kinase-mediated production and autoassembly of Porphyromonas gingivalis fimbriae. Journal of Bacteriology, 192, 1975–1987.
Nishikawa, K., Yoshimura, F., & Duncan, M. J. (2004). A regulation cascade controls expression of Porphyromonas gingivalis fimbriae via the FimR response regulator. Molecular Microbiology, 54, 546–560.
Nowicki, E. M., Shroff, R., Singleton, J. A., Renaud, D. E., Wallace, D., Drury, J., Zirnheld, J., Colleti, B., Ellington, A. D., Lamont, R. J., Scott, D. A., & Whiteley, M. (2018). Microbiota and metatranscriptome changes accompanying the onset of gingivitis. MBio, 9, e00575.
Opoku-Temeng, C., Zhou, J., Zheng, Y., Su, J., & Sintim, H. O. (2016). Cyclic dinucleotide (c-di-GMP, c-di-AMP, and cGAMP) signalings have come of age to be inhibited by small molecules. Chemical Communication (Cambridge), 52, 9327–9342.
Ortet, P., Whitworth, D. E., Santaella, C., Achouak, W., & Barakat, M. (2015). P2CS: Updates of the prokaryotic two-component systems database. Nucleic Acids Research, 43, D536–D541.
Palmer, R. J., Jr., Kazmerzak, K., Hansen, M. C., & Kolenbrander, P. E. (2001). Mutualism versus independence: Strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infection and Immunity, 69, 5794–5804.
Peng, X., Michalek, S., & Wu, H. (2016a). Effects of diadenylate cyclase deficiency on synthesis of extracellular polysaccharide matrix of Streptococcus mutans revisit. Environmental Microbiology, 18, 3612–3619.
Peng, X., Zhang, Y., Bai, G., Zhou, X., & Wu, H. (2016b). Cyclic di-AMP mediates biofilm formation. Molecular Microbiology, 99, 945–959.
Periasamy, S., & Kolenbrander, P. E. (2009). Mutualistic biofilm communities develop with Porphyromonas gingivalis and initial, early, and late colonizers of enamel. Journal of Bacteriology, 191, 6804–6811.
Perry, J. A., Jones, M. B., Peterson, S. N., Cvitkovitch, D. G., & Levesque, C. M. (2009). Peptide alarmone signalling triggers an auto-active bacteriocin necessary for genetic competence. Molecular Microbiology, 72, 905–917.
Prisic, S., Dankwa, S., Schwartz, D., Chou, M. F., Locasale, J. W., Kang, C. M., Bemis, G., Church, G. M., Steen, H., & Husson, R. N. (2010). Extensive phosphorylation with overlapping specificity by Mycobacterium tuberculosis serine/threonine protein kinases. Proceedings of the National Academy of Sciences of the United States of America, 107, 7521–7526.
Rickard, A. H., Palmer, R. J., Jr., Blehert, D. S., Campagna, S. R., Semmelhack, M. F., Egland, P. G., Bassler, B. L., & Kolenbrander, P. E. (2006). Autoinducer 2: A concentration-dependent signal for mutualistic bacterial biofilm growth. Molecular Microbiology, 60, 1446–1456.
Romling, U., Galperin, M. Y., & Gomelsky, M. (2013). Cyclic di-GMP: The first 25 years of a universal bacterial second messenger. Microbiology and Molecular Biology Reviews, 77, 1–52.
Sankhe, G. D., Dixit, N. M., & Saini, D. K. (2018). Activation of bacterial histidine kinases: Insights into the kinetics of the cis autophosphorylation mechanism. mSphere, 3(3).
Sarkar, J., Frederick, J., & Marconi, R. T. (2010). The Hpk2-Rrp2 two-component regulatory system of Treponema denticola: A potential regulator of environmental and adaptive responses. Molecular Oral Microbiology, 25, 241–251.
Sarkar, J., Miller, D. P., Oliver, L. D., Jr., & Marconi, R. T. (2018). The Treponema denticola PAS domain-containing histidine kinase Hpk2 is a heme binding sensor of oxygen levels. Journal of Bacteriology, 200(18), JB.00116–JB.00118.
Sato, K., Naito, M., Yukitake, H., Hirakawa, H., Shoji, M., McBride, M. J., Rhodes, R. G., & Nakayama, K. (2010). A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proceedings of the National Academy of Sciences of the United States of America, 107, 276–281.
Scott, J. C., Klein, B. A., Duran-Pinedo, A., Hu, L., & Duncan, M. J. (2013). A two-component system regulates hemin acquisition in Porphyromonas gingivalis. PLoS One, 8, e73351.
Semmelhack, M. F., Campagna, S. R., Federle, M. J., & Bassler, B. L. (2005). An expeditious synthesis of DPD and boron binding studies. Organic Letters, 7, 569–572.
Shi, L., Pigeonneau, N., Ravikumar, V., Dobrinic, P., Macek, B., Franjevic, D., Noirot-Gros, M. F., & Mijakovic, I. (2014). Cross-phosphorylation of bacterial serine/threonine and tyrosine protein kinases on key regulatory residues. Frontiers in Microbiology, 5, 495.
Simionato, M. R., Tucker, C. M., Kuboniwa, M., Lamont, G., Demuth, D. R., Tribble, G. D., & Lamont, R. J. (2006). Porphyromonas gingivalis genes involved in community development with Streptococcus gordonii. Infection and Immunity, 74, 6419–6428.
Smalley, J. W., & Olczak, T. (2017). Heme acquisition mechanisms of Porphyromonas gingivalis—strategies used in a polymicrobial community in a heme-limited host environment. Molecular Oral Microbiology, 32, 1–23.
Smalley, J. W., Byrne, D. P., Birss, A. J., Wojtowicz, H., Sroka, A., Potempa, J., & Olczak, T. (2011). HmuY haemophore and gingipain proteases constitute a unique syntrophic system of haem acquisition by Porphyromonas gingivalis. PLoS One, 6, e17182.
Stacy, A., Everett, J., Jorth, P., Trivedi, U., Rumbaugh, K. P., & Whiteley, M. (2014). Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proceedings of the National Academy of Sciences of the United States of America, 111, 7819–7824.
Stacy, A., Fleming, D., Lamont, R. J., Rumbaugh, K. P., & Whiteley, M. (2016). A commensal bacterium promotes virulence of an opportunistic pathogen via cross-respiration. MBio, 7(3), e00782–e00716.
Stancik, I. A., Sestak, M. S., Ji, B., Axelson-Fisk, M., Franjevic, D., Jers, C., Domazet-Loso, T., & Mijakovic, I. (2018). Serine/threonine protein kinases from bacteria, archaea and eukarya share a common evolutionary origin deeply rooted in the tree of life. Journal of Molecular Biology, 430, 27–32.
Stock, A. M., Robinson, V. L., & Goudreau, P. N. (2000). Two-component signal transduction. Annual Review of Biochemistry, 69, 183–215.
Takeuchi, H., Hirano, T., Whitmore, S. E., Morisaki, I., Amano, A., & Lamont, R. J. (2013). The serine phosphatase SerB of Porphyromonas gingivalis suppresses IL-8 production by dephosphorylation of NF-kappaB RelA/p65. PLoS Pathogens, 9, e1003326.
Tan, K. H., Seers, C. A., Dashper, S. G., Mitchell, H. L., Pyke, J. S., Meuric, V., Slakeski, N., Cleal, S. M., Chambers, J. L., McConville, M. J., & Reynolds, E. C. (2014). Porphyromonas gingivalis and Treponema denticola exhibit metabolic symbioses. PLoS Pathogens, 10, e1003955.
Tribble, G. D., Mao, S., James, C. E., & Lamont, R. J. (2006). A Porphyromonas gingivalis haloacid dehalogenase family phosphatase interacts with human phosphoproteins and is important for invasion. Proceedings of the National Academy of Sciences of the United States of America, 103, 11027–11032.
Vincent, M. S., Durand, E., & Cascales, E. (2016). The PorX response regulator of the Porphyromonas gingivalis PorXY two-component system does not directly regulate the type IX secretion genes but binds the PorL subunit. Frontiers in Cellular and Infection Microbiology, 6, 96.
Wang, B. Y., Alvarez, P., Hong, J., & Kuramitsu, H. K. (2011a). Periodontal pathogens interfere with quorum-sensing-dependent virulence properties in Streptococcus mutans. Journal of Periodontal Research, 46, 105–110.
Wang, B. Y., Deutch, A., Hong, J., & Kuramitsu, H. K. (2011b). Proteases of an early colonizer can hinder Streptococcus mutans colonization in vitro. Journal of Dental Research, 90, 501–505.
Wang, Q., Wright, C. J., Dingming, H., Uriarte, S. M., & Lamont, R. J. (2013). Oral community interactions of Filifactor alocis in vitro. PLoS One, 8, e76271.
Waters, C. M., & Bassler, B. L. (2005). Quorum sensing: Cell-to-cell communication in bacteria. Annual Review of Cell and Developmental Biology, 21, 319–346.
Whiteley, M., Diggle, S. P., & Greenberg, E. P. (2017). Progress in and promise of bacterial quorum sensing research. Nature, 551, 313–320.
Whitmore, S. E., & Lamont, R. J. (2012). Tyrosine phosphorylation and bacterial virulence. International Journal of Oral Science, 4, 1–6.
Wright, C. J., Xue, P., Hirano, T., Liu, C., Whitmore, S. E., Hackett, M., & Lamont, R. J. (2014). Characterization of a bacterial tyrosine kinase in Porphyromonas gingivalis involved in polymicrobial synergy. Microbiology, 3, 383–394.
Wu, J., Lin, X., & Xie, H. (2007). Porphyromonas gingivalis short fimbriae are regulated by a FimS/FimR two-component system. FEMS Microbiology Letters, 271, 214–221.
Wu, J., Lin, X., & Xie, H. (2009). Regulation of hemin binding proteins by a novel transcriptional activator in Porphyromonas gingivalis. Journal of Bacteriology, 191, 115–122.
Yamada, M., Ikegami, A., & Kuramitsu, H. K. (2005). Synergistic biofilm formation by Treponema denticola and Porphyromonas gingivalis. FEMS Microbiology Letters, 250, 271–277.
Yan, W., Qu, T., Zhao, H., Su, L., Yu, Q., Gao, J., & Wu, B. (2010). The effect of c-di-GMP (3′-5′-cyclic diguanylic acid) on the biofilm formation and adherence of Streptococcus mutans. Microbiological Research, 165, 87–96.
Zhang, Y., Wang, T., Chen, W., Yilmaz, O., Park, Y., Jung, I. Y., Hackett, M., & Lamont, R. J. (2005). Differential protein expression by Porphyromonas gingivalis in response to secreted epithelial cell components. Proteomics, 5, 198–211.
Zhu, L., & Kreth, J. (2010). Role of Streptococcus mutans eukaryotic-type serine/threonine protein kinase in interspecies interactions with Streptococcus sanguinis. Archives of Oral Biology, 55, 385–390.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Miller, D.P., Lamont, R.J. (2019). Signaling Systems in Oral Bacteria. In: Belibasakis, G.N., Hajishengallis, G., Bostanci, N., Curtis, M.A. (eds) Oral Mucosal Immunity and Microbiome. Advances in Experimental Medicine and Biology, vol 1197. Springer, Cham. https://doi.org/10.1007/978-3-030-28524-1_3
Download citation
DOI: https://doi.org/10.1007/978-3-030-28524-1_3
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-28523-4
Online ISBN: 978-3-030-28524-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)