Abstract
Microbial communities colonizing in and around the plants are essential for their survival and act as key determinants for plant’s holistic health to make the dynamic plant microbiome. The microbiome comprises of trillions of bacteria, fungi, viruses and other microorganisms interacting with each other as well as with the plants. Metagenomics is a powerful tool that enables rapid analysis of microbial heterogenicity, thus helping us to understand the association of microorganisms within their environment and the overall functioning of microbiome. Herein, an overview of culture-independent methods to explore the unculturable/yet to culture microbial diversity of plant microbiome is addressed. This chapter focuses on the different constituents of plant-microbe interface and the metagenomic studies related to them.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
12.1 Introduction: Plant Microbiome
A vast diversity of microorganisms present in nearby microenvironment of either outside or inside of plants constitute the plant microbiome. Plant microbiome consists of almost all groups of microbes including virus, archaea, bacteria, oomycetes and fungi; in spite of many decades of long studies, details of the composition and their interrelationship related to microbial diversity and richness of species that comprise the plant microbiome are not yet fully explored (Raaijmakers et al. 2009; Singh et al. 2019). To understand the dynamics and functioning of microbiome of a plant, it is divided in three different fractions, i.e. (1) microorganisms that reside outside the plants in rhizosphere (soil surrounding the roots and root surface); (2) endophytes that exist inside the plants tissues, and (3) epiphytes, colonizing the outer surface of plants mostly on phyllosphere (the aerial parts of plants). It is assumed that thousands of epiphytes and endophytic species exist on a single plant species.
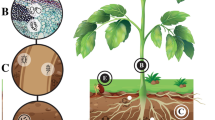
Microorganisms constituting the plant microbiome are part of a complex food web; they utilize the nutrients released from plants and in return help in nutrient cycling and detoxification of harmful compounds and induce resistance in plants against both types of stresses, be it abiotic or biotic, and protect plants from plant pathogens, thus imposing a significant impact on plant productivity (Yadav et al. 2018). Exudates released from both plants and microorganisms are reported to act as inducer molecules which play a major role in signalling each other through which plants and microorganism communicate. Due to the interaction of plants with microbe, the whole plant microbiome is referred as extension of genetic compendium of plants and coined as plant’s ‘second genome’ (Berendsen et al. 2012). Microbiome regulates several physiological processes in the host. Figure 12.1 explains the composition of plant microbiome and their specific functions.
Rhizosphere acts as a strong componential pillar of the plant microbiome. The complex microbial diversity in rhizosphere is influenced by climatic conditions, viz. temperature, salinity etc., of soil and physical factors including presence of metal ions and organic compounds, as well as biotic factors. Extensive researches reveal that plants also contribute to design their own designer microbiome as per their requirement. Exudates released from plant roots are composed of carbohydrates, proteins, lipids, phenolic compounds, organic acid and enzymes. These molecules are utilized by selective groups of microorganisms, thus proving that the composition of microbial community in any given plant microbiome is highly influenced by the released root exudates from the existing plant species. The presence of nitrogen-fixing bacteria and phosphate-solubilizing bacteria and production of plant-growth hormones are few examples of selective induction of microbial diversity in plant vicinity (Mendes et al. 2011). Similarly, natural suppressive soils are also an example of plant-driven stimulation of antibiotic-producing bacteria. Hence it can be said that plants have evolved themselves in such a way that they know how to build their designer rhizosphere communities which aid in protection from various stresses.
Endophytes constitute the other important component of plant microbiome. In the ancient time, microorganisms existing inside the plants were considered as only disease agents. The revelation of the presence of non-pathogenic microorganisms inside the plant led to the concept that microbes can colonize inside the plants as nonpathogens without posing any threat to their health. Later on these endophytes were studied by several groups, and it was found that no plant is free from endophytes (Rosenblueth and Martínez-Romero 2006). They help host plants in managing their pathogens and also promote plant growth. Endophytic Burkholderia spp. is known to control the growth of the pathogen Fusarium moniliforme. Endophytic diazotrophs from sugarcane roots produce amino acids and other plant growth-promoting substances which aid in improving their health (Suman et al. 2001). Genomic studies show the vast diversity in endophytic community and suggest that the ecology and genome size of entophytic population depend on environmental conditions.
Epiphytes or microorganism colonizing the outer surface of plants is the third important component of plant microbiome. It comprises of bacteria, fungi and algae. Protozoa and nematodes have also been reported at lower frequencies in as epiphytes (Lindow and Brandl 2003). As per rough estimate, plant leaves surface could harbour approximately 1026 bacterial cells (Vorholt 2012). The structure and composition of epiphytic microbial diversity is largely influenced by the nutritional heterogeneity of plant surfaces and also with the environmental interaction with plants. Epiphytes play a major role in plant development by acting as soldiers combating against invading pathogens; some of the epiphytes also help in phytohormones biosynthesis, performing nitrogen fixation, etc. (Padhi et al. 2013). Phyllosphere-associated fungi also interact with pathogenic fungi and help to control pathogenic invasion on leaves and leaf litter degradation.
12.2 Metagenomics: Effective Tool to Explore Plant Host Interface
Plant microbiome comprises of relatively diverse yet under-characterized microbial community. Exploring it can potentially enrich our understanding of plant-microbial ecology and their interaction within the community. Phylogenetic surveys show that the unknown prokaryotic microbial species outnumber the known cultured prokaryotes in any single plant microbiome. In recent past decades, several studies have compared the phylogenetic presence and abundance of different microbes in the phyllosphere region of various plants like spinach, apple, lettuce, rice and Arabidopsis by traditional and culture-independent methods revealing that the phyllosphere comprised more of the unknown uncultivable microbial population which tallied to a percentage of 90–99 (Rastogi et al. 2012).
Fortunately, the recent developments in metagenomics, viz. next-generation sequencing technologies and other culture-independent approaches, have enabled the investigations of the functional genetic diversity of various microorganisms without the inherent biases of manual cultivation, competition amongst microbes and plants, parasitism and other biotic/abiotic stress (e.g. salinity, temperature, humidity, etc.) and have helped us to have a deeper knowledge of microbial ecology (Oulas et al. 2015). The term metagenomics is based on the concept of meta-analysis (the statistical process of combining separate analyses) and genomics (the comprehensive analysis of an organism’s genetic material). Figure 12.2 clearly depicts the work methodology of metagenomic analysis from any environmental sample be it from rhizosphere, endosphere or phyllosphere. Metagenomics is a combination of all modern techniques of the field of genomics that have metamorphosized our understanding of the microbial population and their interactions with the environment. It has opened a magnificent door to the biotechnology field especially based on the exploitation of uncultivated microbial species. Initially, metagenomic studies were focused on only uncultured microflora and ancient DNA findings, but nowadays the technology has reached to another level and is applied to study the whole plant microbiome and gastrointestinal ecosystems of human and animals as well (Müller and Ruppel 2014).
Although the new metagenomic techniques allow us to conclude changes in microbial communities at the genetic level, few challenges have to be fought like heterogeneity of the scales used for sampling and the connectivity between those scales. Selecting a good site for sampling and methods used thereafter are important factors to contemplate when beginning a metagenomic analysis of a microbiome. Microbial activity and population are affected by its physical, chemical and biological properties. The minute changes in any condition which affects plant growth such as increased or decreased nutrient concentration or major changes like drought all have profound impacts on the structure and functions of epiphytic and endophytic microbial communities.
Metagenomic methodology starts with isolation of environmental DNA. A library of clones is constructed and screened followed by sequencing and analysis of isolated metagenomic DNA which renders informative data on various aspects of the studied sample, allowing to typify the microbial life in any given environment extensively. It not only identifies the species of the microbiome but also provides a glimpse about the metabolomic activities related to the functional aspects of the cultivable and unculturable microbes of a given population (Langille et al. 2013).
12.3 Rhizosphere and Its Components
The physically, chemically and biologically agile zone of the soil around the plant roots is referred to as rhizosphere. It comprises of the soil adhering to plant roots which has great importance and exhibits complex interrelationships between microbial confraternity and their mutual interactions with the plant. The diversity and complexity shown by any rhizospheric microbial community is greatly influenced by root exudation and physiochemical properties reflected by soil owing to the agronomic operations and practices used (Shrivastava et al. 2014). It harbours all the soil-borne microbes including protozoans, fungi, archaea and bacteria, which have a great impact on the roots and its exudates through their biological, physical and chemical interactions. Hence, it is imperative to study the interactions between plants and these soil microorganisms for cognizing various plant-related processes (Amann et al. 1995). Most of the microorganisms residing in the soil are not culturable in the standard laboratory conditions. Different plant species may be biased in supporting various microbial communities in their rhizospheric zones owing to their root exudates. The plant rhizosphere profusely secretes a large number of compounds that are utilized by the soil microbial communities in many different ways. These phytochemical exudates act as selective nutritional sources for stimulation and enrichment of specific groups of soil microorganisms which in turn help in the growth and development of the plants (Larkin et al. 1993; Mendes et al. 2013). This hot spot is considered to be one of the most aggressively enterprising interfaces on earth (Philippot et al. 2013).
12.4 Metagenomics to Explore Rhizosphere Environment
Rhizosphere is the most active interface in which plants and microorganisms establish a complex and varied molecular dialogue, involving nutrient transfer as well as specific interactions mediated by the release of signalling molecules from plant roots, thereby resulting in enhanced plant productivity (Prasad et al. 2015). The rhizosphere microbiome is a dynamic blend of beneficial and pathogenic (plant, human) microorganisms. Kumar et al. (2018) studied the rhizosphere of alfalfa and explored the structure and diversity of microbial community using 16S rRNA metagenome analysis. Metabolic network approaches also find their usage in exploring the associations between structure and functions of environment in complex microbial rhizosphere microbiome. The DNA data from two agricultural crops, viz., wheat and cucumber, were extracted using the same techniques (Ofaim et al. 2017).
Hypergeometric enrichment tests have been used to study enriched pathways (metabolites/enzymes) and possible functional significance for observed co-occurrence patterns of various taxonomic combinations and their complementary metabolite profiles. The soil plays a prominent role in the structural composition of microbial communities residing in it. Many novel members of Crenarchaeota group displaying resistance to different metals were discovered using these techniques from Tinto river (Mirete et al. 2007). The microbially incongruent communities in the rhizosphere showed more complexity than those in the river and mainly represented heterotrophic acidophiles suggesting that the soil composition rendered the diverse resistance to the microbiome.
Results in the effects of fertilizers and other agricultural practices on rhizosphere microbiome have been studied revealing that some genes for phytic acid utilization were upregulated after the incorporation of fertilizers. Plants start harbouring those microbes which are beneficial for their growth in varied conditions. There was a clear demarcation in the taxonomic profiles of the samples collected from rhizosphere and bulk soil again suggesting a role of plants and soil environment on the microbial species present there (Uroz et al. 2010). Similarly some crops like soybean have been shown to allow some selected microbial communities to inhabit their rhizosphere based on beneficial functional traits aiding in their growth promotion and nutrition. Techniques like stable-isotope probing (SIP) along with metagenomics of fungal communities have led to the discovery of many new OTUs belonging to Basidiomycota and Ascomycota (Gkarmiri et al. 2017). Metagenomic analysis of many citrus rhizospheres have concluded that functional properties involved in host-microbe interactions are significantly critical for the microbiome-inhabiting plant root surfaces and are influenced remarkably by the availability of plant exudates. These rhizoplane-enriched functional properties are advantageous to the plant host. Thereby, determining genetic and microbial intricacy in the citrus rhizoplane microbiome compared to that in the rhizosphere communities, indicating the filter effect of plant hosts on the closely associated rhizoplane microbiome assembly (Zhang et al. 2017). Pyrosequencing analysis has been used to analyse the shift in microbial communities as an effect of addition of various fertilizers (Li et al. 2016). Table 12.1 enlists the dominating phylum’s explored metagenomically in different rhizospheres.
12.5 Endosphere
12.5.1 Endosphere and Its Components
Endosphere is defined as the region present inside the plant. Within the endosphere, microbes inhabit various microenvironments like the intra- and intercellular spaces inside the plant body, and each microenvironment presents an unmatched and diverse biochemical profile. Plants as metaorganisms populate microbes showing heterogeneity residing in different habitats (such as endosphere, rhizosphere and phyllosphere), situated inside or on the surface of vegetative parts (roots, stems and leaves) and reproductive parts (flowers, fruits and seeds) of the host plant (Truyens et al. 2015). Endophytic microbes refer to microbial population that reside within the tissues of plants without resulting in any visible adverse effects on their host (Knief 2014). Hence, endophytic microbes are mostly facultative rhizospheric microorganisms and/or accidental passengers in the root, suggesting that the overall composition of the various taxonomic and phylogenetic profiles of the dominant residing microbes will have homology and similarity. Bacterial colonies extensively colonize the internal plant tissues, found in almost every plant worldwide. Both culture-dependent and culture-independent techniques have shown the diversity of endophytic bacteria that include various bacterial taxa across a broad range of different plant species. Studies suggest that endophytes originated from the rhizosphere (soil) and/or are maternally transferred to future generations (vertical transmission through seeds). Several microbes residing in endosphere have the potential to affect plant growth either directly or indirectly by helping them in the production of siderophores and procurement of different macronutrients by mineral phosphate solubilization and biological nitrogen fixation. The direct routes also involve production of phytohormones such as auxin (indole-3-acetic acid, IAA) by microbes which stimulates plant growth, especially of roots (Bulgarelli et al. 2013). The mechanisms of promoting plant growth used by endophytic bacteria are similar to the mechanisms used by rhizospheric bacteria. Similar to rhizospheric plant growth-promoting bacteria, endophytic bacteria can also act to facilitate plant growth in horticulture, agriculture and silviculture as well as in strategies for environmental cleanup (viz. phytoremediation). Understanding these mechanisms is crucial to determine the principles governing structure, function and robustness of microbial community. Bacterial endophytes may have a benefit over bacteria inhabiting the rhizosphere, since living inside plant’s tissues gives them a chance to always be in direct contact with the plant’s cells, and thus, they can more easily exert an enhanced beneficial effect. Bacteria residing within the rhizosphere also have potential chances to enter and colonize the plant roots. This microecosystem is one most commonly studied primary route of endophytic colonization (Hallmann et al. 1997). More and more extensive studies suggest that endophytic bacterial diversity can be considered a subset of the rhizosphere and/or root-associated bacterial population, and rhizospheric and endophytic bacterial communities sometimes exhibit different overall patterns of relative sufficiency of the major groups at the phylum level (Kent and Triplett 2002; Cocking 2003). Fungi with different morphological characteristics were isolated from both rhizosphere and endosphere fungi of C. japonicum. The genus Trichoderma is most often isolated and deeply studied endospheric fungi, and the distribution of fungi is similar between rhizosphere and endosphere.
12.5.2 Metagenomics of Endosphere Environment
It has been surmised that endophytic root bacterial communities comprehend a subset of colonists originating from the encircling rhizosphere soil (Cocking 2003), and the resulting community framework is affected by the surrounding soil and environmental properties. The term metagenomics incorporates the analysis of an assemblage of similar but non-identical items (Glass 1976). It basically involves isolating DNA from an environmental sample which is called a metagenome, cloning the environmental DNA into a suitable vector, transforming the clones into a host bacterium and screening the resulting transformants. The resultant clones are then screened for phylogenetic markers or ‘anchors’, for example, conserved sequences, viz. 16S rRNA and recA, or any other conserved genes by hybridization or multiplex PCR or for any function like expression of specific traits like enzyme activity or antibiotic production (Courtois et al. 2003) or they can be sequenced randomly. Traditional and metagenomic approach both have certain benefits over each other along with certain limitations. Therefore it is advisable to use these approaches together to enrich the understanding of the uncultured world, providing insight into the vast microbial population that is still unrevealed and entirely unknown. Metagenomic analysis has unveiled substantial microheterogeneity in apparently uniform populations where the challenge lies in linking the genomic information with the organism or ecosystem from which the DNA was isolated. Culture-based techniques allow to study isolated microbes in depth, and the modern molecular techniques like metagenomics and metabolomics help to explore the unidentified microbial communities in situ. These studies hold an important place in core areas like plant breeding and microbiology apart from allied field of agriculture and healthcare system as plant microbiome is a decisive determinant of plant health and productivity and has received substantial attention in recent years (Bulgarelli et al. 2013). Comparisons between endogenome and rhizogenome with emphasis on plant growth-promoting bacteria have disclosed potential genetic factors involved in an endophytic lifestyle, which facilitates a better cognizance of the functioning of bacterial endophytes. Competition for resources among community members is based on the usage of diverse survival tactics, like antagonism and mutualism among the members. Metagenomics has redefined the concept of a genome which has enhanced the prospects of solving many problems and given a momentum to the rate of gene discovery. The potential for application of metagenomics to biotechnology seems endless. Table 12.2 shows the list of microbes metagenomically isolated and identified from endosphere region.
Usually, endophytic bacteria are known to be non-pathogenic, causing no visible symptoms, but sometimes they may include latent pathogens that may cause disease depending on the availability of favourable environmental conditions and host genotypes. Model organisms like Burkholderia, Herbaspirillum and Azoarcus spp., residing in the non-leguminous plants, mainly grasses, have been extensively studied for extracting information about the taxonomic diversity and mechanisms of infection and colonization of endophytic microbes within plant system (Thomas 2017). Culture-independent methods, such as analyses of 16S rRNA and nifH transcripts and metagenome analyses, have paved a way for exploring huge melange of endophytes in the economically important crops like sugarcane and rice. The studies suggest that rhizobia (and other α-Proteobacteria) are very common endophytes, as are β-proteobacteria, γ-proteobacteria and Firmicutes. The core endophytic bacterial microbiome of A. thaliana was studied using high-throughput sequencing (HTS) of 16S rRNA. These studies showed that although various soil types altered the bacterial endophyte microbiome, some species of prokaryotes were persistently present in endosphere as compared with the rhizosphere environment and included Actinobacteria and some families from the Proteobacteria.
12.6 Phyllosphere
12.6.1 Phyllosphere and Its Components
Phyllosphere represents the microbial flora and fauna dwelling on and in aerial plant organs, which constitute the total part of living plant above the ground (Newton et al. 2010). It is further categorised into caulosphere (stems), carosphere (fruits) and anthosphere (flowers) (Berlec 2012). The major part of this surface is provided by green leaves, and it is believed to represent one of the largest dwelling sites on earth. There exist little information about the bacterial communities which reside in the above said categories apart from leaves; therefore, the maximum information about phyllosphere consists of the knowledge pertaining to leaves.
Recent cultivation-independent studies have helped us to examine the composition of microbial phyllosphere communities in a better way. It is evident that these communities do not represent random assemblies of microorganisms, but instead undergo selection that results, at least partially, in predictable microbial communities with few dominant phyla and their subgroups. Diverse communities of microorganisms including bacteria, fungi, archaea and protists are known to exist in harmony in the phyllosphere region. Actinobacteria, Firmicutes, Bacteroidetes and Proteobacteria dominate the phyllosphere community along with few bacterial genera including Bacillus, Pseudomonas, Massilia, Sphingomonas, Arthrobacter, Methylobacterium and Pantoea, which reside as the core phyllosphere microbial communities (Delmotte et al. 2009).
Most studies done on the abundance of organisms in the phyllosphere region have focused on bacteria and a lesser range to fungi as archaea are apparently not abundant in the phyllosphere (Knief et al. 2012; Finkel et al. 2011). Most bacteria on leaf surfaces do not occur as single cells or small groups of cells, as fungi tend to, but form larger assemblages which are particularly common at the depressions formed at the boundary of epidermal cells, along the veins and at the bases of trichomes, and in these depressions, they are generally lodged within extracellular polymeric substances (EPS). The EPS helps in providing a hydrated area to the bacterial surrounding and also concentrates detoxifying enzymes (Baldotto and Olivares 2008; Lindow and Brandl 2003).
The microbial communities of phyllosphere play a vital role in remediation of pesticides, hydrocarbon pollutants from atmosphere and cycling of nutrients as saprophytes, which are important for plant growth and healthy development serving as phytostimulators, biofertilizers and biopesticides to combat plant pathogens (Zhou et al. 2011; Ali et al. 2012) and affect global carbon and nitrogen cycles (Whipps et al. 2008)
The phyllosphere is an ephemeral or short-lived environment as compared to the rhizosphere, as the annual plants complete their life cycle within a single growth season, whereas perennial deciduous plants spontaneously form and shed leaves every year and evergreen plants do so sequentially throughout the year. Successful phyllosphere inhabitants can be expected to multiply and occupy newly formed niches while the leaves are expanding. Moreover, the waxy cuticle covering the plant epidermal cells is hydrophobic and reduces evaporation of water as well as leaching of plant metabolites, thus resulting in an oligotrophic environment.
Microorganisms dwelling within the phyllosphere live as commensals on their hosts; they can either be endophytic or epiphytic. Presently the extent to which plants are benefited by colonization of these commensal microbiota in their aerial parts is almost unknown (Innerebner et al. 2011; Knief et al. 2012). Although the exact extent of benefits which the plants receive from the endophytic microbes is not fully explored, the presence of surface appendages, comprising of trichomes and hydathodes, veins and stomata alter nutrient availability in addition to the environmental factors which affect them such as fluctuations in UV, temperature, humidity, water availability and light irradiation (Innerebner et al. 2011; Knief et al. 2012).
Consequently, the frequency of occurrence and multiplication of these microorganisms is uneven over the leaf surface (Remus-Emsermann et al. 2012) owing to the environmental variabilities and their encounter to the antimicrobial compounds produced by plants and other microbes. Trees adapted to xerophytic conditions have the capacity to secrete some soluble compounds which consequently result in alkaline and saline leaf surfaces, thus leading to saline or alkaline stress of phyllosphere microbes (Finkel et al. 2011). Figure 12.3 shows the interactions of the factors governing the different components of phyllosphere.
12.6.2 Metagenomics of Phyllosphere
The need for exploration of microbial life within the phyllosphere is crucially important for two reasons—first, understanding the survival strategies of disease-causing pathogens and developing methods to prevent their spread, thereby improving plant health to improve biomass production and prevent biomass losses. Second, there is an alarming rise in the food poisoning cases associated with vegetables, fruits and salads contaminated with food-borne pathogenic microbes especially bacteria, Salmonella enterica and Escherichia coli (Teplitski et al. 2011). Proper safety methods and decontamination strategies are important to prevent any outbreak affecting public health. Another interesting area of potential is phytoremediation, using microorganisms for removal of volatile pollutants such as phenol or benzene from the air using phyllosphere also called phylloremediation (De Kempeneer et al. 2004; Sandhu et al. 2007).
The triangular relationship between host, environment and pathogenic phyllospheric microbiota can give valuable insights into the population biology and genetics of phylloplane pathogens leading to more effective and sustainable disease management practices (Montarry et al. 2008).
The realization that a huge percentage of the microorganisms associated with plants, as those in other natural environments too, is metabolically active, but nonculturable in commonly used media and culture conditions, has had important accompaniment for plant microbiology and has brought about the beginning of culture-independent detection methods into phyllosphere research.
The recent developments in the area of exploration of microbiome in the phyllosphere, especially with the advances in metagenomics, environmental genomics, have greatly extended our understanding about the contribution of phyllosphere microbiome in plant-environment interactions along with the ecosystemic impact of the phyllosphere.
Analysis of the makeup of microbiome in leaf samples without any bias of cultivation based on amplicon sequencing and the 16S rRNA gene amplification has given many milestone results. There is a benefit of accessing a broader range of microbial inhabitants than culture techniques; however, the shortcomings comprise of the defects of PCR amplification, lack of quantitative information, sensitivity to inhibitory compounds, primer mismatch sensitivity and, primarily, the amplification of interfering plant organelle-derived RNA sequences (Saito et al. 2007; Berlec 2012).
The oncome of next-generation DNA sequencing has significantly reduced the experimental costs and allowed multiplexing of hundreds of samples in a single sequencing run. The 454 pyrosequencing platform was among the ‘first’ to be commonly executed in microbiota analysis through rRNA or whole-genome sequencing, shotgun metagenomics, ITS amplicon sequencing and transcriptional profiling (Delmotte et al. 2009; Rastogi et al. 2012). Ultra-high-throughput sequencing of microbial communities by ‘second’ next-generation sequencing technology like the Illumina platform (Degnan and Ochman 2012) yields amounts of sequence data that are of several order magnitude higher than generated by other techniques. Proteogenomics represents another important technical advancement which summates the application of metagenomic with metaproteomic analysis (Delmotte et al. 2009). Combined together, these technological revolutions are nobly helpful in the relative ecological analyses and help provide new introspections into the structure, function and heterogeneity of microbiome in the phyllosphere and different environments. In Table 12.3, many recent examples of phyllosphere studies that used high-throughput molecular methods are listed.
12.7 Conclusion
Plant microbiome is a composite ecosystem that hosts a number of interactions at ‘microbe-soil-microbe-plant-microbe interface’. Earlier it was difficult to study and understand the plant microbiome as a whole due to the unculturability of majority of microorganisms. Advances in latest molecular technologies, culture-independent methodology and next-generation sequencing have rapidly expanded the research in the area of microbial ecology of a particular niche and provided an in-depth knowledge of various genes present within the microbiome. Several studies have proved that the microbes are an integral part of plant genome, but their population is highly diverse and varies with the environmental as well as the biotic elements. Horizontal gene transfer and plant-based selection add to the plant microbiome diversity. Although, in the past decade, understanding of microbial ecology has grown very rapidly but to predict the ecophysical behaviour and to improve the plant productivity using custom-made microbiome, still more research is required.
References
Ali N, Sorkhoh N, Salamah S, Eliyas M, Radwan S (2012) The potential of epiphytic hydrocarbon-utilizing bacteria on legume leaves for attenuation of atmospheric hydrocarbon pollutants. J Environ Manag 93:113–120
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169
Baldotto LEB, Olivares FL (2008) Phylloepiphytic interaction between bacteria and different plant species in a tropical agricultural system. Can J Microbiol 54:918–931
Berendsen RL, Pieterse CM, Bakker PA (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17:478–486
Berlec A (2012) Novel techniques and findings in the study of plant microbiota: search for plant probiotics. Plant Sci 193:96–102
Bhatia S, Batra N, Pathak A, Green SJ, Joshi A, Chauhan A (2015) Metagenomic evaluation of bacterial and archaeal diversity in the geothermal hot springs of Manikaran, India. Genome Announc 3:e01544–e01514
Bulgarelli D, Schlaeppi K, Spaepen S, van Themaat EVL, Schulze-Lefert P (2013) Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64:807–838
Cocking EC (2003) Endophytic colonization of plant roots by nitrogen-fixing bacteria. Plant Soil 252:169–175
Compant S, Clément C, Sessitsch A (2010) Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem 42:669–678
Corneo PE, Suenaga H, Kertesz MA, Dijkstra FA (2016) Effect of twenty four wheat genotypes on soil biochemical and microbial properties. Plant Soil 404(1–2):141–155
Courtois S, Cappellano CM, Ball M, Francou FX, Normand P, Helynck G et al (2003) Recombinant environmental libraries provide access to microbial diversity for drug discovery from natural products. Appl Environ Microbiol 69:49–55
Crump BC, Koch EW (2008) Attached bacterial populations shared by four species of aquatic angiosperms. Appl Environ Microbiol 74:5948–5957
De Kempeneer L, Sercu B, Vanbrabant W, Van Langenhove H, Verstraete W (2004) Bioaugmentation of the phyllosphere for the removal of toluene from indoor air. Appl Microbiol Biotechnol 64:284–288
Degnan PH, Ochman H (2012) Illumina-based analysis of microbial community diversity. ISME J 6(1):183
Delmotte N, Knief C, Chaffron S, Innerebner G, Roschitzki B, Schlapbach R, Vorholt JA (2009) Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc Natl Acad Sci U S A 106:16428–16433
Dennis PG, Miller AJ, Hirsch PR (2010) Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol Ecol 72:313–327
Donn S, Kirkegaard JA, Perera G, Richardson AE, Watt M (2015) Evolution of bacterial communities in the wheat crop rhizosphere. Environ Microbiol 17:610–621
Edwards J, Johnson C, Santos-Medellín C, Lurie E, Podishetty NK, Bhatnagar S, Eisen JA, Sundaresan V (2015) Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci U S A 112:E911–E920
El-Badry MA (2016) Bacterial community metagenomic and variation of some medicinal plant rhizosphere collected form Sinai. SCIREA J Agric 1(1):16
Fahimipour AK, Kardish MR, Lang JM, Green JL, Eisen JA, Stachowicz JJ (2017) Global-scale structure of the eelgrass microbiome. Appl Environ Microbiol. https://doi.org/10.1128/AEM.03391-16
Fierer N, Lauber CL, Ramirez KS, Zaneveld J, Bradford MA, Knight R (2012) Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J 6(5):1007–1017
Filippidou S, Junier T, Wunderlin T, Lo CC, Li PE, Chain PS, Junier P (2015) Under-detection of endospore-forming firmicutes in metagenomic data. Comput Struct Biotechnol J 13:299–306
Finkel OM, Burch AY, Lindow SE, Post AF, Belkin S (2011) Geographical location determines the population structure in phyllosphere microbial communities of a salt-excreting desert tree. Appl Environ Microbiol 77:7647–7655
Gadhave KR, Devlin PF, Ebertz A, Ross A, Gange AC (2018) Soil inoculation with Bacillus spp. modifies root endophytic bacterial diversity, evenness, and community composition in a context-specific manner. Microb Ecol 76:741–750
Gang GH, Cho G, Kwak YS, Park EH (2017) Distribution of rhizosphere and endosphere fungi on the first-class endangered plant Cypripedium japonicum. Mycobiology 45:97–100
Gkarmiri K, Mahmood S, Ekblad A, Alström S, Högberg N, Finlay R (2017) Identifying the active microbiome associated with roots and rhizosphere soil of oilseed rape. Appl Environ Microbiol. https://doi.org/10.1128/AEM.01938-17
Glass GV (1976) Primary, secondary, and meta-analysis of research. Educ Res 5:3–8
Gottel NR, Castro HF, Kerley M, Yang Z, Pelletier DA, Podar M et al (2011) Distinct microbial communities within the endosphere and rhizosphere of Populus deltoides roots across contrasting soil types. Appl Environ Microbiol 77:5934–5944
Hallmann J, Quadt-Hallmann A, Mahaffee WF, Kloepper JW (1997) Bacterial endophytes in agricultural crops. Can J Microbiol 43:895–914
Innerebner G, Knief C, Vorholt JA (2011) Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl Environ Microbiol 77:3202–3210
Janssen PH (2006) Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol 72:1719–1728
Jumpponen A, Jones KL (2009) Massively parallel 454 sequencing indicates hyperdiverse fungal communities in temperate Quercus macrocarpa phyllosphere. New Phytol 184:438–448
Kent AD, Triplett EW (2002) Microbial communities and their interactions in soil and rhizosphere ecosystems. Annu Rev Microbiol 56:211–236
Knief C (2014) Analysis of plant microbe interactions in the era of next generation sequencing technologies. Front Plant Sci 5:216–239
Knief C, Delmotte N, Chaffron S, Stark M, Innerebner G, Wassmann R, Von Mering C, Vorholt JA (2012) Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J 6:1378–1390
Kumar V, AlMomin S, Al-Aqeel H, Al-Salameen F, Nair S, Shajan A (2018) Metagenomic analysis of rhizosphere microflora of oil-contaminated soil planted with barley and alfalfa. PLoS One 13:e0202127
Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Thurber RL, Knight R, Beiko RG (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821
Larkin RP, Hopkins DL, Martin FN (1993) Effect of successive watermelon plantings on Fusarium oxysporum and other microorganisms in soils suppressive and conducive to fusarium wilt of watermelon. Phytopathology 83:1097–1105
Lauber CL, Strickland MS, Bradford MA, Fierer N (2008) The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol Biochem 40:2407–2415
Leveau JHJ, Tech JJ (2010) Grapevine microbiomics: bacterial diversity on grape leaves and berries revealed by high-throughput sequence analysis of 16S rRNA amplicons. In: International symposium on biological control of postharvest diseases: challenges and opportunities, vol 90, pp 31–42
Li JG, Shen MC, Hou JF, Li L, Wu JX, Dong YH (2016) Effect of different levels of nitrogen on rhizosphere bacterial community structure in intensive monoculture of greenhouse lettuce. Sci Rep 6:25305–25314
Lindow SE, Brandl MT (2003) Microbiology of the phyllosphere. Appl Environ Microbiol 69:1875–1883
Lopez-Velasco G, Welbaum GE, Boyer RR, Mane SP, Ponder MA (2011) Changes in spinach phylloepiphytic bacteria communities following minimal processing and refrigerated storage described using pyrosequencing of 16S rRNA amplicons. J Appl Microbiol 110:1203–1214
Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, Del Rio TG, Edgar RC (2012) Defining the core Arabidopsis thaliana root microbiome. Nature 488:86–90
Mendes R, Kruijt M, De Bruijn I, Dekkers E, van der Voort M, Schneider JH, Piceno YM, DeSantis TZ, Andersen GL, Bakker PA, Raaijmakers JM (2011) Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332(6033):1097–1100
Mendes R, Garbeva P, Raaijmakers JM (2013) The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev 37:634–663
Mirete S, De Figueras CG, González-Pastor JE (2007) Novel nickel resistance genes from the rhizosphere metagenome of plants adapted to acid mine drainage. Appl Environ Microbiol 73:6001–6011
Montarry J, Cartolaro P, Delmotte F, Jolivet J, Willocquet L (2008) Genetic structure and aggressiveness of Erysiphe necator populations during grapevine powdery mildew epidemics. Appl Environ Microbiol 74:6327–6332
Müller T, Ruppel S (2014) Progress in cultivation-independent phyllosphere microbiology. FEMS Microbiol Ecol 87:2–17
Nakashima Y, Egami Y, Kimura M, Wakimoto T, Abe I (2016) Metagenomic analysis of the sponge Discodermia reveals the production of the cyanobacterial natural product kasumigamide by ‘Entotheonella’. PLoS One 11:e0164468
Newton AC, Gravouil C, Fountaine JM (2010) Managing the ecology of foliar pathogens: ecological tolerance in crops. Ann Appl Biol 157:343–359
Ofaim S, Ofek-Lalzar M, Sela N, Jinag J, Kashi Y, Minz D, Freilich S (2017) Analysis of microbial functions in the rhizosphere using a metabolic-network based framework for metagenomics interpretation. Front Microbiol 8:1606–1620
Oulas A, Pavloudi C, Polymenakou P, Pavlopoulos GA, Papanikolaou N, Kotoulas G, Arvanitidis C, Iliopoulos L (2015) Metagenomics: tools and insights for analyzing next-generation sequencing data derived from biodiversity studies. Bioinf Biol Insights 9:75–88
Padhi L, Mohanta YK, Panda SK (2013) Endophytic fungi with great promises: a review. J Adv Pharm Educ Res 3:152–170
Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, Buckler ES, Ley RE (2013) Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci U S A 110(16):6548–6553
Philippot L, Raaijmakers JM, Lemanceau P, Van Der Putten WH (2013) Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol 11:789–799
Prasad R, Kumar M, Varma A (2015) Role of PGPR in soil fertility and plant health. In: Egamberdieva D, Shrivastava S, Varma A (eds) Plant growth-promoting rhizobacteria (PGPR) and medicinal plants. Springer International Publishing, Cham, pp 247–260
Raaijmakers JM, Paulitz TC, Steinberg C, Alabouvette C, Moënne-Loccoz Y (2009) The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 321:341–361
Rahman MM, Flory E, Koyro HW, Abideen Z, Schikora A, Suarez C, Schnell S, Cardinale M (2018) Consistent associations with beneficial bacteria in the seed endosphere of barley (Hordeum vulgare L.). Syst Appl Microbiol 41:386–398
Rastogi G, Sbodio A, Tech JJ, Suslow TV, Coaker GL, Leveau JH (2012) Leaf microbiota in an agroecosystem: spatiotemporal variation in bacterial community composition on field-grown lettuce. ISME J 6:1812–1822
Remus-Emsermann MN, Tecon R, Kowalchuk GA, Leveau JH (2012) Variation in local carrying capacity and the individual fate of bacterial colonizers in the phyllosphere. ISME J 6:756–765
Rosenblueth M, Martínez-Romero E (2006) Bacterial endophytes and their interactions with hosts. Mol Plant-Microbe Interact 19:827–837
Saito A, Ikeda S, Ezura H, Minamisawa K (2007) Microbial community analysis of the phytosphere using culture-independent methodologies. Microbes Environ 22:93–105
Sandhu A, Halverson LJ, Beattie GA (2007) Bacterial degradation of airborne phenol in the phyllosphere. Environ Microbiol 9:383–392
Schlaeppi K, Dombrowski N, Oter RG, van Themaat EVL, Schulze-Lefert P (2014) Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc Natl Acad Sci U S A 111:585–592
Shakya M, Gottel N, Castro H, Yang ZK, Gunter L, Labbé J et al (2013) A multifactor analysis of fungal and bacterial community structure in the root microbiome of mature Populus deltoides trees. PLoS One 8:e76382
Shrivastava S, Prasad R, Varma A (2014) Anatomy of root from eyes of a microbiologist. In: Morte A, Varma A (eds) Root engineering, vol 40. Springer, Berlin, pp 3–22
Singh D, Raina TK, Kumar A, Singh J, Prasad R (2019) Plant microbiome: a reservoir of novel genes and metabolites. Plant Gene. https://doi.org/10.1016/j.plgene.2019.100177
Suman A, Shasany AK, Singh M, Shahi HN, Gaur A, Khanuja SPS (2001) Molecular assessment of diversity among endophytic diazotrophs isolated from subtropical Indian sugarcane. World J Microbiol Biotechnol 17:39–45
Teplitski M, Warriner K, Bartz J, Schneider KR (2011) Untangling metabolic and communication networks: interactions of enterics with phytobacteria and their implications in produce safety. Trends Microbiol 19:121–127
Thomas P (2017) Potential applications of endophytic microorganisms in agriculture. Biotechnol Dev 19:3–23
Toju H, Tanabe AS, Yamamoto S, Sato H (2012) High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS One 7:e40863
Truyens S, Weyens N, Cuypers A, Vangronsveld J (2015) Bacterial seed endophytes: genera, vertical transmission and interaction with plants. Environ Microbiol Rep 7:40–50
Uroz S, Buée M, Murat C, Frey-Klett P, Martin F (2010) Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil. Environ Microbiol Rep 2:281–288
Varon-Lopez M, Dias ACF, Fasanella CC, Durrer A, Melo IS, Kuramae EE, Andreote FD (2014) Sulphur-oxidizing and sulphate-reducing communities in Brazilian mangrove sediments. Environ Microbiol 16:845–855
Vorholt JA (2012) Microbial life in the phyllosphere. Nat Rev Microbiol 10:828–840
Whipps J, Hand P, Pink D, Bending GD (2008) Phyllosphere microbiology with special reference to diversity and plant genotype. J Appl Microbiol 105:1744–1755
Yadav AN, Kumar V, Dhaliwal HS, Prasad R, Saxena AK (2018) Microbiome in crops: diversity, distribution, and potential role in crop improvements. In: Prasad R, Gill SS, Tuteja N (eds) Crop improvement through microbial biotechnology. Elsevier, pp 303–327
Zhang Y, Xu J, Riera N, Jin T, Li J, Wang N (2017) Huanglongbing impairs the rhizosphere-to-rhizoplane enrichment process of the citrus root-associated microbiome. Microbiome 5:97–114
Zhou Y, Qiao X, Li W, Xu J, Wang W, Chen X (2011) Phyllosphere bacterial communities associated with the degradation of acetamiprid in Phaseolus vulgaris. Afr J Biotechnol 10:3809–3817
Acknowledgement
PB thanks DST-SERB: SB/YS/LS-213/2013 for financial support.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Bhargava, P. et al. (2019). Metagenomics as a Tool to Explore New Insights from Plant-Microbe Interface. In: Varma, A., Tripathi, S., Prasad, R. (eds) Plant Microbe Interface. Springer, Cham. https://doi.org/10.1007/978-3-030-19831-2_12
Download citation
DOI: https://doi.org/10.1007/978-3-030-19831-2_12
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-19830-5
Online ISBN: 978-3-030-19831-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)