Abstract
Geminiviruses are a major threat to world agriculture, and breeding resistant crops against these viruses is one of the major challenges faced by both plant pathologists and biotechnologists. In the past, most of these strategies follow the conceptual development ranging from coat protein-mediated restricted viral propagation to the expression of mutant or truncated viral proteins that interfere with virus infection, or RNA molecule-mediated gene silencing approach transcription of viral RNA sequences that silence the expression of virus genes. However much of the progress has been made so far in this direction observes limited success in field, but still research is running and new approaches such as CRISPR/Cas9 have found space in laboratories. To date, no comparative data has been published or available that examines the merit of different approaches which have been used against this class of viruses. There is a common belief among the geminivirologists across the globe about the recombination and mutation capacity as the main reason for the appearance of new species and breaking resistance. This chapter deals with different strategies which have been used to curb geminivirus spread.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
1 Introduction
Plant viruses are important pathogens causing enormous losses to agricultural crops, thus affecting the economy of a country. Any plant which is grown by humans for food, fiber, or any other use is virtually infected by one or more viruses. Although the direct effects of plant viruses on human beings are not substantial as compared to animal viruses, the damage caused by the former is considerable when it comes to food supply.
Geminiviruses, grouped into family Geminiviridae, are widely distributed plant viruses infecting a wide range of plants from monocots such as maize to dicots such as cassava and tomato (Hanley-Bowdoin et al. 1999). They all share two distinctive features: (1) the geminate morphology of the virion particle and (2) the nature of their genetic material that consists of one or two single-stranded DNA (ssDNA) molecules (2.5–3.0 kb in length), depending on the genera. Differences in the genetic organization of their genomes as well as their host range and insect vectors serve as criteria to recognize nine different genera.
The single-stranded (ss) circular DNA genome of geminiviruses is packed into twin-shaped virions (Zhang et al. 2001). Virions are twinned, geminate, icosahedral, non-enveloped, 38-nm long, and 22 nm in diameter and contain 22 capsomeres per nucleocapsid. Genomic DNA is either mono- or bipartite, circular ssDNA. There are coding regions in both the virion sense (+) and complementary (−) sense strands. They have a single coat protein (CP) of 28–34 kDa and a replication (Rep) protein of 41 kDa, which initiates rolling circle replication. There is a potential stem-loop structure in the intergenic region that includes a conserved nonanucleotide sequence (TAATATTAC), where single-stranded DNA synthesis is initiated.
1.1 Replication of Geminivirus
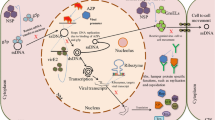
Geminivirus DNA replication follows a rolling circle strategy (Saunders et al. 1992), which resembles that of prokaryotic ssDNA replicons (Fig. 1). The initial stage encompasses the conversion of the ssDNA genome into a dsDNA intermediate product (Saunders et al. 1992). It requires a priming step at the so-called (−)- strand or c-strand origin. In the second stage, ds DNA intermediate acts as a template for genome amplification through a RCR mechanism, a mechanism common among bacterial systems, where it is used frequently for viral and plasmid DNA replication (de la Campa et al. 1990). The initial priming event depends on the interaction of the viral initiator protein (rep protein) with cis-acting signals of the genetically defined origin. A 9-nt sequence (TAATATT↓AC) is present invariably in all the geminivirus sequences to date. This sequence contributes to form a stem-loop structure to which Rep can get access and carry out the initiation reaction, a single-stranded, site-specific, endo-nucleolytic cleavage that provides a free 3′-OH primer terminus for further elongation during the RCR stage (Khan and Dijkstra 2006).
A mastrevirus, transmitted by leafhopper, generally infects monocots and possesses monopartite genome (type species: Maize streak virus). A curtovirus is leafhopper-transmitted, infects dicots, and has monopartite genome (type species: Beet curly top virus). Begomoviruses are whitefly-transmitted, infect dicots, and have either monopartite or bipartite genome (type species: Bean golden mosaic virus-Puerto Rico). The members of genus Topocuvirus are transmitted by treehoppers, infect dicots, and have monopartite genome (type species: Tomato pseudo curly top virus). Mastreviruses mostly infect monocots in the family Poaceae; two species, namely Bean yellow dwarf virus (BeYDY, Liu et al. 1997) reported from South Africa and Tobacco yellow dwarf virus (Morris et al. 1991) from Australia, are also known to infect dicots. Chickpea chlorotic dwarf virus from India is transmitted by leafhopper and is considered to be a tentative mastrevirus species (Horn et al. 1993). Different leafhopper species transmit mastreviruses in a persistent circulative manner and the genus includes about a dozen species. MSV causes maize streak disease, one of the oldest known plant viral diseases reported about a century ago (Fuller 1901). It is one of the three economically most important plant virus diseases in Africa (Shepherd et al. 2009). Other mastreviruses occur in Asia, Australia, or Pacific islands and Wheat dwarf virus is prevalent in Europe.
Curtoviruses are transmitted by leafhoppers and possess a monopartite genome; Begomoviruses are spread by whiteflies and most of them have a bipartite genome referred to as DNA-A and DNA-B. Sequence comparison between the genera has led to the suggestion that Curtoviruses have evolved from a recombination of ancient mastreviruses and begomoviruses (Chen et al. 2010).
Type species of genus Topocuvirus is Tomato pseudo curly top virus. Its genome encodes six proteins and members of these genera are known to spread by treehoppers. Like most of the geminiviruses, these genera also infect dicotyledonous plant.
The name of genus Begomovirus is derived from the first two letters in the name of the type species, Bean golden mosaic virus (Padidam et al. 1996). DNA-A and DNA-B components of bipartite Begomoviruses are each 2.6 Kb in size and share a noncoding intergenic or common region (CR) of 180–200 nucleotides (nt) that is typically identical for cognate components of the same virus species. The CR contains modular cis-acting elements of the origin of replication (ori) (Fontes et al. 1994), while five ORFs capable of encoding proteins >10 kilodaltons (Kd) in size are conserved among the DNA-A component of Begomoviruses. The coat protein, CP, is encoded by the ORF (AV1) and is the most highly conserved gene among Begomoviruses. The replication-associated protein (Rep) encoded by the AC1 ORF initiates viral DNA replication, and specificity of replication is mediated through interactions of REP with cis-acting elements of the ori (Jupin et al. 1994; Laufs et al. 1995; Bisaro 2006). DNA B encodes two polypeptides, BV1 and BC1; both are essential for systemic movement and have been shown to influence host range (Lazarowitz et al. 1992).
Genus Becurtovirus contains two recognized species, namely Beet curly top Iran virus and Spinach curly top Arizona virus. Members of both species unusually have the “TAATATTAC” nonanucleotide instead of “TAAGATTCC” nonanucleotide. Becurtoviruses infect dicotyledonous plants. Members of the genus Eragrovirus have been reported to infect monocotyledonous Eragrostis curvula (weeping love grass) in the Kwa-Zulu Natal region of South Africa.
Turnip curly top virus (TCTV) is currently the only species within the genus Turncurtovirus. All the known TCTV isolates reported so far possess “TAATATTAC” nonanucleotide sequence motif. It infects dicotyledonous plants.
Genus Capulavirus contains four species, transmitted by an aphid, and genus Grablovirus has only one species. Not much information is available so far.
2 Economic Losses Due to Geminivirus Disease
Though geminivirus may be the subject of interest in molecular virology because of their relatively small genome, and their ability to manipulate and reprogram host cellular processes to their advantage (Rojas et al. 2005), they cause significant economic losses to several crops across the globe. Some notable examples are losses in cotton in Asian countries such as India and Pakistan (Briddon and Markham 2001; Khan and Ahmad 2005). In Pakistan, the cotton leaf curl disease causes loss of 5 billion dollars, and it seriously challenged national economy. In India, every year cotton leaf curl disease caused severe losses, ranging between 10 and 50%, and in 2015, a serious epidemic break caused yield loss up to 100% in several cotton growing areas in western parts of India. Cassava and maize in Africa are prone to geminiviral infection (Thresh and Cooter 2005; Shepherd et al. 2009). The loss observed due to this is several million dollars every year. Legumes in India are infected with begomovirus that cause annual yield losses estimated at $300 million (Varma and Malathi 2003). An annual loss of $140 million is estimated due to Tomato leaf curl viruses (ToLCVs) in Florida, USA, and continues to be a major constraint to tomato production worldwide.
3 Geminivirus Management Strategies
3.1 Conventional Approaches
Conventional management of geminivirus diseases is based on the applications of different insecticides to control insect vectors (Singh 1990; Lapidot and Friedmann 2002). The insect control required massive use of insecticides and chemicals every year resulting in the development of resistance in the vector. Furthermore, chemicals and insecticides cause environmental pollution, health hazards, and phytotoxicity besides their high cost. Physical barriers such as fine mesh screens have been used in the Mediterranean basin to protect crops (Cohen and Antignus 1994). UV-absorbing plastic screens have shown to inhibit penetration of whiteflies into greenhouses (Antignus et al. 2001). However, use of physical barriers is not the best solution owing to high cost and creates problems of shading, overheating, and poor ventilation (Lapidot and Friedmann 2002).
Cultural practices such as crop-free periods, altering dates, crop rotation, weed and crop residue disposal, high planting densities, floating row cover, mulches, trap crop, or living barriers performed well against several plant viruses in general. This control is based on the removal of infected plants, production, and using of virus-free planting stock. Use of virus-free seed is advisable, as seed-borne viruses are transmitted in the embryo of the seed, but some viruses, such as Tobacco mosaic virus (TMV) in tomato, are transmitted through contamination of the seed coat. In this case, seed transmission may be controlled by sterilizing the seed coat in hydrochloric acid or sodium hypochlorite. The suggested methods of cultural practices are, however, not much successful against geminiviruses. Alternatively, it was suggested that cultural practices when combined with insecticides are more effective (Hilje et al. 2001).
3.1.1 Control Through Resistant Cultivars
Development of resistant plant varieties against virus or its vector through breeding techniques is yet another attractive approach for controlling viral diseases. Virus-resistant crops increase profitability for the breeders, as this approach requires no extra input toward production of virus-free planting materials or control of virus vectors (Valkonen 1998).
There seems to be disagreement between plant breeders and plant pathologists. On one end, breeders are interested in improving the performance of a plant variety under field conditions, but on the other end, plant pathologists focus more on the fate of the plant virus.
Another challenge to the breeders are emergence of new strains of geminiviruses due to frequent recombination and changes in cultivation habits (Padidam et al. 1999). Co-infection by two or more viruses to a crop plant is another issue. The different challenges posed by viruses have necessitated the development of plants, which confers multi-virus resistance. Some of the approaches addressed by several authors involve in assembling of different resistance genes, which will ultimately give multi-virus resistance (Lapidot and Friedmann 2002). However, this idea poses another problem to the breeders in distinguishing two different traits/genes to be combined and a continuous system to check the resistance against newly emerged virus.
3.1.2 Cross-Protection
It has been long observed that plants infected by mild strain can be protected against infection by more severe strain of the related virus, a biological term called cross-protection. The cross-protection test has been previously regarded as an important means for identifying the same strain or a distinct species of plant virus. This method is also quoted as plant vaccination by some authors (Nicaise 2014). This phenomenon was first discovered by McKinney in 1926; since then several reports of cross-protection have been identified (McKinney 1926; Crowdy and Posnette 1947; Fletcher 1978; Wang 1991; Wen et al. 1991; Hugues and Ollennu 1994; Nakazono-Nagaoka et al. 2009; Kurth et al. 2012).
In detail, this strategy is dependent on the prowess of the primary virus, whose infection is weak (either symptomless with low viral load or mild symptom with low virus titer). This triggers virus-induced gene silencing (VIGS), which targets both mild infection virus and challenge viruses (Nishiguchi and Kobayashi 2011). Primary virus acts as vaccines and is further classified as the attenuated and the engineering viruses. An attenuated virus corresponds to a weak isolate that triggers cross-protection against virulent isolates of the same virus or closed related viruses (Ziebell and Carr 2010; Nishiguchi and Kobayashi 2011). However, cross-protection strategy also bears some disadvantages. Infection by the mild strain might cause significant yield loss sometimes. Another fear is that a severe strain might evolve from the mild strain leading to more serious disease incidence.
Conventional methods are effective but also protracted and expensive. To overcome these challenges and limitations, nonconventional methods of genetic engineering are practiced.
3.2 Nonconventional Methods
3.2.1 Pathogen-Derived Resistance
Concept of pathogen-derived resistance or parasite-derived resistance was developed by Sanford and Johnston in 1985. Subsequently, Powell-Abel et al. (1986) applied this approach and opened new horizons for plant pathologists following the development of virus-resistant crops. Since then, several attempts were made to develop transgenic plants using virus-derived genes or genome fragments and led to the development of virus-resistant plants for commercial application (Beachy 1993; Wilson 1993; Baulcombe 1994, Lomonossoff 1995).
3.2.1.1 Coat Protein
CP gene was the first and one of the most widely used genes to confer pathogen-derived resistance (PDR) against plant viruses (Prins 2003). Virus resistance has been achieved by transforming the plants with viral CP gene. Remarkable success has been achieved in transformed tobacco showing resistance to (TMV) (Powell-Abel et al. 1986) and transgenic papaya plants resistant to Papaya ringspot virus (Gonsalves 1998).
The CP gene of tobacco mosaic virus TMV was manipulated for the first time demonstrating virus-derived resistance in transgenic plants (Powell-Abel et al. 1986). In these experiments, transgenic tobacco plants expressing high levels of TMV CP were more resistant to TMV virions than to TMV RNA inocula. Based on the observation, they suggested that CP-mediated protection against TMV was through the inhibition of virion disassembly in the initially infected cells (Register and Beachy 1988). CP-mediated resistance has been successfully applied to numerous crop species (Beachy 1997) (see Table 1).
3.2.1.2 Movement Protein
Cell-to-cell movement of plant viruses in host plant is associated with the movement protein encoded by viruses. MP interacts with the plasmodesmata and thus modifies it to facilitate cell-to-cell movement of virus. Transgenic tobacco plants that expressed a gene encoding a defective TMV movement protein showed resistance to Tobacco rattle virus, Tobacco ringspot nepovirus, Alfalfa mosaic alfamovirus, and Cucumber mosaic virus (Cooper et al. 1995). Resistance shown by transgenic expression of a dysfunctional TMV MP is probably due to competition for plasmodesmatal binding sites between the mutant MP and the wild-type MP of the inoculated virus (Lapidot et al. 1993). An interesting and potentially useful attribute of MP-mediated protection is the broad spectrum efficacy of the resistance mechanism (Table 1).
3.2.1.3 Replication-Associated Protein
Rep-mediated approach is the second most widely used method to control plant viruses (Lomonossoff 1995; Wintermantel et al. 1997). The engineering of geminivirus resistance using Rep gene has been achieved in model host species against Begomoviruses. Expression of full-length or a truncated N-terminal portion of Rep gene of African cassava mosaic virus (ACMV) inhibits replication of ACMV in Nicotiana tabacum protoplasts. A modest degree of ACMV resistance was achieved by the expression of full-length Rep gene in experimental plant tobacco. None of transgenic tobacco plants was resistant to distantly related viruses TGMV and Beet curly top virus (sharing Ca. 60% Rep amino acid sequence identity with ACMV), suggesting that resistance was probably ACMV specific or to its closely related viruses (Hong and Stanley 1996).
Rep gene has been successfully manipulated to engineer resistance against Tomato yellow leaf curl Sardinia virus (TYLCSV) in N. benthamiana (Noris et al. 1996) and tomato (Brunetti et al. 1997) against cotton leaf curl disease in experimental plant tobacco (Asad et al. 2003) (Table 1).
3.3 RNA Silencing
In recent years, resistance-mediated by RNA silencing seems to be one of the most promising approaches. RNA silencing is an ancient mechanism involved in different fundamental processes, such as gene regulation, de novo histone and DNA methylation, establishment of heterochromatin, defense against viruses, and control of transposon mobility (Baulcombe 2004; Voinnet 2005). RNA silencing involves suppression of gene expression by sequence-specific degradation of mRNA in diverse eukaryotes. The RNA silencing phenomena was first discovered and termed post-transcription gene silencing (PTGS) in plants (Napoli et al. 1990), quelling in fungi (Cogoni and Macino 1997), and RNA interference (RNAi) in animals (Cogoni et al. 1996; Fire et al. 1998).
The key molecules which are involved in the RNA silencing pathways are ribonuclease Dicer (RNA-dependent RNA polymerase, RDR) and Argonaute (AGO). The RNA silencing machinery in plants is more evolved than in fungal and animal systems. Its pathway follows a dsRNA trigger: a processor called Dicer or a Dicer-like (DCL) protein generating small RNAs (siRNAs or miRNAs) of 21–24 nt in length in an effector complex called RISC (RNA-induced silencing complex) in which the AGO protein plays a key role. The siRNA-guided AGO actually cleaves target RNA, which is recognized by RDR. The Arabidopsis genome encodes four DCL enzymes, 6 RDRs, and 10 AGO proteins.
There are three different pathways in the gene silencing mechanism: (1) cytoplasmic short interfering (siRNA) silencing, (2) silencing of endogenous mRNAs by microRNAs (miRNAs), and (3) DNA methylation and suppression by transcription (Vanitharani et al. 2005). The siRNA silencing is actually post-transcriptional gene silencing (PTGS) (Bisaro) in 2006 resulting in the production of 21–25 nucleotide siRNA from inducing dsRNA leading to the degradation of mRNA (Hamilton and Baulcombe 1999). Practically, linking the sense and antisense sequences by an intron, which is eventually spliced, resulted in efficient silencing in plants (Smith et al. 2000; Wesley et al. 2001). The mechanism is now better understood and widely used to engineer plant against virus infection (Tenllado et al. 2003). Several vectors have been developed for the efficient expression of such hairpin dsRNA in plants (Wesley et al. 2001; Khatoon et al. 2016; Table 1). The dsRNA region is processed into small interfering RNAs (siRNAs), which guide silencing complexes to target regions on RNA or DNA (Fig. 2).
Geminiviruses have an ability to suppress the induced RNA silencing. About 35 RNA silencing suppressor proteins have been identified in recent years from several plant and animal viruses. There are mainly three distinct phases reported in the RNA silencing process: initiation, maintenance, and systemic signaling (Llave et al. 2002). These suppressor proteins do not share homology at either sequence or viral functional levels; it is assumed that these suppressor proteins might target similar or different steps of the RNA silencing pathway.
A study on cassava-infecting geminiviruses demonstrated that AC4 region has the capacity to suppress the induced post-transcriptional gene silencing (Vanitharani et al. 2004).
It is a well-known fact that geminiviruses have no dsRNA stage in their replication cycle, but they do induce the production of virus-specific siRNA and have been shown to trigger PTGS in infected plants, as demonstrated by Chellappan et al. (2004). Another observation is that an increased accumulation of cassava-infecting geminivirus-derived siRNAs in infected cassava is associated with a corresponding decrease in disease symptom severity (Chellappan et al. 2004), providing a clue for RNAi as an adaptive defense against geminiviral infection in plants. According to the reports of Vanitharani et al. (2005), both silencing mechanism PTGS and TGS are applicable to geminiviruses, whereas for RNA viruses only TGS is applicable.
Betasatellite molecule is associated with a number of monopartite begomoviruses, and it induces symptom. The betasatellite associated with Tomato yellow leaf curl China virus-Y10 has shown to behave as a silencing suppressor in N. benthamiana 16c plants (Cui et al. 2005). It was shown that TYLCCV along with betasatellite DNA could prevent silencing in newly emerging leaves of infected plants.
Intergenic region (IR) is also a potential candidate which could play a crucial role for the generation of siRNA-mediated resistance. IR contains origin of replication and divergent promoter (Zulma et al. 2002). Methylation of this region can hamper the virus’s ability to thrive in the plant host. The region of homology ranges from 80 to 100 bp in length and includes some common motifs found in Begomoviruses, such as nonanucleotide sequence (TAATATTAC). This finding is successfully applied in the generation of transgenic cotton, which showed complete protection against CLCuD (Khatoon et al. 2016). In an attempt to generate siRNA-mediated African cassava mosaic virus (ACMV, genus Begomovirus) resistant transgenic plants, a 360 nt fragment corresponding to IR of ACMV DNA-A was cloned in sense and antisense orientation, interrupted with a synthetic plant intron. N. benthamiana plants were stably transformed with an intron-spliced dsRNA construct cognate to bidirectional promoter of ACMV DNA. The transgenic lines expressed multiple siRNAs species upon ACMV inoculation. It was demonstrated that the mRNA transcribed from ACMV genome was degraded by 21–22 nt siRNA and the begomoviral genomic DNA appeared to be methylated by 24–25 nt siRNA. It was demonstrated that silencing was associated with hypermethylation of promoter sequence and did not occur with heterologous begomovirus infection (Dogar 2006).
PTGS occurs as a natural defense mechanism against virus infection. Virus or its derivative or replication intermediate acts as a pathogenic agent by the host. This triggers a response responsible for the progressive slowdown in virus accumulation (Ratcliff et al. 1997, 1999). However, in this situation, viruses counteract the host response by encoding suppressors of PTGS (Voinnet et al. 1999; Hamilton et al. 2002). PTGS in plants can be triggered due to the presence of an inverted repeat in the transcribed region of a transgene (Jones et al. 1999). Tobacco plants transformed with constructs that produce RNAs capable of duplex formation induced virus immunity or gene silencing when targeted against virus or endogenous genes (Smith et al. 2000; Waterhouse et al. 1998). There are strong evidences in support of dsRNA as an inducer of PTGS in both the plant and animal kingdoms.
3.4 MicroRNA-Based Resistance
MicroRNA is a small noncoding sequences, thought to be useful in gene regulation during development and in the stress conditions in eukaryotes. They are about 18–25 nucleotides in length and play a major role in the negative regulation at post-transcriptional level for the normal activity of the organisms under stress. These miRNAs sharing the homology with the target mRNAs in plant are capable of causing the RNA-induced silencing lead to mRNA cleavage (Fig. 3).
3.4.1 Origin and Evolutionary Role of miRNA
The animal miRNA is believed to be originated about 420 million years ago from the metazoans as common ancestors (Pasquinelli et al. 2003). The hypothesis regarding origin of plant miRNAs is not clear. Many experimental and computational predictions of miRNA and its targets led to identification of a large number of miRNA families that are evolutionarily conserved across all major lineages of plants including bryophytes, lycopods, ferns, and seed plants (Axtell and Bartel 2005; Zhang et al. 2005, 2006; Jones-Rhoades et al. 2006), and even many are reported to be conserved between monocots and dicots (Sunkar and Jagadeeswaran 2008). There are reports of 21 miRNA families (predominantly 156, 159, 160, 162, 164, 166–169, 171, 172, 319, 390, 393–399, and 408) which are identified. These miRNAs are highly conserved between all three sequenced plant genomes: Arabidopsis, Oryza sativa, and Populus trichocarpa (Axtell and Bowman 2008). Several plant miRNAs are believed to be universal among land plants; they are less conserved than animal miRNA (Axtell and Bowman 2008). Recent advanced techniques of DNA sequencing and research into miRNA gene complements of individual species have listed several “non-conserved” miRNAs (nearly 48 in Arabidopsis), which outnumbered the “conserved” miRNAs. On the other hand, several (16 out of 48) non-conserved Arabidopsis miRNA genes are believed to exhibit significant sequence similarity outside the miRNA binding sites within their putative target genes. These features signify that non-conserved miRNAs originated recently with high birth and death rates (Rajagopalan et al. 2006; Fahlgren et al. 2007). Various roles of microRNAs are described in the recent past, but plant defense mechanism to viruses is one of the several anticipated roles, which is yet to be explored in full potential. There has been a successful application of this strategy in recent times against cotton leaf curl virus (CLCuV) (Akmal et al. 2017; Shweta et al. 2018)
3.4.2 Biogenesis and Mechanism of miRNAs
MicroRNAs are small non-protein coding RNAs consisting of 21–24 nucleotides present in intergenic regions of genome. The extensive complementarily between the target mRNA and the miRNA leads to target mRNA cleavage and gene silencing in plants. Biogenesis of plant miRNA occurs in multiple steps to form mature miRNAs from miRNA genes. Initially, the miRNA genes are transcribed by their own promoters, resulting in primary transcripts (pri-miRNAs) (Tang et al. 2003). Further, these pri-miRNAs fold up into unique stem-loop structures. This structure is further identified and cleaved by the Dicer-like (DCL) enzyme of the RNase III family (Tang et al. 2003). In the plant nucleus, DCL-1, in association with HYL1 protein, processes the pri-miRNAs. The mature miRNA then unwinds into single-strand miRNA by helicase (Bartel 2004) and assembled into the RISCs complex, which contains Arganoute (AGO1) protein to carryout silencing reactions. The plant miRNA will have the complementarity with the target mRNA and lead to cleavage of it (Llave et al. 2002; Bartel 2004; Dugas and Bartel 2004). The high base pairing requirement of plant miRNAs results in limited number of targets compared to animal miRNA. The mRNA cleavage is considered to be predominant mechanism used by plant miRNAs, but reports also prove the presence of translational inhibition by plant miRNA and other siRNAs (Chen et al. 2010; Aukerman and Sakai 2003). The method of overexpression of miRNAs has proved to a promising approach against several pathogens including geminivirus also (Baldrich and Segundo 2016). Recently, overexpression of Gossypium hirsutum miRNAs (miR398/miR2950) in G. hirsutum has been demonstrated to suppress symptoms of cotton leaf curl disease (CLCuD) caused by Cotton leaf curl Multan virus (genus Begomovirus, family Geminiviridae) in association with circular, single-stranded DNA molecule satellite molecule (Akmal et al. 2017).
3.5 Artificial microRNA
The amiRNA acts as a specific, powerful, and robust tool that can be applied to study metabolic pathways, gene functions, and for improving favorable traits. The AmiRNAs have also been used as a powerful tool to produce antiviral transgenic plants. The transgenic Arabidopsis expressing amiRNAs targeting the viral mRNA sequences encoding gene silencing suppressor P69 of turnip yellow mosaic virus (TYMV) and HC-Pro of turnip mosaic virus (TuMV) are specifically resistant to TYMV and TuMV (Niu et al. 2006). There have also been reports of developing resistant tobacco and Arabidopsis by expressing amiRNAs. These amiRNAs were also used for generating resistance against Watermelon Silver Mottle Virus in tobacco (Kung et al. 2012). The idea of amiRNA was also successfully used against monopartite and bipartite Tomato leaf curl virus (Vu et al. 2013).
In principle, artificial microRNA (amiRNA) technology is based on designing miRNA or engineering miRNA artificially by mimicking the intact secondary structure of endogenous miRNA precursors (Ossowski et al. 2008; Sablok et al. 2011). It was demonstrated that altering several nucleotides within sense and antisense strands of miRNA has no effect on its biogenesis and maturation, as long as secondary structure of its precursor remains unaltered. It was also demonstrated that amiRNAs when expressed under constitutive or tissue-specific promoters can downregulate a number of endogenous genes without affecting the expression of other unrelated genes (Alvarez et al. 2006).
3.6 Antisense RNA Approach
Antisense RNA approach, based on the manipulation of potential gene sequence of targeted virus, would prove to be a significant approach against the virus. It ends up into degradation of mRNA in a processing body inside the cytoplasm of the cell. Antisense technology involves the cloning of a gene in reverse orientation with respect to the promoter such that the coding strand acts as a template and the sequence of mRNA is the same as the opposite strand or the coding “sense” strand. The gene cloned in reverse orientation when transcribed gives rise to mRNA having the sequence complementary to the sense mRNA. The RNA-RNA binding of the sense-antisense RNA strand leads to inhibition of sense mRNA expression.
Naturally occurring antisense RNAs are known to regulate gene expression in plants too. Antisense RNA arises when transcription of a gene proceeds in the strand opposite to template in the absence of a strong transcription termination site in the short intergenic region. On the basis of certain basic regulatory mechanisms, they are classified into three classes:
-
Class I–antisense RNAs are directly complementary to the coding region of target gene, resulting in direct inhibition of translation or mRNA destabilization.
-
Class II–RNAs include those that bind to the non-coding regions of target RNA resulting in indirect effects produced by, e.g., alternative secondary structure formations that sequesters the ribosome binding site.
-
Class III–antisense RNA regulates transcription of target mRNA by a mechanism similar to transcription attenuation.
The antisense inhibition may also take place at translational phase as antisense transcript would compete with the ribosomes to bind 5′ end of the sense RNA, hence inhibiting the translation. Antisense RNA technology is a proven strategy that has been widely used for crop improvement. The most commercial example is the development of Flavr Slavr tomatoes in which tomato plants were transformed with antisense Polygalacturonase gene (gene responsible for cell wall degradation and fruit softening) (Kumria et al. 1998 and references therein). The resultant transgenic plants showed longer shelf life. Other examples are antisense inhibition of chitinase gene expression which resulted in enhancement of fungal disease susceptibility in Arabidopsis plants and alteration of lignin composition by inhibiting lignin biosynthetic enzymes in tobacco (Kumria et al. 1998 and references therein). Its application is not limited in enhancing quality of food crops, but it has also been used to confer resistance to viral plant infections. Transgenic potato plants expressing antisense RNA to potato leaf roll luteovirus coat protein were resistant to the infection (Kumria et al. 1998 and references therein). Antisense RNA construct against cotton leaf curl virus has been successfully developed by Asad et al. (2003). Antisense construct targeting coat protein region of the begomovirus isolated from leaf curl disease affected papaya plant was also developed by Sinha et al. (2017).
3.7 Application of CRISPR/cas9 in the Generation of Geminivirus Resistance
CRISPR/Cas9 is a molecular immunity system, exclusively found in prokaryotic system (Fig. 4). This system actually acts against invading nucleic acids, following methods of horizontal gene transfer and phages, as suggested by Marraffini and Sontheimer (2008). This molecular memory concept further follows the process in which bacteria and archaea acquire short pieces or even spacers from these invading nucleic acids and incorporate them within their genome (Bolotin et al. 2005). In case of subsequent infection(s), these short pieces are transcribed as part of CRISPR array. Further, after transcription and maturation, the CRISPR RNA (CrRNA) can help guide the Cas9 endonuclease to scan the invading DNA and cleave the target sequence (Nunez et al. 2016).
A simplified CRISPR mechanism: In the acquisition phase (I phase), foreign DNA is incorporated into the bacterial genome at the CRISPR loci. CRISPR loci is then transcribed and processed into crRNA during crRNA biogenesis (II phase). During interference, Cas9 endonuclease form a complex with crRNA, separate tracrRNA cleaves foreign DNA containing a 20-nucleotide crRNA complementary sequence adjacent to the PAM sequence (III phase)
Some recent studies demonstrated the efficiency of this system against geminiviruses. In one of the studies (Ali et al. 2015), N. benthamiana plants expressing CRISPR/Cas9 exhibited resistance against Tomato yellow leaf curl virus, Beet curly top virus, Merremia mosaic virus. Other studies (Ji et al. 2015) demonstrated interfere in virus activities in N. benthamiana against Bean yellow dwarf virus (BeYDV) and BCTV, respectively. Also, later it was suggested that catalytically inactive Cas9 can be used to mediate virus interference. This action eliminates concerns of off-target activities in the plant genome. In model plant N. benthamiana targeting conserved nonanucleotide sequence of Cotton leaf curl Kokhran virus (CLCuKoV), broad spectrum resistance was achieved (Ali et al. 2016). However, all the studies used N. benthamiana, which is a model plant system. According to Woo et al. (2015), CRISPR/Cas9 system can be used to engineer “non-transgenic” virus-resistant varieties. Major advantage to use this system is that progenies of genome-edited plant carrying desired edits can be selected easily (Kanchiswamy 2016).
4 Conclusion
There is a fine and balanced battle occurring between plants and pathogens which consistently attack them. The role of genetic engineering should be in favor of plant. According to Harrison and Robinson (2002), resistant genes against a particular virus should ideally have the following characteristics: (1) they should provide protection against at least the entire range of virus variants, strains, and species that cause the disease; (2) they should provide robust protection that will require the virus to accumulate multiple mutations to overcome the resistance; and (3) they should confine incoming viruses to cells into which they are inoculated. Geminiviruses have received much attention, as this group of virus is one of the most important and studied. Their main prevalence in the tropics and subtropics is due to climatic factors favoring the multiplication and ability of vectors for transmitting them in a more composed way. The incidence and severity of the disease is increasing every year due to the emergence of new geminiviruses through recombination or pseudo-recombination among strains and/or species in various crops.
Conventional control strategies are generally effective, but they are time-consuming, and sound knowledge of agronomic practices, chemicals, and their effects on environment is must. A serious limitation of breeding program is the availability of resistance traits. It is often difficult to transfer the new character(s) from one species or variety to another while maintaining the agronomical qualities of the target cultivar. There are several other methods such as field sanitation, eradication of infected plants serving as primary source of virus inoculum, removal of weeds and alternate host plants from fields, plantation practices, spraying of insecticides, and use of virus-free planting material. The genetic engineering approach has many advantages, as it can give significant protection against viruses. Application of pathogen-derived resistance in various disciplines of biological science has raised a number of questions on its possible impact on the environment and human health. In the light of available knowledge, there is less or no environmental hazard, it seems. Moreover, PDR, if applied responsibly, is a powerful and safe means for combating plant pathogens, which cannot be controlled otherwise.
The gene silencing approach targeted against virus genes is nowadays a major approach against most of the plant viruses. It occurs either through repression of transcription (TGS) or through mRNA degradation (PTGS). PTGS results from a marked decrease in transcription and hypermethylation of the gene occurs. In TGS, mRNA synthesis is greatly reduced or absent. In addition to genetic engineering, genome engineering has recently emerged as a potential tool to improve various eukaryotic species, which also include variety of plant species. This concept follows introduction of trait of interest through the site-specific modification of the genome (Sovova et al. 2016). Genome engineering refers to the use of site-specific nucleases (SSNs), which can be designed to bind and cleave a specific nucleic acid sequence by introducing double-stranded breaks (Stella and Montoya 2016). CRISPR/Cas9 is one of the classes of SSNs. However, potential of this technique is yet to be tested in open field. The coming years will provide and witness more details on these technologies and development of marketable crops.
References
Akmal M, Baig MS, Khan JA (2017) Suppression of cotton leaf curl disease symptoms in Gossypium hirsutum through over expression of host-encoded miRNAs. J Biotechnol 10(263):21–29
Ali Z, Abulfaraj A, Idris A, Ali S, Tashkandi M, Mahfouz MM (2015) CRISPR/Cas9-mediated viral interference in plants. Genome Biol 16:238
Ali Z, Ali S, Tashkandi M, Zaidi SS, Mahfouz MM (2016) CRISPR/Cas9-mediated immunity to geminiviruses: differential interference and evasion. Sci Rep 6:26912. https://doi.org/10.1038/srep30223
Alvarez JP, Pekker I, Goldshmidt A, Blum E, Amsellem Z, Eshed Y (2006) Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 18:1134–1151
Antignus Y, Nestel D, Cohen S, Lapidot M (2001) Ultraviolet-deficient greenhouse environment affects whitefly attraction and flight-behavior. Environ Entomol 30:394–399
Asad S, Haris WA, Bashir A, Zafar Y, Malik KA, Malik NN, Lichtenstein CP (2003) Transgenic tobacco expressing geminiviral RNAs are resistant to the serious viral pathogen causing cotton leaf curl disease. Arch Virol 148:2341–2352
Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15:2730 2741
Axtell MJ, Bartel DP (2005) Antiquity of microRNAs and their targets in land plants. Plant Cell 17:1658–1673
Axtell MJ, Bowman JL (2008) Evolution of plant microRNAs and their targets. Trends Plant Sci 13:343–349
Baldrich P, Segundo BS (2016) MicroRNAs in rice innate immunity. Rice (NY) 9(6). https://doi.org/10.1186/s12284-016-0078-5
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Baulcombe DC (1994) Nove1 strategies for engineering virus resistance in plants. Curr Opin Biotechnol 5:117–124
Baulcombe DC (2004) RNA silencing in plants. Nature 431:356–363
Beachy RN (1993) Transgenic resistance to plant viruses. Arch Virol 4:327–416
Beachy RN (1997) Mechanisms and application of pathogen-derived resistance in transgenic plants. Curr Opin Biotechnol 8:215–220
Bisaro DM (2006) Silencing suppression by geminivirus proteins. Virology 344:158–168
Bolotin A, Quinquis B, Sorokin A, Ehrlich SD (2005) Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extra chromosomal origin. Microbiol 151:2551–2561
Briddon RW, Markham PG (2001) Cotton leaf curl virus disease. Virus Res 71:151–159
Brunetti A, Tavazza M, Noris E, Tavazza R, Caciagli P, Ancora G, Crespi S, Accotto GP (1997) High expression of truncated viral rep protein confers resistance to tomato yellow leaf curl virus in transgenic tomato plants. Mol Plant-Microbe Interact 10:571–579
Chellappan P, Masona MV, Vanitharani R, Taylor NJ, Fauquet CM (2004) Broad spectrum resistance to ssDNA viruses associated with transgene-induced gene silencing in cassava. Plant Mol Biol 56:601–611
Chen LF, Brannigan K, Clark R, Gilbertson RL (2010) Characterization of curtoviruses associated with curly top disease of tomato in California and monitoring for these viruses in beet leafhoppers. Plant Dis 94:99–108
Cogoni C, Macino G (1997) Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. Proc Natl Acad Sci USA 94:10233–10238
Cogoni C, Irelan JT, Schumacher M, Schmidhauser TJ, Selker EU, Macino G (1996) Transgene silencing of the al-1 gene invegetative cells of Neurospora is mediated by a cytoplasmiceffector and does not depend on DNA-DNA interactions or DNA methylation. EMBO J 15:3153–3163
Cohen S, Antignus Y (1994) Tomato yellow leaf curl virus, a whitefly-borne geminivirus of tomatoes. Adv Dis Vec Res 10:259–288
Cooper B, Lapidot M, Heick JA, Dodds JA, Beachy RN (1995) A defective movement protein of TMV in transgenic plants confers resistance to multiple viruses whereas the functional analog increases susceptibility. Virology 206:307–313
Crowdy SH, Posnette AF (1947) Virus diseases of cacao in West Africa II. Cross immunity experiments with virus 1A, 1B and 1C. Ann Appl Biol 34:403–411. https://doi.org/10.1111/j.1744-7348.1947.tb06373.x
Cui X, Li G, Wang D, Hu D, Zhou X (2005) A begomovirus DNAbencoded protein binds DNA, functions as a suppressor of RNA silencing, and targets the cell nucleus. J Virol 79:10764–10775
de Campa l, del Solar AG, Espinosa M (1990) Initiation of replication of plasmid pLS1: the initiator protein RepB acts on two distant DNA regions. J Mol Biol 213:247–262
Dogar AM (2006) RNAi dependent epigenetic marks on geminivirus promoter. Virol J 3:5
Dugas DV, Bartel B (2004) MicroRNA regulation of gene expression in plants. Curr Opin Plant Biol 7:512–520
Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL et al (2007) High-throughput sequencing of Arabidopsis microRNAs: Evidence for frequent birth and death of MIRNA genes. PLoS One 2:e219
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double stranded RNA in Caenorhabditis elegans. Nature 391:806–811
Fletcher JT (1978) The use of a virulent virus strain to protect plants against the effects of virulent strains. Ann Appl Biol 89:110–114. https://doi.org/10.1111/j.1744-7348.1978.tb02581.x
Fontes EPB, Gladfelter HJ, Schaffer RL, Petty ITD, Hanley-Bowdoin L (1994) Geminivirus replication origins have a modular organization. Plant Cell 6:405–416
Fuller C (1901) First Rep. Gov Entomol Natal 1899–1901:17–19
Gazal W, Jawaid AK (2018) Overexpression of ghr-miR166b generates resistance against Bemisia tabaci infestation in Gossypium hirsutum plants. Planta 247:1175–1189
Gonsalves D (1998) Control of papaya ringspot virus in papaya: a case study. Annu Rev Phytopathol 36:415–437
Hamilton AJ, Baulcombe DC (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286:950–952
Hamilton AJ, Voinnet O, Chappell L, Baulcombe DC (2002) Two classes of short interfering RNA in RNA silencing. EMBO J 21:4671–4679
Hanley-Bowdoin L, Settlage SB, Orozco BM, Nagar S, Robertson D (1999) Geminiviruses: models for plant DNA replication, transcription, and cell cycle regulation. Crit Rev Plant Sci 18:71–106
Harrison BD, Robinson DJ (2002) Green shoots of geminivirology. Physiol Mol Plant Pathol 60:215–218
Hilje L, Costa HS, Stansly PA (2001) Cultural practices for managing Bemisia tabaci and associated viral diseases. Crop Prot 20:801–812
Hong Y, Stanley J (1996) Virus resistance in Nicotiana benthamiana conferred by African cassava mosaic virus replication associated (ACI) transgene. Mol Plant Micro Interact 9:219–225
Horn NM, Reddy SV, Roberts IM, Reddy DVR (1993) Chickpea chlorotic dwarf virus, a new leafhopper-transmitted geminivirus of chickpea in India. Ann Appl Biol 122:467–479
Hugues JA, Ollennu LAA (1994) Mild strain protection of cocoa in Ghana against cocoa swoollen shoot virus—a review. Plant Pathol 43:442–457. https://doi.org/10.1111/j.1365-3059.1994.tb01578.x
Ji X, Zhang H, Zhang Y, Wang Y, Gao C (2015) Establishing a CRISPR-Cas-like immune system conferring DNA virus resistance in plants. Nat Plants 1:15144
Jones L, Hamilton AJ, Voinnet O, Thomas CL, Maule AJ, Baulcombe DC (1999) RNA-DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell 11:2291–2301
Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57:19–53
Jupin I, De Kouchkovsky F, Jouanneau F, Gronenborn B (1994) Movement of tomato yellow leaf curl geminivirus (TYLCV): involvement of the protein encoded by ORF C4. Virology 204:82–90
Kanchiswamy CN (2016) DNA-free genome editing methods for targeted crop improvement. Plant Cell Rep 35:1469–1474
Khan JA, Ahmad J (2005) Diagnosis, monitoring and transmission characteristics of Cotton leaf curl virus. Curr Sci 88:1803–1809
Khan JA, Dijkstra J (2006) Plant viruses as molecular pathogens. The Haworth Press, New York
Khatoon S, Kumar A, Sarin NB, Khan JA (2016) RNAi-mediated resistance against cotton leaf curl disease in elite Indian cotton Gossypium hirsutum cultivar. Virus Genes 52(4):530–537
Kumria R, Verma R, Rajam MV (1998) Potential applications of antisense RNA technology in plants. Curr Sci 74:35–41
Kung YJ, Lin SS, Huang YL, Chen TC, Harish SS, Chua NH et al (2012) Multiple artificial microRNAs targeting conserved motifs of the replicase gene confer robust transgenic resistance to negative-sense single-stranded RNA plant virus. Mol Plant Pathol 13(3):303–317
Kunik T, Salomon R, Zamir D, Navot N, Zeidan M, Michelson I et al (1994) Transgenic tomato plants expressing the tomato yellow leaf curl virus capsid protein are resistant to the virus. Nat Biotechnol 12(5):500–504
Kurth EG, Peremyslov VV, Prokhnevsky AI, Kasschau KD, Miller M, Carrington JC et al (2012) Virus-derived gene expression and RNA interference vector for grapevine. J Virol 86:6002–6009. https://doi.org/10.1128/JVI.00436-12
Lapidot M, Friedmann M (2002) Breeding for resistance to whitefly-transmitted geminiviruses. Ann Appl Biol 140:109–127
Lapidot M, Gafny R, Ding E, Wolf S, Lucas WJ, Beachy RN (1993) A dysfunctional movement protein of tobacco mosaic virus that partially modifies the plasmodesmata and limits virus spread in transgenic plants. Plant J 4:959–970
Laufs J, Traut W, Heyraud F, Matzeit V, Rogers SG, Schell J, Gronenborn B (1995) In vivo cleavage and joining at the viral origin of replication by the replication initiator protein of tomato yellow leaf curl virus. Proc Natl Acad Sci USA 92:3879–3883
Lazarowitz SG, Wu LC, Rogers SG, Elmer JS (1992) Sequence-specific interaction with the viral AL1 protein identifies a geminivirus DNA replication origin. Plant Cell 4:799–809
Liu L, van Tonder T, Pietersen G, Davies JW, Stanley J (1997) Molecular characterisation of a subgroup I geminivirus from a legume in South Africa. J Gen Virol 78:2113–2117
Llave C, Xie Z, Kasschau KD, Carrington JC (2002) Cleavage of scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297:2053–2056
Lomonossoff GP (1995) Pathogen-derived resistance to plant viruses. Annu Rev Phytopathol 33:323–343
Marraffini LA, Sontheimer EJ (2008) CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843–1845
McKinney HH (1926) Virus mixtures that may not be detected in young tobacco plants. Phytopathology 16:883
Morris B, Richardson KA, Eddy P, Zhan X, Haley A, Gardner R (1991) Mutagenesis of the AC3 open reading frame of African cassava mosaic virus DNA a reduces DNA B replication and ameliorates disease symptoms. J Gen Virol 72:1205–1213
Nakazono-Nagaoka E, Takahashi T, Shimizu T, Kosaka Y, Natsuaki T, Omura T et al (2009) Cross-protection against bean yellow mosaic virus (BYMV) and clover yellow vein virus by attenuated BYMV isolate M11. Phytopathology 99:251–257. https://doi.org/10.1094/PHYTO-99-3-0251
Napoli C, Lemieux C, Jorgensen R (1990) Introduction of a chimeric chalcone synthase gene into Petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2:279–289
Nicaise V (2014) Crop immunity against viruses: outcomes and future challenges. Front Plant Sci 5:660. https://doi.org/10.3389/fpls.2014.00660
Nishiguchi M, Kobayashi K (2011) Attenuated plant viruses: preventing virus diseases and understanding the molecular mechanism. J Gen Plant Pathol 77:221–229
Niu Q-W, Lin S-S, Reyes JL, Chen K-C, Wu H-W, Yeh S-D, Chua N-H (2006) Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat Biotechnol 24:1420–1428
Noris E, Accotto GP, Tavazza R, Brunetti A, Crespi S, Tavazza M (1996) Resistance to tomato yellow leaf curl geminivirus in Nicotiana benthamiana plants transformed with a truncated viral C1 gene. Virology 224:130–138
Nunez JK, Harrington LB, Doudna JA (2016) Chemical and biophysical modulation of Cas9 for tunable genome engineering. ACS Chem Biol 11:681–688
Ossowski S, Schwab R, Weigel D (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53:674–690
Padidam M, Beachy RN, Fauquet CM (1996) The role of AV2 (“precoat”) and coat protein in viral replication and movement in tomato leaf curl geminivirus. Virology 224:390–404
Padidam M, Sawyer S, Fauquet CM (1999) Possible emergence of new geminiviruses by frequent recombination. Virology 265:218–225
Pasquinelli AE, McCoy A, Jiménez E, Saló E, Ruvkun G, Martindale MQ et al (2003) Expression of the 22 nucleotide let-7 heterochronic RNA throughout the Metazoa: a role in life history evolution. Evol Develop 5:372–378
Powell-Abel P, Nelson RS, De B, Hoffman N, Rogers SG, Fraley RT, Beachy RN (1986) Delay of disease development in transgenic plants that express the tobacco mosaic virus coat protein gene. Science 232:738–743
Prins M (2003) Broad virus resistance in transgenic plants. Trends Biotechnol 21:373–375
Rajagopalan R, Vaucheret H, Trejo J, Bartel DP (2006) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 20:3407–3425
Ratcliff F, Harrison BD, Baulcombe DC (1997) A similarity between viral defense and gene silencing in plants. Science 276(5318):1558–1560
Ratcliff FG, MacFarlane SA, Baulcombe DC (1999) Gene silencing without DNA: RNA-mediated cross-protection between viruses. Plant Cell 11(7):1207–1215
Register JC 3rd, Beachy RN (1988) Resistance to TMV in transgenic plants results from interference with an early event in infection. Virology 166:524–532
Rojas MR, Hagen C, Lucas WJ, Gilbertson RL (2005) Exploiting chinks in the plant’s armor. Evolution and emergence of geminiviruses. Annu Rev Phytopathol 43:361–394
Sablok G, Pérez-Quintero ÁL, Hassan M, Tatarinova TV, López C (2011) Artificial microRNAs (amiRNAs) engineering–on how microRNA-based silencing methods have affected current plant silencing research. Biochem Biophy Res Comm 406(3):315–319
Saunders K, Lucy A, Stanley J (1992) RNA-primed complementary-sense DNA synthesis of the geminivirus African cassava mosaic virus. Nucleic Acids Res 20:6311–6315
Shepherd DN, Martin DP, Van der Walt E, Dent K, Vasrani A, Rybicki EP (2009) Maize streak virus: an old and complex emerging pathogen. Mol Plant Pathol 11:1–12
Shweta, Akhter Y, Khan JA (2018) Genome wide identification of cotton (Gossypium hirsutum)-encoded microRNA targets against cotton leaf curl Burewala virus. Gene 638:60–65
Singh I (1990) Papaya. IBH Publishing, New Delhi
Sinha V, Sarin NB, Bhatnagar D (2017) The efficacy of antisense-based construct for inducing resistance against Croton yellow vein mosaic virus in Nicotiana tabacum. Virus Genes 53(6):906–912
Smith NA, Singh SP, Wang MB, Stoutjesdijk P, Green A, Waterhouse PM (2000) Total silencing by intron-spliced hairpin RNAs. Nature 407:319–320
Sovova T, Kerins G, Demnerova K, Ovesna J (2016) Genome editing with engineer ednuclease sin economically important animals and plants: state of the art in the research pipeline. Curr Issues MolBiol 21:41–62
Stella S, Montoya G (2016) The genome editing revolution: a CRISPR- Cas TALE off-target story. BioEssays 38:S4–S13
Sunkar R, Jagadeeswaran G (2008) In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol 8:37
Tang G, Reinhart BJ, Bartel DP, Zamore PD (2003) A biochemical framework for RNA silencing in plants. Genes Dev 17:49–63
Tenllado F, Martinez-Garcia B, Vargas M, Diaz-Ruiz JR (2003) Crude extracts of bacterially expressed dsRNA can be used to protect plants against virus infections. BMC Biotechnol 3:3
Thresh JM, Cooter RJ (2005) Strategies for controlling cassava mosaic virus disease in Africa. Plant Pathol 54:587–614
Valkonen J (1998) Virus disease control in plants using natural and engineered resistance and some consideration regarding biosafety. Currents 17:51–55
Vanitharani R, Chellappan P, Pita JS, Fauquet CM (2004) Differential roles of AC2 and AC4 of cassava geminiviruses in mediating synergism and suppression of posttranscriptional gene silencing. J Virol 78(17):9487–9498
Vanitharani R, Chellappan P, Fauquet CM (2005) Geminiviruses and RNA silencing. Trends Plant Sci 10:144–151
Varma A, Malathi VG (2003) Emerging geminivirus problems: a serious threat to crop production. Ann Appl Biol 142:145–164
Voinnet O (2005) Induction and suppression of RNA silencing: insights from viral infections. Nat Rev Genet 6:206–220. https://doi.org/10.1038/nrg1555
Voinnet O, Pinto YM, Baulcombe DC (1999) Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc Natl Acad Sci USA 96:14147–14152
Vu TV, Choudhury NR, Mukherjee SK (2013) Transgenic tomato plants expressing artificial microRNAs for silencing the pre-coat and coat proteins of a begomovirus, Tomato leaf curl New Delhi virus, show tolerance to virus infection. Virus Res 172:35–45. https://doi.org/10.1016/j.virusres.2012.12.008
Wang HL (1991) Effectiveness of cross protection by a mild strain of zucchini yellow mosaic virus in cucumber, melon, and squash. Plant Dis 75:203
Waterhouse PM, Graham MW, Wang MB (1998) Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc Natl Acad Sci U S A 95:13959–13964
Wen F, Lister RM, Fattouh FA (1991) Cross-protection among strains of barley yellow dwarf virus. J Gen Virol 72:791–799
Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, Robinson SP, Gleave AP, Green AG, Waterhouse PM (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27:581–590
Wilson TMA (1993) Strategies to protect crop plants against viruses: pathogen-derived resistance blossoms. Proc Natl Acad Sci USA 90:3134–3141
Wintermantel WM, Banerjee N, Oliver JC, Paolillo DJ, Zaitlin M (1997) Cucumber mosaic virus is restricted from entering minor veins in transgenic tobacco exhibiting replicase-mediated resistance. Virology 231:248–257
Woo JW, Kim J, Kwon SI, Corvalan C, Cho SW, Kim H et al (2015) DNA- free genome editing in plants with pre-assembled CRISPR- Cas9 ribonucleoproteins. Nat Biotechnol 33:1162–1164
Yang Y, Sherwood T, Patte C, Hiebert E, Polston J (2004) Use of Tomato yellow leaf curl virus (TYLCV) Rep gene sequences to engineer TYLCV resistance in tomato. Phytopathology 94(5):490–496
Zhang SC, Wege C, Jeske H (2001) Movement proteins (BC1 and BV1) of Abutilon mosaic geminivirus are cotransported in and between cells of sink but not of source leaves as detected by green fluorescent tagging. Virology 290:249–260
Zhang BH, Pan XP, Wang QL, Cobb GP, Anderson TA (2005) Identification and characterization of new plant microRNAs using EST analysis. Cell Res 15:336–360
Zhang BH, Pan XP, Cannon CH, Cobb GP, Anderson TA (2006) Conservation and divergence of plant microRNA genes. Plant J 46:243–259
Ziebell H, Carr JP (2010) Cross-protection: a century of mystery. Adv Virus Res 76:211–264
Zulma IM, Argüello-Astorga GR, Bustamante RFR (2002) Geminivirus replication and gene expression. In: Khan JA, Dijkstra J (eds) Plant viruses as molecular pathogens. The Haworth Press, New York, p 537
Acknowledgements
AK is funded by Science and Engineering Research Board (SERB), Govt of India, and is thankful to Mr. Ajay Pratap Singh, Sr Director, IILM-CET Greater Noida for facilities and keen interest.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Kumar, A., Khan, J.A. (2019). Geminivirus Resistance Strategies. In: Kumar, R. (eds) Geminiviruses. Springer, Cham. https://doi.org/10.1007/978-3-030-18248-9_11
Download citation
DOI: https://doi.org/10.1007/978-3-030-18248-9_11
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-18247-2
Online ISBN: 978-3-030-18248-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)