Abstract
In this article, we propose the use of a new simple Bayesian classifier (SBND) that quickly learns a Markov boundary of the class variable and a network structure relating class variables and the said boundary. This model is compared with other Bayesian classifiers, then experimental tests are carried out for which 31 well-known ICU databases and two bases of artificial variables have been used. With these databases we compare the results obtained by such algorithms studied in the state of the art such as Naive Bayes, TAN, BAN, RPDag, CRPDag, SBND and combinations with different metrics such as K2, BIC, Akaike, BDEu. The experimental work was done in Elvira software.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Learning can be defined as ‘any process, through which a system improves its efficiency’ [6]. The ability to learn is considered a central feature of intelligent systems [9], and this is why a lot of effort and dedication have been put on the research and development of this topic. The development of knowledge based systems has motivated research in the area of learning with the aim of automatizing the knowledge acquisition process, what is considered one of the main problems in the building of these systems. For some time, algorithms for learning without Bayesian networks restrictions, especially those based on the metric + search paradigm have been considered inadequate for competitive construction of classifiers based on Bayesian networks [1]. This perception is being changed due to the development of generic networks learning methods, which are very competitive [1]. Bayesian networks (without structural restrictions of any kind) can also be used for classifying. In this case, classifiers are referred to as non-restricted Bayesian net-works. These will be used in this paper too. Any Bayesian network can be used in supervised classification, for which it is enough to use the Markov blanket of the case variable. It is necessary to consider that a non-restricted Bayesian classifier has a higher expressive power than a structurally restricted model [8].
In this paper we present a new classifier, which we have called Simple Bayesian classifier, consisting in a generic Bayesian network, but learned from a voracious techniques.
2 Simple Bayesian Classifier
SBND is a new simple Bayesian classifier designed to simplify this activity. To start using this classifier, we will need a PARENTS function, which, given a variable \(X_i\) and a set of candidates, can calculate the best parents set in \(X_i\) among that set of candidates. The parents set is returned in \(\varPi _i\) and at the same time gives back a numerical value constituting the Score of this variable, given that set of parents meas-ured by a Bayesian Score.
This PARENTS function is the one making a heuristic search for the best set of parents among a set of candidates and this occurs by adding and removing parents as long as the score improves. The idea is to start introducing C as a root node in the Bayesian network \(\mathscr { B}\) and keep a set of nodes \(\mathbf {X'} \) of the attributes already introduced in the net (initially empty) [13].
Different Score metrics \(Score(X_i,\mathbf {A}|\mathscr { D})\) measuring the suitability of \(\mathbf {A}\) as \(X_i\) parents set can be used (these metrics can be BDEu, BIC, K2 o Akaike).
Since we assume to have a procedure PARENTS(\(X_i\), CANDIDATOS, \(\varPi _i\)), which calculates the best set of parents \(\varPi _i\) from \(X_i\) using the selected metric, and returns the value of this optimum metric, when implementing this function we will have a voracious algorithm, which starts with an empty \(\varPi _i\) and keeps adding and removing form \(\varPi _i\) the variable producing the highest metric increase, until there is not any possible improvement. In these conditions the values for each \(X_i \in \mathbf {X}\setminus \mathbf {X'}\) variable is calculated:
Infor \(Infor(X_i,C)\) calculates the differences among the best \(X_i\) metrics with parents set chosen between \(\mathbf {X'}\) including C and without including C in the candidates. Intuitively, it is a measure of \(X_i\) and C conditional dependency, given the already included variables. This value is always theoretically higher or equal to zero, but it could be negative since the best parents set is calculated approximately [13].
Once this value has been calculated for each \(X_i \in {\mathbf {X}}\setminus {\mathbf {X'}}\) variable, \(X_{max} = \arg \hbox {m}\acute{\mathrm{a}}\mathrm{x}_{X_i \in {\mathbf {X}}\setminus {\mathbf {X'}}}\) \(Infor(X_i,C)\) is selected. This would be the variable providing most information about the C class according to the already introduced variables. If \(Infor(X_{max},C)>0\), then this variable provides additional information about C and is inserted in the network and in \(\mathbf {X'}\). Its parents set is calculated with \(\text{ PARENTS }(X_{max},\mathbf {X'} \cup \{C\},\varPi _i)\). In theory, \(C \in \varPi _i\), always, since otherwise \(Infor(X_{max},C)=0\), although due to the voracious nature of the procedure, \(C \notin \varPi _i\), what is a remote possibility.
In other words the variable giving most information is added to the network provided that the information is positive, and is considered, as well as its parents set, the best parents set provided by this function. Since the information is positive, the class variable should be supposed to be included in the parents set. The algorithm ends if the function \(Infor(X_{max},C) \le 0\) [13].
The main characteristics of this classifier are:
-
Learns an arbitrary Bayesian network with a subset of initial variables directly influencing this variable. In this sense, it can be considered an algorithm, which calculates a Markov boundary, because it intends to obtain a set of such variables that, once obtained, the rest of the variables are independent.
-
The class variable is always a root node and there are links from this node to the rest of the attributes (except for very few occasions due to the approximate nature of the parents calculation). It is, in this sense, similar to other Bayesian classifiers, where there is always a link from the class to each of the attributes.
-
The arrangement of the attributes in based on selecting in a voracious way those providing most information on the class, given the selected attributes. In this way the most relevant attributes are introduced first. Obtaining the network with the best metrics is not based on the space of the attributes order, but in obtaining the maximum information for the class. It can even be some network quality lose in this sense, but the algorithm gains speed [13].
3 Experimentation
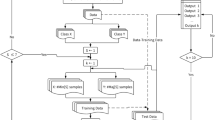
In this section experimental tests are carried out through using 31 well - known to ICU databases [11] and two bases including artificial variables. The databases can be seen in Table 1. With these two databases. The results obtained by the algorithms mentioned in the state of art, namely Naive Bayes [15], TAN [7], BAN [3], SBND [12], RPDAG and C-RPDAG [1, 9] are compared to other combinations with different metrics, namely K2 [4], BIC [14], Akaike [2], BDEu. These methods build up classifiers constituting generic Bayesian networks equivalent in independence and equivalent in classification. The experimental work was made at Elvira [5].
Table 1 provides a brief description of each database characteristics, including the number of instances, attributes, and the states for the class variable. These data sets have been preprocessed as follows: continuous variables have been discretized using the procedure proposed by [10], and the instances having non definite or missing values were removed. For this pre-processing stage, the results obtained by [1] have been used.
4 Results
The results obtained by each classifier and its combinations with the studied metrics can be observed in Tables 2, 3, 4 and 5 (due to their size, they have been divided in 4).
Below some non-parametric tests are made of the differences among the different methods in determining the best classifying algorithm, It is important to indicate that the means value for each of the algorithms has been included, as well. The best means is obtained by CRPDAG-BDEu with an 88.354 value, followed by SBND Akaike with 88.156. The basic non parametric test used is Friedman, since it has more than 2 associated samples.
The null hypothesis \((H_{0})\) being contrasted is that the answers associated to each of the treatments have the same probability distribution or distributions with the same means against the alternative hypothesis stating that at least the distribution of one of the means differs from the others.
The values that will be used in these tests can be seen in Table 6, where the average order of the algorithms are presented. The best performance is shown by SBND K2 algorithm.
Friedman test’s results are shown in Table 7, where a value lower than 0.05 is seen, thus rejecting the null hypothesis and it is determined that the differences measure the statistically significant distributions of the different methods.
When the differences detected are significant, Holm’s test is applied for comparing the control algorithm (the best classified) to the rest. Holm’s is a multiple comparison test, by means of which we confront SBND with K2, the best classifying value, with the rest of the algorithms.
Table 8 shows Holm test’s results for 0.05 significance level and Table 9 for 0.10 significance level.
In the first place it was considered \(\alpha =0.05\). P values in Holm test is \(P\le 0.0045\). This value is compared to the rest of the algorithms based on the right column of Table 8. It can be observed that this algorithm is significantly better than Naive Bayes, SBND BIC, RPDag Learning BIC and there are no significant differences with the rest of the algorithms.
In the second place, \(\alpha =0.10\) significance level is considered, P value in Holms test is \(P\le 0.01\). With this value multiple comparisons with the values in the right column of Table 9 are made. It can be determined that our control algorithm SBND with K2 classifies better than Naive Bayes, SBND BIC, RPDag Learning BIC, CRPDag Learning BIC algorithms and there are no significant differences with the rest of the algorithms.
5 Conclusions
In this article we have introduced a Bayesian classifier known as SBND which is based in quickly obtaining an easy to learn and very competitive Markov’s boundary. This classifiers is fast to learn and very competitive as compared to other classifiers of the state of art. Various experiments were made using 31 well known in the ICU databases and two bases of artificial variables.
SBND classifier’s performance in some examples is dependent on the metric being used. With BIC the result is not good, Akaike gives good results in reference to the means, and K2 shows good results in non-parametric tests.
For future research, it is important to include the costs of wrong classifications in the problem, since a false positive is not the same as a false negative. If the cost of a false negative was considered better than that of a false positive, it could be detected that more students would drop out, although the number of students at risk of abandoning would increase.
References
Acid, S., De Campos, L., Castellano, J.: Learning Bayesian network classifiers: searching in a space of partially directed acyclic graphs. Mach. Learn. 59(3), 213–235 (2005)
Akaike, H.: A new look at the statistical model identification. IEEE Trans. Autom. Control 19, 716–723 (1974)
Cheng, J., Russell, G.: Comparing Bayesian network classifiers. In: Proceedings of the Fifteenth Conference on Uncertainty in Artificial Intelligence, pp. 101–108. Morgan Kaufmann Publishers Inc. (1999)
Cooper, G., Herskovits, E.: A Bayesian method for the induction of probabilistic networks from data. Mach. Learn. 9(4), 309–347 (1992)
Elvira, C.: Elvira: an environment for probabilistic graphical models. In: Gámez, J., Salmerón, A. (eds.) Proceedings of the 1st European Workshop on Probabilistic Graphical Models, pp. 222–230 (2002)
Felgaer, P., Britos, P., Sicre, J., Servetto, A., García-Martínez, R., Perichinsky, G.: Optimización de redes bayesianas basada en técnicas de aprendizaje por instrucción. In: Proceedings del VIII Congreso Argentino de Ciencias de la Computación, vol. 1687 (2003)
Friedman, N., Michal, L., Iftach, N., Dana, P.: Using Bayesian networks to analyze expression data. Comput. Biol. 7(3–4), 601–620 (2000)
García, F.: Modelos Bayesianos para la clasificación supervisada: aplicaciones al análisis de datos de expresión genética, Tesis Doctoral, Universidad de Granada (2009)
García-Martínez, R., Borrajo, D.: An integrated approach of learning, planning, and execution. J. Intell. Robot. Syst. 29(1), 47–78 (2000)
Irani, K., Jie, C., Usama, F., Zhaogang, Q.: Applying machine learning to semiconductor manufacturing. iEEE Expert 8(1), 41–47 (1993)
Lichman, M.: UCI machine learning repository. University of California, Irvine, School of Information and Computer Sciences (2013). http://archive.ics.uci.edu/ml
Oviedo, B., Moral, S., Puris, A.: A hierarchical clustering method: applications to educational data. Intell. Data Anal. 20(4), 933–951 (2016)
Oviedo Bayas, B.W.: Modelos gráficos probabilisticos aplicados a la predicción del rendimiento en educación (2016)
Schwarz, G.: Estimating the dimension of a model. Ann. Stat. 6(2), 461–464 (1978)
Webb, G.I., Pazzani, M.J.: Adjusted probability Naive Bayesian induction. In: Antoniou, G., Slaney, J. (eds.) AI 1998. LNCS, vol. 1502, pp. 285–295. Springer, Heidelberg (1998). https://doi.org/10.1007/BFb0095060
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Oviedo, B., Zambrano-Vega, C., León-Acurio, J., Martinez, A. (2019). Simple Bayesian Classifier Applied to Learning. In: Botto-Tobar, M., Pizarro, G., Zúñiga-Prieto, M., D’Armas, M., Zúñiga Sánchez, M. (eds) Technology Trends. CITT 2018. Communications in Computer and Information Science, vol 895. Springer, Cham. https://doi.org/10.1007/978-3-030-05532-5_29
Download citation
DOI: https://doi.org/10.1007/978-3-030-05532-5_29
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-05531-8
Online ISBN: 978-3-030-05532-5
eBook Packages: Computer ScienceComputer Science (R0)