Abstract
Biological macromolecules, such as proteins, nucleic acids, and complexes thereof, are characterized by specific structural and dynamic features that are the basis of their respective biological activity, and define their dynamic personalities [29]. Understanding macromolecular activity thus requires studying structural changes over time and on various time-scales, such as equilibrium fluctuations and conformational changes orchestrating enzyme catalysis or enabling signal transduction. The first step in human vision, for instance, is the sub-picosecond time-scale photoisomerization of the retinal pigment in rhodopsin [73], which within microseconds leads to the conformational changes required for activation of transducin, the regulatory protein that initiates the signaling cascade beyond the macromolecular level.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
11.1 Introduction
Biological macromolecules, such as proteins, nucleic acids, and complexes thereof, are characterized by specific structural and dynamic features that are the basis of their respective biological activity, and define their dynamic personalities [29]. Understanding macromolecular activity thus requires studying structural changes over time and on various time-scales, such as equilibrium fluctuations and conformational changes orchestrating enzyme catalysis or enabling signal transduction. The first step in human vision, for instance, is the sub-picosecond time-scale photoisomerization of the retinal pigment in rhodopsin [73], which within microseconds leads to the conformational changes required for activation of transducin, the regulatory protein that initiates the signaling cascade beyond the macromolecular level.
Complementary biophysical techniques each open a window in time on macromolecular dynamics occurring from femtoseconds to minutes (Fig. 11.1). Among those, kinetic X-ray crystallography permits trapping of macromolecular conformational intermediates along a reaction pathway, and their characterization at the atomic level of spatial resolution [8]. The basic concept underlying this ensemble of techniques is that macromolecular activity can be triggered within a crystal, and the structure of intermediate states characterized, provided that (1) the macromolecule is active in the crystalline state; (2) an efficient and synchronous triggering of activity can be attained (at room-temperature) for all (or most) probed molecules; and (3) the structural information can be recorded on a time scale shorter than the lifetime of the intermediate state of interest. As within a macromolecular crystal neighboring molecules are bathed in solvent (on average around 50% of the crystal volume) and generally make limited direct contacts, they most often preserve their functionally relevant structural dynamics and therefore remain active in the crystalline state [55]. After triggering of the biological reaction within macromolecular crystals, functionally relevant conformational changes are either arrested by flash-cooling the crystal, allowing for characterization of the structure by conventional cryo-crystallography, or followed in real time by time-resolved crystallography. The temporal resolution of the latter is limited to 100 ps if carried out in the form of Laue crystallography at synchrotrons. The advent of X-ray free electron lasers (X-ray FEL) has pushed the resolution to the sub-ps regime, allowing for ultrafast changes to be studied by time-resolved serial femtosecond crystallography. Below, we introduce the principles of time-resolved crystallography and summarize its implementation at synchrotron sources, before offering a review of time-resolved structural studies carried out so far using serial femtosecond crystallography (SFX [11]) at X-ray FELs. Challenges, limitations, and perspectives of time-resolved SFX round out this chapter.
11.2 Principles of Time-Resolved Crystallography
In time-resolved crystallography, a reaction in the crystalline macromolecule is triggered (pump) at t = 0 and a diffraction pattern (probe) collected after a well-defined lapse of time ∆t 1 (pump–probe delay). The pump–probe sequence is repeated sufficiently enough for a complete diffraction data set to be produced from which a macromolecular structure can be determined that features conformational changes characteristic of the time delay ∆t 1 after reaction initiation. The pump–probe delay is then changed to ∆t 2 and the procedure repeated, etc. The structure determined at each ∆t i represents a frame in a molecular movie featuring conformational changes along the reaction pathway (Fig. 11.2). A complete and intelligible time-resolved study of the mode of action of the rhodopsin protein mentioned above, for instance, would require structural data to be collected over ten orders of magnitude, from the fs time scale where the actinic photon is absorbed, up to the μs timescale where transduction is activated.
Principle of pump–probe time-resolved crystallography. A crystalline macromolecule is probed by X-ray pulses (“probe,” red beam) generated by a synchrotron or an X-ray FEL source. The time domain in the diffraction experiment is defined by the synchronization of X-ray pulses with optical laser pulses (“pump,” blue beam). The interaction of optical laser pulses with the sample triggers a reaction and defines the time t = 0. The evolution of the protein structure at a generic time ∆t i is monitored by variation of the ∆t delay between pump and probe. Every pump–probe sequence is repeated N times to produce a complete dataset from which a structural model can be determined
Efficient and synchronous reaction triggering is key to a successful pump–probe experiment. The pump needs to trigger the reaction faster than the process of interest (synchrony) and in a large fraction of the crystalline molecules (efficiency) so that structural changes can be observed by ensemble-averaged crystallography. The pump method to be used depends on the reaction to be triggered: UV–visible light activates inherently light-sensitive proteins or so-called caged compounds complexed with light-insensitive proteins; diffusion of substrates initiates enzyme catalysis (see Chap. 12 in this book); a temperature jump shifts conformational equilibria [44]; X-ray irradiation can provide electrons to trigger redox processes [69] or the breakdown of strained intermediate states [12]; electric field pulses allow for studying protein mechanics [28]. By far the most widespread method is optical triggering because of its straightforward technical implementation, the availability of crystals for a large number of light-sensitive proteins with cyclic reactions to be studied, and the accessibility of ultrafast (fs–ps) time scales.
The X-ray probe produces diffraction patterns that allow for structure determination at ∆ti after reaction initiation. The probe needs to be pulsed so that structural changes can be captured, analogous to shutter opening and closing in photography. The probe-pulse length must be shorter than the process of interest and sets, together with the pump-pulse length, the best time resolution that can be attained. At synchrotron sources, the pulse length cannot be shorter than about 100 ps, making it impossible to study processes faster than that limit. Also, time-resolved crystallography at synchrotrons is in practice limited to cyclic reactions because the pump–probe sequence is repeated multiple times on one or several macro-sized crystals. Using time-resolved (TR) SFX at X-ray FELs, however, cyclic as well as noncyclic reactions can be studied since each microcrystal is probed only once by a single X-ray shot. Furthermore, with their femtosecond X-ray pulses, X-ray FELs have extended the time resolution to the sub-ps regime, permitting the structural study of ultrafast processes such as those immediately following photon absorption in light-sensitive processes. Last but not least, the risk of X-ray radiation damage to biological macromolecules, known to compromise structural data collected at synchrotrons [21], is abolished in most SFX studies because diffraction data are collected before chemical and structural damage has had the time to develop [57]. Hence, the advent of X-ray FELs has revived and extended the reach of time-resolved crystallography.
11.3 Time-Resolved Crystallography at Synchrotron Sources
The advent of third-generation synchrotron sources and undulator beamlines, poised with 1018 and 106 higher peak brilliance than X-ray tubes and second-generation bending magnet sources, respectively, permitted the introduction of time-resolved crystallography by means of the so-called Laue technique [54]. In this approach, crystals of inherently photosensitive proteins or of non-photosensitive proteins complexed with photolabile precursors of their substrates or products—the so-called caged compounds—are activated by exposure to a femtosecond or nanosecond laser (pump) at room temperature. A highly intense polychromatic X-ray beam is then used to probe the structure, at various ∆t i. The advantage of using a white (∼1–5% bandwidth) or pink (∼0.05 to 0.1% bandwidth) polychromatic beam is that enough photons are elastically scattered, even from a single X-ray bunch, to enable collection of a diffraction pattern on which enough information is present to derive a partial dataset. Combining multiple such diffraction images, typically 10–30 exposures to the polychromatic X-ray beam of one or more crystals in different orientations, experimentalists can then produce a full dataset for each ∆t i. The time resolution of Laue crystallography at synchrotrons is inherently limited to 100 ps, corresponding to the pulse length of a single electron bunch generating the X-rays. Furthermore, the methodology requires large (0.5 mm), highly ordered, radiation-resistant protein crystals (to enable multiple exposures to the polychromatic beam without loss of resolution due to X-ray damage) with small unit cells (to minimize Bragg-peak overlaps), and works best with proteins undergoing cyclic photoreactions (see above). In the case of highly radiation sensitive samples, nonreversible photoreactions, or in studies where non-inherently photosensitive proteins are light-functionalized by complexation with caged compounds [68], more crystals are indeed needed. In the case of large proteins with correspondingly large unit cell parameters (e.g., photosystems I and II, or cytochrome c oxidase), data processing is complicated, due to overlaps between diffracted spots even when using a pink beam. Hence the methodology has failed to enable time-resolved crystallography on a large variety of proteins, and has mostly remained limited to a handful of proteins including the small GTPase Ras [68], myglobin [6, 7, 75, 79], the photoactive yellow protein [23, 34, 36, 74], and the photosynthetic reaction center [3, 96]. The largest protein to have been successfully studied by time-resolved Laue crystallography is hemoglobin [39]. The advent of X-ray FELs and monochromatic SFX has allowed for the resolution of time-resolved crystallography to be improved to the fs timescale, and to extend the feasibility of time-resolved studies to larger and radiation sensitive proteins.
11.4 Time-Resolved Serial Femtosecond Crystallography at X-Ray FELs: An Inventory
In time-resolved serial femtosecond crystallography (TR-SFX) following a pump–probe data collection scheme, macromolecular crystals are streamed across the pulsed X-ray FEL beam typically by means of a liquid jet [78, 93], a high-viscosity injector [5, 84, 92] or by acoustic droplet injection coupled with a conveyer-belt drive [20]. When the protein is inherently photosensitive or has been photosensitized by complexation with a caged-compound, crystals can be activated by an optical laser pulse (pump) prior to interaction with the X-ray pulse (probe), generating a diffraction pattern (Fig. 11.3). Repetition of this process on many crystals provides a full data set at a well-defined ∆ti from which the corresponding structural snapshot can be determined. Varying ∆t i then allows for generating frames of a molecular movie. If macromolecular activity is triggered by diffusion of a small molecule into the crystal, two solutions containing the crystals and the small molecule are mixed prior to interaction with an X-ray pulse. This so-called mix-and-inject strategy [71] is extensively discussed in Chap. 12 of this book and has been successfully applied to study ligand-binding in adenine riboswitches [80] and ß-lactamase [46].
General setup for TR-SFX experiments at X-ray FELs. There are a variety of ways to present crystals to the (probe) X-ray beam in SFX experiments. In the present example, crystals are jetted across the pulsed X-ray beam, and thousands of diffraction patterns are collected for each dataset. In TR-SFX, a pump laser is used to trigger conformational changes in the crystalline protein, and data are collected by the X-ray probe beam at various pump–probe delays. The experiment may require photoconversion of crystals within a pre-illumination device [67], as in the case of the photoswitchable rsEGFP2 that needed to be switched by 488 nm light from the fluorescent on-state (resting state) to the nonfluorescent off-state that was subsequently activated by a pump laser at 400 nm [15]
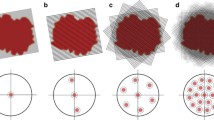
Time-resolved crystallography experiments at X-ray FELs remedy several shortcomings that limit those carried out at synchrotron sources. Firstly, the achievable time resolution of maximal 100 ps at a synchrotron is improved to 10 fs, owing to the short length of FEL pulses. Secondly, the high peak brilliance of X-ray FEL pulses generates exploitable diffraction from micron-sized crystals that can be fully activated by the optical pump laser. Indeed, absorption by optically dense protein crystals (Fig. 11.4) limits light penetration to outer layers in the case of large crystals (at least several tens of microns in size) such as those required by time-resolved synchrotron studies. And third, the very nature of (TR)-SFX, wherein the sample is replenished after each shot in a serial manner, enables time-resolved structural studies on nonreversible reactions, provided that enough sample can be produced for the envisaged delivery system.
Low pump-light penetration depth in chromophore-containing protein crystals results in only a fraction of the probed molecules being excited and effectively undergoing conformational changes. The figure shows the calculated transmission of pump light across crystals of the three proteins that have so far been studied by TR-SFX on the ps or sub-ps time scale, that is, myoglobin (Mb; [2]), the photoactive yellow protein (PYP; [63]), and the reversibly photoswitchable fluorescent protein rsEGFP2 [15] as a function of crystal thickness. In these studies, concentrations of the crystalline Mb, PYP, and rsEGFP2 were 53, 63, and 27 mM, respectively. The molar extinction coefficients of Mb, PYP, and rsEGFP2 in its off state are 13,950, 45,500, and 22,000 M−1 cm−1 at 532, 450, and 400 nm, respectively. Of note, the molar extinction coefficients used for these calculations were those of the ground states and assumed to be constant across the crystal over the pulse length. A more sophisticated calculation that takes into account the change of penetration depth as time progresses and molecules bleach has been presented for PYP [87]
The first TR-SFX study was carried out on crystals of a photoactive complex of photosystem I (PS I) with ferredoxin [1] with pump–probe delays of 5 μs and 10 μs. Only virtual powder patterns were used in the analysis, since the low amount of indexed patterns per time-delay precluded extraction of structure factors. At 5 μs, a slight increase in diffracted intensities was observed, which was interpreted as a correlated structural change induced by electron transfer in the PSI—ferredoxin complex. At 10 μs, diffraction intensities dropped, rationalized as originating from the disordering of crystals upon dissociation of ferredoxin from the binding pocket, in line with time-resolved optical spectroscopy. Since this proof-of-principle study, several pump–probe TR-SFX studies have been carried out on various proteins (myoglobin, photoactive yellow protein, bacteriorhodopsin, photosystem II, a reversibly photoswitchable green fluorescent protein, cytochrome c oxidase, nitrite oxide reductase), and on timescales ranging from seconds to sub-picoseconds (Fig. 11.6). These results are briefly presented below.
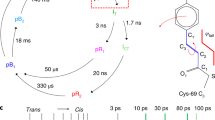
Three TR-SFX studies entered the ultrafast time scale of photochemical reactions with pump–probe delays of a picosecond and shorter. (a) Structural changes in myoglobin (Mb) occurring 0.5 ps after photoexcitation [2]. The Fo light − Fo dark difference electron density map is contoured at +3σ (green) and −3σ (red). (b) Oscillatory side-chain dynamics, excited by photodissociation of the heme, reflect fast coherent motions that ultimately couple with larger-scale displacement of entire helices. (c) A structural transition at around 590 fs in the photoactive yellow protein (PYP) has been associated with trans-to-cis isomerization of the chromophore [63]. (d, e) Structural changes in the reversibly photoswitchable fluorescent protein rsEGFP2 occurring 1 ps (red model) after photoexcitation of the nonfluorescent off-state (grey model) feature a fully twisted chromophore in the electronically excited state. The Fo light − Fo dark difference electron density map is overlaid in (d) and contoured at +3.5σ (green) and −3.5σ (red). (e) Chromophore twisting at 1 ps is accompanied by a shift in the entire helix that carries the chromophore. Panels (a, b) and (e) were extracted from published work ([2, 15], respectively). Panels (c) and (d) were provided by Marius Schmidt and Nicolas Coquelle, respectively
The first high-resolution (1.6 Å) TR-SFX study was carried out on the photoactive yellow protein (PYP) with pump–probe delays of 1 μs and 10 ns using optical pump laser pulses on the ns regime [87]. The difference Fourier maps, computed between data sets of illuminated (light) and non-illuminated (dark) crystals, showed structural changes qualitatively similar to those observed in TR synchrotron Laue studies at comparable time delays, demonstrating validity of the TR-SFX method. Yet peaks in the TR-SFX difference maps were higher than in the corresponding Laue difference maps, owing to the more extensive pump-laser activation of the micron-sized crystals used in the TR-SFX studies, as compared to those used in Laue experiments. As mentioned previously, usability of μm- and sub-μm-sized crystals is one of the advantages of X-ray FEL time-resolved studies. Further steps into the uncharted regime of ps resolution were subsequently taken, as discussed in detail in the next section.
It is of important note that with much of the biological dynamics of interest being in the ns or slower time scales, many TR-SFX studies utilize the unique advantages of X-ray FEL based methods to study relatively slow dynamics compared to the X-ray FEL pulse duration, such as those involved in photosystem II (PS II) activity. The high scientific interest in the water-splitting cycle of PS II has made it so far the macromolecular system most studied by TR-SFX (Fig. 11.6). PSII is a large membrane-bound protein complex (700 kDa) involved in photosynthesis that produces dioxygen by catalyzing light-driven water splitting at a so-called oxygen evolving complex (OEC) containing a Mn4CaO5 cluster. The photocycle of PSII is a reversible reaction that involves sequential absorption of four photons generating four consecutive redox states of the cluster that cycles through five S-states (S0 to S4; the so-called “Kok cycle”) on the μs to ms time scale [40]. Each of the four steps is characterized by a non-unitary quantum yield, so that a mixture of states has accumulated at the end of one photocycle; therefore, the reaction is in practice irreversible and each crystal may be used only once. A high-resolution (1.9 Å) structure of PSII has been solved by synchrotron macromolecular crystallography [90], but determination of the accurate OEC geometry had been hampered by the high X-ray sensitivity of this metalloenzyme [97]. The high-radiation sensitivity, combined with the experimentally nonreversible reaction, makes TR-SFX the method of choice to study the Kok cycle. Using femtosecond FEL pulses, it was indeed possible to determine the radiation-damage free structure of PS II and its OEC at 1.95 Å resolution by rotating and translating a large single crystal at 100 K in a stepwise fashion [82]. Comparison of the synchrotron and X-ray FEL structures revealed a more compact OEC with bond lengths 0.1–0.2 Å shorter in the X-ray FEL structure. Characterizing the radiation-damage free high-resolution structure of PS II [82] is yet only the first step in understanding the atomic details of water splitting, and TR-SFX studies that aimed at resolving time-dependent structural changes following light absorption were then carried out. In the first published TR-SFX study on PS II, simultaneous SFX and X-ray emission spectroscopy (XES) data were collected on crystals in the dark-adapted S1 state, and in the S2 state generated by light illumination with a pump–probe delay of 0.4–0.5 s [37]. Electron density maps of the S2 structure at 5.7 Å resolution did not show differences with respect to the S1 structure and it was concluded that structural changes, if present, are too small to be seen at that resolution. The simultaneous collection of SFX and XES data not only allowed for assigning the dominant S-state to the structural data collected but also provided evidence that the fs pulses probed the intact electronic structure of metal centers in the OEC. Subsequent studies focused on structural changes that accompany the transition to the S3 state. While Kupitz and coworkers reported changes in the structure of an S3 enriched state at 5.5 Å resolution compared to S1 [45], a similar study by Kern and coworkers came to the conclusion that any structural changes related to the transitions between S1 and S3 must be smaller than what can be detected at the resolution achieved (between 4.5 and 5.2 Å) [38, 66]. The resolution of S3 enriched structures was then improved to 2.25 Å [99] and 2.35 Å [83], yet without reaching a consensus on the nature of associated conformational changes. In one case [99], ∼0.1 Å changes in metal distances in the OEC were reported, that is, changes small enough to be at the limit of uncertainty. In the other [83], peaks were observed on the OEC in Fourier difference maps that were interpreted as the appearance of a new oxygen atom and the displacement of a water molecule. Awaiting further improvements in diffraction resolution, the jury is thus still out as to time-resolved changes in the OEC during water splitting.
Another molecular mechanism actively studied by TR-SFX is the structural sequence of photo-intermediates in the light-driven proton pump bacteriorhodopsin (bR). Despite being a well-studied model for membrane proteins, certain structural details of the bR photocycle remain controversially discussed because of a possible convolution with radiation-induced changes in cryo-trapped structural intermediates solved at synchrotron sources [95]. A proof-of-principle that TR-SFX on bR is possible has been provided by Nogly and coworkers [60], using a thick high-viscosity jet to present lipidic-cubic phase (LCP) grown bR crystals to the X-ray beam. Using femtosecond excitation and a pump–probe delay of 1 ms, the structure of the M photointermediate could be solved at 2.3 Å resolution. As multiple consecutive excitations are not possible during the fs pump-pulse duration (see detailed discussion in Sect. 11.5), the intermediate-state occupancy was limited to 13%. The first extensive TR-SFX study on bR presented diffraction data collected following nanosecond excitation and spanning five orders of magnitude in time, from ns to ms [56]. Transient structures solved at 2.1 Å resolution covered photointermediates K to M, and provided a three-dimensional view of the structural changes underlying unidirectional proton pumping. In the earliest time-point studied (16 ns), the chromophore (retinal) had already photoisomerized, so that ultrafast changes right after photon absorption are still to be uncovered.
Two heme proteins other than myoglobin (see below) have been studied by time-resolved crystallography at an X-ray FEL, namely cytochrome c oxidase [77] and NO reductase [89]. In the former, CO release has been followed 20 ns and 100 μs after photolysis with a ns pump-laser, and the characterized structural changes allowed for establishing a mechanism for functionally relevant closing of a water channel [77]. Similar to the characterization of the radiation-damage free high-resolution structure of PS II [82], and at variance with most TR-SFX studies published so far, cytochrome c oxidase crystals were not presented as micron-sized samples in random orientations within a jet or by a conveyer-belt drive, but rather large (up to 500 μm) macrocrystals were rotated and translated through the FEL beam (serial femtosecond rotational crystallography (SF-ROX) [31]). The achievement standing out in the study on NO reductase is the first and successful use of caged compounds in TR-SFX. The enzyme was indeed rendered photosensitive by complexation with a photolabile precursor of NO (caged NO), which was cleaved on the μs time scale by means of ns laser irradiation at 308 nm. X-ray data were collected at a pump–probe delay of 20 ms, allowing for determination of the structure of the ferric NO complex at 2.1 Å resolution. Unlike corresponding synchrotron structures, the TR-SFX structure was devoid of X-ray radiation damage, as corroborated by accompanying QM/MM studies [89].
11.4.1 Ultrafast TR-SFX on the fs—ps Time Scale
Three TR-SFX studies have so far been published beyond the 100 ps resolution-limit of synchrotron-based Laue crystallography. Pump–probe delays of a picosecond and shorter time allowed for entering the time scale of photochemical reactions—a regime inaccessible by X-ray sources other than X-ray FELs—enabling visualization of ultrafast light-induced motions in carbonmonoxy myoglobin [2] and characterization of excited-state photoisomerization intermediates in PYP [63] and in a reversibly photoswitchable variant of the green fluorescent protein (GFP, [15]). In carbonmonoxy (CO) myoglobin (Mb), structural changes following light-activated cleavage of the Fe-CO bond were captured at nominal delays of 0.5 ps, 1 ps, 3 ps, 10 ps, 50 ps and 150 ps [2]. In line with results from other methods, the crystallographic maps showed that within 1 ps after photodissociation, CO has moved to its primary docking site, the heme has domed, and side-chains of neighboring residues have moved (Fig. 11.5a). Significant main-chain conformational changes were observed on the ps time scale with helices moving essentially as rigid bodies, in line with time-resolved solution scattering experiments [47]. The jitter between pump laser and X-ray FEL pulses caused the nominally 500 fs time delay data to contain images with various true time delays, so that they could be sorted into datasets with average time delays of −100 to +600 fs, with bin widths of ∼100 fs. Refinement against these ultrafast datasets revealed that residues in the heme-binding pocket showed an oscillatory behavior with a 500 fs period upon cleavage of the CO. These oscillations were noted to fit very well inside the general normal mode model for protein dynamics [53], in which fast coherent motions ultimately couple with larger-scale displacements of entire helices (Fig. 11.5b). In PYP, structural changes of the p-coumaric acid chromophore after femtosecond illumination at 450 nm have been followed at nominal pump–probe delays of 0.3 ps, 0.6 ps and 3 ps [63]. A structural transition has been proposed to occur at around 0.59 ps, associated with a trans-to-cis isomerization of the chromophore that involves some atoms moving by as much as 1.3 Å, on the sub-ps time scale (Fig. 11.5c). At delays shorter than 0.59 ps, the chromophore displays a trans, yet distorted, conformation, suggesting that it is still in the electronic excited state. At longer pump–probe delays, a cis chromophore is observed, indicating relaxation to the ground state. In the reversibly photoswitchable fluorescent protein rsEGFP2, used for nanoscale imaging of living cells by means of REversible Saturable Optical Linear Fluorescence Transitions (RESOLFT) microscopy [26], photoisomerization intermediates have been characterized 1 and 3 ps after femtosecond illumination of the crystalline protein in its nonfluorescent off-state with the hydroxybenzylidene imidazolinone chromophore in the planar trans configuration [15]. One ps after illumination, the chromophore is fully twisted, with its two rings oriented perpendicular to each other (Fig. 11.5d). Quantum-mechanics/molecular-mechanics (QM/MM) and excited-state molecular dynamics simulations and time-resolved absorption spectroscopy in solution indicated that the twisted chromophore conformation after 1 ps corresponds to that of the electronic excited S1 state, close to the conical intersection where the system relaxes back to the ground state. The entire helix carrying the chromophore shifts along its axis, so as to permit chromophore twisting (Fig. 11.5e). Three ps after illumination, the twisted chromophore conformation is less occupied than at 1 ps, and features indicative of the presence of the cis chromophore are present in the difference Fourier map. The twisted chromophore (Fig. 11.5d, e) thus represents a photoisomerization intermediate, half-way between trans and cis isomers. The maximum displacement of a chromophore atom (phenol OH group) is 4.5 Å in rsEGFP2—that is, much larger than in the case of PYP (1.3 Å)—possibly explaining why photoisomerization takes longer in rsEGFP2 than in the PYP (at 1 ps, rsEGFP2 is still in the excited state, while PYP is already in the ground state again). The excited-state structure of rsEGFP2 (Fig. 11.5d) was the basis for rationally improving this protein used as a molecular label in super-resolution fluorescence microscopy. The twisted chromophore conformation indicated that mutating Val151 (Fig. 11.5d) into an alanine residue should facilitate photoswitching by enlarging the pocket at the tip of the chromophore. Indeed, the V151A mutant turned out to have a twofold increased photoswitching quantum yield [15].
Since TR-SFX on the picosecond time scale requires pumping with a femtosecond pulse, reaction initiation is limited by the primary quantum yield. As a consequence, reaction initiation in the PYP study was low (structurally estimated to be 13% [63]) and the intermediate-states in rsEGFP2 were present at a total occupancy of maximum 7% [15]. An additional caveat of femtosecond excitation is the requirement of using high pump laser power densities (380 GW/cm2 were used for Mb [2], 570 GW/cm2 for PYP [63] and 400 GW/cm2 for rsEGFP2 [15]), which carries the risk of creating artifacts (see more extensive discussion below). Consequently, all three ps TR-SFX studies relied on complementary QM/MM and optical spectroscopy experiments to interpret and validate the intermediate-state structures obtained.
11.5 Challenges and Limitations of TR-SFX
One might face several challenges and limitations when attempting to carry out TR-SFX. The first and most obvious is the necessity to obtain well-diffracting crystals of the protein of interest, or a functional fraction thereof. When crystals are not available, or when crystal packing hinders or even prevents some of the macromolecular motions required for biological activity, time-resolved solution scattering [10] can be used to follow conformational changes at low resolution after triggering macromolecular activity (see Chap. 15 of this book for further discussions on this matter). Below, we discuss those challenges and limitations that arise when well-diffracting crystals have already been obtained.
11.5.1 Spectroscopic Characterization of Protein Microcrystals
A prerequisite for TR-SFX on photosensitive proteins is to characterize the kinetics of photoproduct formation by spectroscopic methods. The major limiting factor in spectroscopic studies of optically dense protein crystals is the difficulty to grow them in a suitable shape for spectroscopy. Special approaches to produce large, flat crystals for spectroscopic studies were developed in some cases [24, 41] but they cannot be easily generalized to every crystallization condition. Furthermore, it may well be that the crystalline form most suited for spectroscopic studies is not that which diffracts to the highest resolution or is best suited for the TR structural studies, in terms of solvent content, nature of crystal contacts, presence of precipitating agents, unit cell dimensions and space group, radiation sensitivity, etc. An alternative strategy is to characterize the kinetics of photoproduct formation by the protein in solution, and assume the results to be valid for the crystalline state. However, there are several examples showing that not only the time constants but also the kinetic schemes can be altered in protein crystals [18, 58, 89, 98]. A recent work showed the feasibility of single-wavelength spectroscopic characterization of PYP microcrystals after optical femtosecond excitation [33].
11.5.2 Pump Light Penetration in Protein Crystals
The protein concentration in crystals can be as high as 30–60 mM, resulting in a very high optical density in the wavelength range where the chromophore moiety absorbs. As shown in Fig. 11.4 for photosensitive proteins recently studied by TR-SFX, the penetration depth for pump-laser light at the wavelengths used in TR structural studies is limited to only a few microns or less. In case of optical activity, the light penetration can also be dependent on the crystal orientation that is generally randomly distributed in the case of TR-SFX. It is recommended to tune experimental parameters like crystal size, pump-light wavelength, and pump-light polarization in order to ensure sufficient light penetration into the crystal—notably when using an experimental setup where the pump and probe beams are not collinear, that is, when the probability increases of probing a poorly illuminated part of the crystal.
11.5.3 Excitation Efficiency and Pump Power Density Issues
An important parameter to be taken into account when designing a TR-SFX experiment is the expected excitation efficiency by the pump light. The excitation efficiency is defined as the ratio between the number of protein molecules wherein the photoreaction under study has effectively been triggered and the total number of photons impinging on the crystal. This efficiency is the product of the absorption cross-section at the given pump-light wavelength and polarization, and the primary quantum yield of the photoreaction. The primary quantum yield is the probability that an absorbed photon triggers the photoreaction. When the pump-pulse length is shorter than the excited-state lifetime, the extent of reaction initiation is limited by the primary quantum yield. For pump-pulses being longer than the lifetime of the excited state, a protein molecule that after excitation has decayed back to the ground state (instead of progressing further along the photoreaction pathway) can be excited more than once, resulting in an extent of reaction initiation that exceeds the primary quantum yield. For example, in recent TR-SFX experiments on PYP, whose excited state lifetime is about 500 fs [48], the extent of reaction initiation was 40% with nanosecond excitation [87]. With femtosecond excitation, the extent of reaction initiation was only 13%, comparable to the primary quantum yield [63].
To study excited-state structural dynamics on the femtosecond/picosecond time scale, only femtosecond excitation can be used. The usual practice is to excite the sample with a photon density corresponding to an average of at least one absorbed photon per chromophore. Considering typical values of protein concentrations in a crystal, of microcrystal sizes and of extinction coefficients, one absorbed photon per chromophore corresponds to a peak power density of the order of tens of GW/cm2 for femtosecond excitation. Such a high value is not only far beyond illumination power densities in any biologically relevant condition, but can also produce parasitic processes, like photobleaching, ionization, and radical formation, which will compete with the photophysical process under study and may produce structural artifacts. Moreover, it is common practice to exceed the regime of one absorbed photon per chromophore, in order to compensate for possible pump-laser intensity fluctuations and position drifts affecting the pump–probe spatial overlap. Illustratively, the three recent femtosecond experiments discussed above used power densities of the order of 380 GW/cm2 [2], 400 GW/cm2 [15], and 570 GW/cm2 [63]. The limited access to X-ray FEL beam time has so far precluded the possibility of routinely performing pump power titrations, as required to define the linear regime within which excitation efficiency is maximized. Therefore, it is recommended to perform at least a preliminary thorough spectroscopic characterization of the system upon femtosecond excitation at various pump-power densities, prior to engaging in TR-SFX studies.
11.5.4 Time Resolution and Pump–Probe Synchronization Diagnostics in Ultrafast Experiments
The time resolution of an ultrafast TR-SFX experiment is given by the convolution of the femtosecond pump pulse temporal profile with the femtosecond probe pulse temporal profile, and should in principle also be in the femtosecond range. However, the effective time resolution also depends on the precision of the pump–probe synchronization. Optical laser pulses are synchronized to the X-ray FEL pulses by radiofrequency phase-locking technology. Currently, the precision of temporal synchronization with this technology is limited by the shot-to-shot jitters of optical and X-ray FEL pulses, which is on the order of more than 100 fs [25], and by thermal drifts of the order of a few picoseconds after several hours, which is the typical time scale of a TR-SFX data collection. Although there is an effort to improve the pump–probe synchronization precision and stability [16, 50], the approach currently used is to directly measure, on a shot-to-shot basis, the relative arrival time delay of optical and X-ray pulses close to the experimental interaction region. This real-time diagnostic approach allows for re-sorting data recorded for a given pump–probe delay, by substituting the nominal time delay assigned to each image with the time delay corrected after post-processing. This method was shown to allow for sub-10 fs precision in time delay determination [27] and was successfully applied to process TR-SFX data with time resolution of ∼100 fs [2, 63], limited by crystallographic data redundancy rather than by the timing diagnostic itself.
11.5.5 Structure Refinement of Low-Occupancy States
In pump–probe TR-SFX experiments on photosensitive proteins, limited pump-light penetration into protein crystals (Fig. 11.4) and non-unity excitation efficiency together result in only a fraction of the probed molecules being excited and effectively undergoing conformational changes. The probed diffraction signal is thus a mixture of that arising from molecules within which the reaction has been triggered, and from molecules that are still in the resting state. Most often, the 2mFo-DFc experimental electron density maps for the datasets without (dark) and with (light) pump excitation hardly differ and straightforward building of a model for the transient structure is not possible. Consequently, insights into conformational changes are obtained by examination of structure-factor amplitude Fourier difference maps, calculated by subtracting observed structure factor amplitudes for the dark data set (Fo dark) from those of the light data set (Fo light), and then phasing this difference with phases calculated from the dark model. Experimental Fo light − Fo dark differences can be weighted prior to map calculation based on the Bayesian likelihood that each Fo light − Fo dark structure factor amplitude difference is observed at a given resolution within measurement errors (sigmas)—an approach termed Q-weighting [91]. Q-weighting was introduced as a means to improve the estimates of difference amplitudes and to reduce the noise in difference maps calculated from time-resolved polychromatic Laue data, but it also increases the signal-to-noise ratio of amplitude differences from monochromatic datasets. In Fo light − Fo dark maps, negative and positive peaks indicate positions from which electron density has vanished and appeared upon pumping, respectively. Using this information, and prior knowledge from the resting-state model, the crystallographer may build a model for the transient structure, include it as an alternate conformer in the light structure, and then refine this new model in terms of coordinates and occupancy in the reciprocal space. An initial guess for the validity and occupancy of the transient conformer in the light structure can be obtained by computing Fc light − Fc dark maps for various relative occupancies of the resting-state and transient states in the light structure, and comparing peak heights (on residues of interest) in these and in the experimental Fo light − Fo dark map. Another approach is to compute extrapolated structure factors for the transient structure at 100% occupancy (Fext light), and then to perform difference refinement against these [88]. By this method, 2mFext light − DFc dark and mFext light − DFc dark electron density maps can be obtained that will guide model-building for the transient structure. The transient structure is then refined by minimizing the residual between the extrapolated and calculated structure factor amplitudes [19, 79, 88]. Again, an initial estimate of the occupancy of the transient state in the light dataset is needed to generate extrapolated structure factors, using the following equation: Fext light = α * Q * (Fo light − Fo dark) + Fo dark, where Q is the optional Bayesian-based weighting factor and 1/α is the occupancy of the time-evolved state in the light structure. This estimate may again be obtained by computing Fc light − Fc dark maps and comparing peak heights on residues of interest in these and in the experimental Fo light − Fo dark map [17]. Alternatively, occupancy-scaling can rely on a similar comparison between the experimental Fo light − Fo dark map, and the mFext light − DFc dark maps computed using extrapolated Fext light for various occupancies of the transient state in the light dataset [15].
11.5.6 Kinetic Analysis of Time-Resolved Diffraction Patterns
A TR-SFX experiment produces a time-dependent set of electron density maps spanning a given time range and possibly containing time-dependent changes in density as the reaction proceeds. The strategy to interpret such a time-resolved dataset depends on the time scale explored, which we divide into dynamic (ultrafast, i.e., fs–ps) and kinetic (several ps and longer) regimes. In the dynamic regime, structural changes reflect the coherent dynamics of atomic motions, allowing for a direct comparison in terms of structure and time scale with results from molecular dynamics simulations. Time-dependent maps each provide a snapshot of the protein in motion and allow for tracking non-exponential structural evolutions such as side chain oscillations in Mb [2]. In the kinetic regime, the time evolution of the crystalline protein ensemble is governed by the energy barriers separating structural intermediates. The time-dependence of density maps then arises from the variation in population of the underlying time-independent intermediate structures, each associated with a reaction intermediate. The objective of a kinetic analysis of time-dependent maps is to extract a set of time-independent maps, each corresponding to a different reaction intermediate that can be used to determine the reaction pathway along with rate coefficients connecting the different reaction intermediates. The two main strategies described in the literature for a kinetic analysis are singular value decomposition (SVD) [72] and cluster analysis [42]. An exhaustive description of the two methods goes beyond the scope of this chapter and we refer to the dedicated literature [42, 64, 72]. Briefly, the SVD method, extensively used in the analysis of any kind of time-resolved data, is a mathematical procedure that represents each data set by two sets of vectors, weighted by corresponding so-called singular values. The first set of singular vectors forms a time-independent orthonormal basis on which all time-dependent data can be decomposed; the second set of singular vectors describes the time-dependence of the first set. Since the basis vectors are weighted by their singular values, the data matrix can be approximated by a subset of vectors which contains signal above the noise level. The obtained reduced representation of the data sets allows for interpreting the time evolution of singular vectors in terms of a kinetic model. The cluster analysis is a general method used in statistics to group objects contained in an ensemble, based on a similarity defined by a given mathematical measure. This method has been proposed to deconvolute time-independent reaction intermediates from an intrinsically noisy and time-dependent electron density datasets produced by time-resolved crystallography, via a so-called “analytical trapping procedure.” A complete SVD or cluster analysis has not yet been carried in any of the published TR-SFX studies.
11.5.7 Sample Presentation and Consumption
The ultrashort and highly intense nature of X-ray FEL pulses is what allows for time-resolved studies down to the sub-ps timescale, at the atomic level of resolution. These characteristics are also at the origin of the high sample consumption rate of SFX and TR-SFX experiments. Indeed, because crystals virtually stand still during the fs exposure, all measured reflection intensities are partial, requiring collection from myriads of crystals to obtain a meaningful estimate of structure factor amplitudes. Furthermore, the intense X-ray beam eventually destroys the crystals, requiring the sample to be constantly replenished. Different means to present crystalline samples to the X-ray FEL beam exist, and they are extensively discussed in Chap. 5 of this book. All of them share the prospect of enabling TR-SFX studies, although on different systems and timescales. The first—and to date most successful—approach has been injection of crystalline samples across the X-ray FEL beam by means of gas-focused liquid jets [93]. Sample consumption is the highest when working with liquid jets, with one out of several hundred thousand crystals interacting with an X-ray pulse at a repetition rate of 120 Hz [70], due to the high injection speed (typically 10 m/s) required for the stability and sub-10 μm thickness of the jet (important for minimizing background scattering noise from the jet itself). Liquid jets can be highly stable and enable collection of data at pump–probe delays from sub-ps to a few μs, with pump excitation outside the injector nozzle. Because of the high jet speed, longer pump–probe delays are not easily accessible. Liquid jets remain the first choice in cases where crystalline sample production quantity is not an issue. Illustratively, all three ps timescale pump–probe TR-SFX studies published thus far were conducted using such an injection system, requiring about 1 g of protein crystals for a five-shift experiment at the LCLS [2, 15, 63]. To reduce sample consumption, crystals can be grown or embedded in a viscous carrier matrix prior to injection, enabling a stable jet at a reduced injection speed of typically a few mm/s [5, 84, 92]. The first matrix of the kind was a lipidic cubic phase, in which crystals of the human serotonin receptor where grown before injection across the X-ray FEL beam [49], allowing for structure determination from as little as 0.5 mg of protein. Rapidly after, other inert media were proposed that also form easily extrudable gel-like matrices from crystals that do not grow in a lipidic cubic phase [5, 13, 43, 84,85,86]. High-viscosity jets have been used in pump–probe TR-SFX experiments on bR [56, 60], where pump–probe delays in the ms range were accessed due to the slow jet speed. Weaknesses of high-viscosity jets for TR-SFX include reduced transparency that limits the excitation efficiency [89], the possibility of fluctuating jet speeds that reduce precision of longer pump–probe delays and the inability to address diffusion-based processes that are prohibited by the high viscosity of the matrix. Furthermore, the slow flow leads to concerns about pre-illumination of the sample from the previous laser pulses, a problem that is worse for irreversible reactions. Recently, the design was reported of a conveyer belt on which crystals may ride following acoustic droplet ejection, allowing for multiple illuminations, and/or exposure to solute or vapors prior to interaction with the X-ray beam [20]. Hence, the conveyer belt system is in theory adapted both for pump–probe and diffusion based TR-SFX experiments. Published work to date used only the pump–probe mode, to provide insights into the photocycle of PS II [99], but it is expected that other experiments will soon take advantage of the versatility and completeness of the setup. Weaknesses are that the size of ejected drops remains high and that the device does not run in vacuum, both generating high background. TR-SFX crystallography data can also be collected from a much smaller number of crystals presented sequentially to the X-ray beam in a cryo-loop, as recently shown for the cytochrome c oxidase [77]. About 40 large crystals (0.5 mm) per time point were used, each of which was exposed at multiple angles and positions while maintained in a humid air flow at 4 °C. Chip-based micro-compartmented systems allowing for sequestration of microcrystals and their presentation to the X-ray beam at a speed matching the repetition rate of most X-ray FEL sources have also been proposed and demonstrated, which could enable TR-SFX on microcrystals with minimal background and high hit rates [61, 62, 65]. Solid-support based-approaches thus hold the promise of allowing for a more efficient use of crystalline material and beamtime, during TR-SFX experiments on photosensitive proteins or proteins complexed with caged compounds. In combination with microfluidic devices, solid-support based-approaches could also provide insight into diffusion-based mechanisms, by means of a variation on the mix-and-inject approach [71].
11.6 Perspective on Future Applications of TR-SFX
Since the first study in 2012 [1], an increasing number of different light-sensitive proteins have been studied by TR-SFX (Fig. 11.6). Even if most of them are model systems that have been extensively studied by various biophysical techniques, TR-SFX experiments, in particular those carried out on the ultrafast time scale, provided new insights. For some model systems (e.g., bR and other rhodopsins), ultrafast TR-SFX is yet to come. For others (e.g., PS II), further insight from TR-SFX awaits increased diffraction resolution. Time-resolved changes along the reaction pathway are still to be uncovered by TR-SFX in a large number of inherently photosensitive proteins (e.g., reaction center, phytochromes, and cryptochromes), as well as in photoenzymes. Also, it is expected that more and more inherently light insensitive proteins will be studied by TR-SFX using caged compounds or synthetic photoswitches [9]. In these cases, the functional relevance of light-induced motions will have to be assessed carefully.
With the proven possibility to be carried out on the ultrafast time scale, TR-SFX provides the unique opportunity to link photochemistry (fs–ps motions) and photobiology (μs-ms motions) for the first time on a structural basis. The prospect of understanding the entire chain of reactions, from photon absorption to signal transduction in the case of rhodopsin, or to proton pumping in the case of bR, is now finally within reach. The complementarity of X-ray FELs and synchrotrons, in particular the highly brilliant fourth generation sources to come up in the next years, is likely to gain shape in the near future. Indeed, serial synchrotron crystallography has gained momentum in the past three years [4, 5, 14, 22, 30, 32, 35, 51, 52, 59, 61, 62, 76, 81, 94] and will be able to cover time-resolved serial crystallography on the μs-ms time scale, should fourth generation sources such as diffraction limited storage rings produce the required photon flux. TR-SFX might then mostly focus on ultrafast studies and on cases where X-ray induced changes need to be avoided.
Thus far, all TR-SFX studies but two [46, 80] have been carried out on light sensitive proteins with an optical laser as the pump method (Fig. 11.6). Other triggering methods mentioned in Sect. 11.2 (T-jump, THz irradiation, X-ray irradiation, …) are likely to be incorporated into TR-SFX protocols to study light-insensitive proteins, as will be the mix-and-inject approach discussed in Chap. 12 of this book.
TR-SFX studies published to date “only” provided one or up to a dozen [56] snapshots of a protein structure after reaction initiation, limited by the extremely scarce X-ray FEL beam time and the requirement to collect a large number of indexable diffraction patterns per data set. With three more X-ray FELs having started operation in 2017, on the one hand, and optimization of SFX data collection and data processing schemes, on the other hand, both bottlenecks are expected to be mitigated, so that TR-SFX may soon provide real-time molecular movies of biomolecules in action.
References
Aquila, A., Hunter, M. S., Doak, R. B., Kirian, R. A., Fromme, P., White, T. A., et al. (2012). Time-resolved protein nanocrystallography using an X-ray free-electron laser. Optics Express, 20(3), 2706–2716. https://doi.org/10.1364/OE.20.002706.
Barends, T. R., Foucar, L., Ardevol, A., Nass, K., Aquila, A., Botha, S., et al. (2015). Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation. Science, 350(6259), 445–450. https://doi.org/10.1126/science.aac5492.
Baxter, R. H., Ponomarenko, N., Srajer, V., Pahl, R., Moffat, K., & Norris, J. R. (2004). Time-resolved crystallographic studies of light-induced structural changes in the photosynthetic reaction center. Proceedings of the National Academy of Sciences of the United States of America, 101(16), 5982–5987. https://doi.org/10.1073/pnas.0306840101.
Beyerlein, K. R., Dierksmeyer, D., Mariani, V., Kuhn, M., Sarrou, I., Ottaviano, A., et al. (2017). Mix-and-diffuse serial synchrotron crystallography. IUCrJ, 4(Pt 6), 769–777. https://doi.org/10.1107/S2052252517013124.
Botha, S., Nass, K., Barends, T. R., Kabsch, W., Latz, B., Dworkowski, F., et al. (2015). Room-temperature serial crystallography at synchrotron X-ray sources using slowly flowing free-standing high-viscosity microstreams. Acta crystallographica Section D, Biological crystallography, 71(Pt 2), 387–397. https://doi.org/10.1107/S1399004714026327.
Bourgeois, D., Vallone, B., Arcovito, A., Sciara, G., Schotte, F., Anfinrud, P. A., et al. (2006). Extended subnanosecond structural dynamics of myoglobin revealed by Laue crystallography. Proceedings of the National Academy of Sciences of the United States of America, 103(13), 4924–4929. https://doi.org/10.1073/pnas.0508880103.
Bourgeois, D., Vallone, B., Schotte, F., Arcovito, A., Miele, A. E., Sciara, G., et al. (2003). Complex landscape of protein structural dynamics unveiled by nanosecond Laue crystallography. Proceedings of the National Academy of Sciences of the United States of America, 100(15), 8704–8709. https://doi.org/10.1073/pnas.1430900100.
Bourgeois, D., & Weik, M. (2009). Kinetic protein crystallography: A tool to watch proteins in action. Crystallography Reviews, 15(2), 87–118.
Broichhagen, J., Frank, J. A., & Trauner, D. (2015). A roadmap to success in Photopharmacology. Accounts of Chemical Research, 48(7), 1947–1960. https://doi.org/10.1021/acs.accounts.5b00129.
Cammarata, M., Levantino, M., Schotte, F., Anfinrud, P. A., Ewald, F., Choi, J., et al. (2008). Tracking the structural dynamics of proteins in solution using time-resolved wide-angle X-ray scattering. Nature Methods, 5(10), 881–886.
Chapman, H. N., Fromme, P., Barty, A., White, T. A., Kirian, R. A., Aquila, A., et al. (2011). Femtosecond X-ray protein nanocrystallography. Nature, 470(7332), 73–77.
Colletier, J. P., Bourgeois, D., Sanson, B., Fournier, D., Sussman, J. L., Silman, I., et al. (2008). Shoot-and-trap: Use of specific x-ray damage to study structural protein dynamics by temperature-controlled cryo-crystallography. Proceedings of the National Academy of Sciences of the United States of America, 105(33), 11742–11747.
Conrad, C. E., Basu, S., James, D., Wang, D., Schaffer, A., Roy-Chowdhury, S., et al. (2015). A novel inert crystal delivery medium for serial femtosecond crystallography. IUCrJ, 2(4), 421–430. https://doi.org/10.1107/S2052252515009811.
Coquelle, N., Brewster, A. S., Kapp, U., Shilova, A., Weinhausen, B., Burghammer, M., et al. (2015). Raster-scanning serial protein crystallography using micro- and nano-focused synchrotron beams. Acta Crystallographica Section D, 71(5), 1184–1196. https://doi.org/10.1107/S1399004715004514.
Coquelle, N., Sliwa, M., Woodhouse, J., Schirò, G., Adam, V., Aquila, A., et al. (2018). Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography. Nature Chemistry, 10, 31–37. https://doi.org/10.1038/nchem.2853.
Danailov, M. B., Bencivenga, F., Capotondi, F., Casolari, F., Cinquegrana, P., Demidovich, A., et al. (2014). Towards jitter-free pump-probe measurements at seeded free electron laser facilities. Optics Express, 22(11), 12869–12879. https://doi.org/10.1364/oe.22.012869.
Duan, C., Adam, V., Byrdin, M., Ridard, J., Kieffer-Jaquinod, S., Morlot, C., et al. (2013). Structural evidence for a two-regime photobleaching mechanism in a reversibly switchable fluorescent protein. Journal of the American Chemical Society, 135(42), 15841–15850. https://doi.org/10.1021/ja406860e.
Efremov, R., Gordeliy, V. I., Heberle, J., & Büldt, G. (2006). Time-resolved microspectroscopy on a single crystal of bacteriorhodopsin reveals lattice-induced differences in the photocycle kinetics. Biophysical Journal, 91(4), 1441–1451. https://doi.org/10.1529/biophysj.106.083345.
Fermi, G., Perutz, M. F., Dickinson, L. C., & Chien, J. C. W. (1982). Structure of human deoxy cobalt haemoglobin. Journal of Molecular Biology, 155(4), 495–505. https://doi.org/10.1016/0022-2836(82)90483-1.
Fuller, F. D., Gul, S., Chatterjee, R., Burgie, E. S., Young, I. D., Lebrette, H., et al. (2017). Drop-on-demand sample delivery for studying biocatalysts in action at X-ray free-electron lasers. Nature Methods, 14(4), 443–449. https://doi.org/10.1038/nmeth.4195.
Garman, E. F., & Weik, M. (2017). Radiation damage in macromolecular crystallography. Methods in Molecular Biology, 1607, 467–489. https://doi.org/10.1007/978-1-4939-7000-1_20.
Gati, C., Bourenkov, G., Klinge, M., Rehders, D., Stellato, F., Oberthur, D., et al. (2014). Serial crystallography on in vivo grown microcrystals using synchrotron radiation. IUCrJ, 1(Pt 2), 87–94. https://doi.org/10.1107/S2052252513033939.
Genick, U. K., Borgstahl, G. E., Ng, K., Ren, Z., Pradervand, C., Burke, P. M., et al. (1997). Structure of a protein photocycle intermediate by millisecond time-resolved crystallography. Science, 275(5305), 1471–1475.
Gerwert, K., Hess, B., Michel, H., & Buchanan, S. (1988). FTIR studies on crystals of photosynthetic reaction centers. FEBS Letters, 232(2), 303–307. https://doi.org/10.1016/0014-5793(88)80758-0.
Glownia, J. M., Cryan, J., Andreasson, J., Belkacem, A., Berrah, N., Blaga, C. I., et al. (2010). Time-resolved pump-probe experiments at the LCLS. Optics Express, 18(17), 17620–17630. https://doi.org/10.1364/oe.18.017620.
Grotjohann, T., Testa, I., Reuss, M., Brakemann, T., Eggeling, C., Hell, S. W., et al. (2012). rsEGFP2 enables fast RESOLFT nanoscopy of living cells. eLife, 1, e00248. https://doi.org/10.7554/eLife.00248.
Harmand, M., Coffee, R., Bionta, M. R., Chollet, M., French, D., Zhu, D., et al. (2013). Achieving few-femtosecond time-sorting at hard X-ray free-electron lasers. Nature Photonics, 7, 215. https://doi.org/10.1038/nphoton.2013.11.
Hekstra, D. R., White, K. I., Socolich, M. A., Henning, R. W., Srajer, V., & Ranganathan, R. (2016). Electric-field-stimulated protein mechanics. Nature, 540(7633), 400–405. https://doi.org/10.1038/nature20571.
Henzler-Wildman, K., & Kern, D. (2007). Dynamic personalities of proteins. Nature, 450(7172), 964–972.
Heymann, M., Opthalage, A., Wierman, J. L., Akella, S., Szebenyi, D. M., Gruner, S. M., et al. (2014). Room-temperature serial crystallography using a kinetically optimized microfluidic device for protein crystallization and on-chip X-ray diffraction. IUCrJ, 1(Pt 5), 349–360. https://doi.org/10.1107/S2052252514016960.
Hirata, K., Shinzawa-Itoh, K., Yano, N., Takemura, S., Kato, K., Hatanaka, M., et al. (2014). Determination of damage-free crystal structure of an X-ray-sensitive protein using an XFEL. Nature Methods, 11(7), 734–736. https://doi.org/10.1038/nmeth.2962.
Huang, C. Y., Olieric, V., Ma, P., Panepucci, E., Diederichs, K., Wang, M., et al. (2015). In meso in situ serial X-ray crystallography of soluble and membrane proteins. Acta crystallographica Section D, Biological crystallography, 71(Pt 6), 1238–1256. https://doi.org/10.1107/S1399004715005210.
Hutchison, C. D. M., Kaucikas, M., Tenboer, J., Kupitz, C., Moffat, K., Schmidt, M., et al. (2016). Photocycle populations with femtosecond excitation of crystalline photoactive yellow protein. Chemical Physics Letters, 654, 63–71. https://doi.org/10.1016/j.cplett.2016.04.087.
Ihee, H., Rajagopal, S., Srajer, V., Pahl, R., Anderson, S., Schmidt, M., et al. (2005). Visualizing reaction pathways in photoactive yellow protein from nanoseconds to seconds. Proceedings of the National Academy of Sciences of the United States of America, 102(20), 7145–7150 Epub 2005 May 7143.
Jaeger, K., Dworkowski, F., Nogly, P., Milne, C., Wang, M., & Standfuss, J. (2016). Serial millisecond crystallography of membrane proteins. Advances in Experimental Medicine and Biology, 922, 137–149. https://doi.org/10.1007/978-3-319-35072-1_10.
Jung, Y. O., Lee, J. H., Kim, J., Schmidt, M., Moffat, K., Srajer, V., et al. (2013). Volume-conserving trans-cis isomerization pathways in photoactive yellow protein visualized by picosecond X-ray crystallography. Nature Chemistry, 5(3), 212–220. https://doi.org/10.1038/nchem.1565.
Kern, J., Alonso-Mori, R., Tran, R., Hattne, J., Gildea, R. J., Echols, N., et al. (2013). Simultaneous femtosecond X-ray spectroscopy and diffraction of photosystem II at room temperature. Science, 340(6131), 491–495. https://doi.org/10.1126/science.1234273.
Kern, J., Tran, R., Alonso-Mori, R., Koroidov, S., Echols, N., Hattne, J., et al. (2014). Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy. Nature Communications, 5, 4371. https://doi.org/10.1038/ncomms5371.
Knapp, J. E., Pahl, R., Srajer, V., & Royer Jr., W. E. (2006). Allosteric action in real time: Time-resolved crystallographic studies of a cooperative dimeric hemoglobin. Proceedings of the National Academy of Sciences of the United States of America, 103(20), 7649–7654. https://doi.org/10.1073/pnas.0509411103.
Kok, B., Forbush, B., & McGloin, M. (1970). Cooperation of charges in photosynthetic O2 evolution-I. A linear four step mechanism. Photochemistry and Photobiology, 11(6), 457–475.
Kort, R., Ravelli, R. B., Schotte, F., Bourgeois, D., Crielaard, W., Hellingwerf, K. J., et al. (2003). Characterization of photocycle intermediates in crystalline photoactive yellow protein. Photochemistry and Photobiology, 78(2), 131–137. https://doi.org/10.1562/0031-8655(2003)0780131copiic2.0.co2.
Kostov, K. S., & Moffat, K. (2011). Cluster analysis of time-dependent crystallographic data: Direct identification of time-independent structural intermediates. Biophysical Journal, 100(2), 440–449. https://doi.org/10.1016/j.bpj.2010.10.053.
Kovacsova, G., Grunbein, M. L., Kloos, M., Barends, T. R. M., Schlesinger, R., Heberle, J., et al. (2017). Viscous hydrophilic injection matrices for serial crystallography. IUCrJ, 4(Pt 4), 400–410. https://doi.org/10.1107/S2052252517005140.
Kubelka, J. (2009). Time-resolved methods in biophysics. 9. Laser temperature-jump methods for investigating biomolecular dynamics. Photochemical & Photobiological Sciences, 8(4), 499–512. https://doi.org/10.1039/b819929a.
Kupitz, C., Basu, S., Grotjohann, I., Fromme, R., Zatsepin, N. A., Rendek, K. N., et al. (2014). Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser. Nature, 513(7517), 261–265. https://doi.org/10.1038/nature13453.
Kupitz, C., Olmos, J. L., Holl, M., Tremblay, L., Pande, K., Pandey, S., et al. (2017). Structural enzymology using X-ray free electron lasers. Structural Dynamics, 4(4), 044003. https://doi.org/10.1063/1.4972069.
Levantino, M., Schiro, G., Lemke, H. T., Cottone, G., Glownia, J. M., Zhu, D., et al. (2015). Ultrafast myoglobin structural dynamics observed with an X-ray free-electron laser. Nature Communications, 6, 6772. https://doi.org/10.1038/ncomms7772.
Lincoln, C. N., Fitzpatrick, A. E., & van Thor, J. J. (2012). Photoisomerisation quantum yield and non-linear cross-sections with femtosecond excitation of the photoactive yellow protein. Physical Chemistry Chemical Physics, 14(45), 15752–15764. https://doi.org/10.1039/c2cp41718a.
Liu, W., Wacker, D., Gati, C., Han, G. W., James, D., Wang, D., et al. (2013). Serial femtosecond crystallography of G protein-coupled receptors. Science, 342(6165), 1521–1524. https://doi.org/10.1126/science.1244142.
Löhl, F., Arsov, V., Felber, M., Hacker, K., Jalmuzna, W., Lorbeer, B., et al. (2010). Electron bunch timing with femtosecond precision in a superconducting free-electron laser. Physical Review Letters, 104(14), 144801.
Martin-Garcia, J. M., Conrad, C. E., Nelson, G., Stander, N., Zatsepin, N. A., Zook, J., et al. (2017). Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation. IUCrJ, 4(Pt 4), 439–454. https://doi.org/10.1107/S205225251700570X.
Meents, A., Wiedorn, M. O., Srajer, V., Henning, R., Sarrou, I., Bergtholdt, J., et al. (2017). Pink-beam serial crystallography. Nature Communications, 8(1), 1281. https://doi.org/10.1038/s41467-017-01417-3.
Miller, R. J. D. (1994). Energetics and dynamics of deterministic protein motion. Accounts of Chemical Research, 27(5), 145–150. https://doi.org/10.1021/ar00041a005.
Moffat, K. (1989). Time-resolved macromolecular crystallography. Annual Review of Biophysics and Biophysical Chemistry, 18, 309–332. https://doi.org/10.1146/annurev.bb.18.060189.001521.
Mozzarelli, A., & Rossi, G. L. (1996). Protein function in the crystal. Annual Review of Biophysics and Biomolecular Structure, 25, 343–365.
Nango, E., Royant, A., Kubo, M., Nakane, T., Wickstrand, C., Kimura, T., et al. (2016). A three-dimensional movie of structural changes in bacteriorhodopsin. Science, 354(6319), 1552–1557. https://doi.org/10.1126/science.aah3497.
Neutze, R., Wouts, R., van der Spoel, D., Weckert, E., & Hajdu, J. (2000). Potential for biomolecular imaging with femtosecond X-ray pulses. Nature, 406(6797), 752–757.
Ng, K., Getzoff, E. D., & Moffat, K. (1995). Optical studies of a bacterial photoreceptor protein, photoactive yellow protein, in single crystals. Biochemistry, 34(3), 879–890. https://doi.org/10.1021/bi00003a022.
Nogly, P., James, D., Wang, D., White, T. A., Zatsepin, N., Shilova, A., et al. (2015). Lipidic cubic phase serial millisecond crystallography using synchrotron radiation. IUCrJ, 2(Pt 2), 168–176. https://doi.org/10.1107/S2052252514026487.
Nogly, P., Panneels, V., Nelson, G., Gati, C., Kimura, T., Milne, C., et al. (2016). Lipidic cubic phase injector is a viable crystal delivery system for time-resolved serial crystallography. Nature Communications, 7, 12314. https://doi.org/10.1038/ncomms12314.
Oghbaey, S., Sarracini, A., Ginn, H. M., Pare-Labrosse, O., Kuo, A., Marx, A., et al. (2016). Fixed target combined with spectral mapping: Approaching 100% hit rates for serial crystallography. Acta crystallographica Section D, Structural Biology, 72(Pt 8), 944–955. https://doi.org/10.1107/S2059798316010834.
Owen, R. L., Axford, D., Sherrell, D. A., Kuo, A., Ernst, O. P., Schulz, E. C., et al. (2017). Low-dose fixed-target serial synchrotron crystallography. Acta Crystallographica Section D, 73(4), 373–378. https://doi.org/10.1107/S2059798317002996.
Pande, K., Hutchison, C. D., Groenhof, G., Aquila, A., Robinson, J. S., Tenboer, J., et al. (2016). Femtosecond structural dynamics drives the trans/cis isomerization in photoactive yellow protein. Science, 352(6286), 725–729. https://doi.org/10.1126/science.aad5081.
Rajagopal, S., Kostov, K. S., & Moffat, K. (2004). Analytical trapping: Extraction of time-independent structures from time-dependent crystallographic data. Journal of Structural Biology, 147(3), 211–222. https://doi.org/10.1016/j.jsb.2004.04.007.
Roedig, P., Ginn, H. M., Pakendorf, T., Sutton, G., Harlos, K., Walter, T. S., et al. (2017). High-speed fixed-target serial virus crystallography. Nature Methods, 14(8), 805–810. https://doi.org/10.1038/nmeth.4335.
Sauter, N. K., Echols, N., Adams, P. D., Zwart, P. H., Kern, J., Brewster, A. S., et al. (2016). No observable conformational changes in PSII. Nature, 533(7603), E1–E2.
Schiro, G., Woodhouse, J., Weik, M., Schlichting, I., & Shoeman, R. L. (2017). Simple and efficient system for photoconverting light-sensitive proteins in serial crystallography experiments. Journal of Applied Crystallography, 50(3), 932–939. https://doi.org/10.1107/S1600576717006264.
Schlichting, I., Almo, S. C., Rapp, G., Wilson, K., Petratos, K., Lentfer, A., et al. (1990). Time-resolved X-ray crystallographic study of the conformational change in Ha-Ras p21 protein on GTP hydrolysis. Nature, 345(6273), 309–315.
Schlichting, I., Berendzen, J., Chu, K., Stock, A. M., Maves, S. A., Benson, D. E., et al. (2000). The catalytic pathway of cytochrome p450cam at atomic resolution. Science, 287(5458), 1615–1622.
Schlichting, I., & Miao, J. (2012). Emerging opportunities in structural biology with X-ray free-electron lasers. Current Opinion in Structural Biology, 22(5), 613–626. https://doi.org/10.1016/j.sbi.2012.07.015.
Schmidt, M. (2013). Mix and inject: Reaction initiation by diffusion for time-resolved macromolecular crystallography. Advances in Condensed Matter Physics, 2013, 10. https://doi.org/10.1155/2013/167276.
Schmidt, M., Rajagopal, S., Ren, Z., & Moffat, K. (2003). Application of singular value decomposition to the analysis of time-resolved macromolecular X-ray data. Biophysical Journal, 84(3), 2112–2129. https://doi.org/10.1016/S0006-3495(03)75018-8.
Schoenlein, R. W., Peteanu, L. A., Mathies, R. A., & Shank, C. V. (1991). The first step in vision: Femtosecond isomerization of rhodopsin. Science, 254(5030), 412–415.
Schotte, F., Cho, H. S., Kaila, V. R., Kamikubo, H., Dashdorj, N., Henry, E. R., et al. (2012). Watching a signaling protein function in real time via 100-ps time-resolved Laue crystallography. Proceedings of the National Academy of Sciences of the United States of America, 109(47), 19256–19261. https://doi.org/10.1073/pnas.1210938109.
Schotte, F., Lim, M., Jackson, T. A., Smirnov, A. V., Soman, J., Olson, J. S., et al. (2003). Watching a protein as it functions with 150-ps time-resolved x-ray crystallography. Science, 300(5627), 1944–1947. https://doi.org/10.1126/science.1078797.
Schubert, R., Kapis, S., Gicquel, Y., Bourenkov, G., Schneider, T. R., Heymann, M., et al. (2016). A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution. IUCrJ, 3(6), 393–401. https://doi.org/10.1107/S2052252516016304.
Shimada, A., Kubo, M., Baba, S., Yamashita, K., Hirata, K., Ueno, G., et al. (2017). A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase. Science Advances, 3(7), e1603042. https://doi.org/10.1126/sciadv.1603042.
Sierra, R. G., Laksmono, H., Kern, J., Tran, R., Hattne, J., Alonso-Mori, R., et al. (2012). Nanoflow electrospinning serial femtosecond crystallography. Acta crystallographica Section D, Biological crystallography, 68(Pt 11), 1584–1587. https://doi.org/10.1107/S0907444912038152.
Šrajer, V., T-y, T., Ursby, T., Pradervand, C., Ren, Z., Adachi, S.-i., et al. (1996). Photolysis of the carbon monoxide complex of myoglobin: Nanosecond time-resolved crystallography. Science, 274(5293), 1726.
Stagno, J. R., Liu, Y., Bhandari, Y. R., Conrad, C. E., Panja, S., Swain, M., et al. (2017). Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography. Nature, 541(7636), 242–246. https://doi.org/10.1038/nature20599.
Stellato, F., Oberthur, D., Liang, M., Bean, R., Gati, C., Yefanov, O., et al. (2014). Room-temperature macromolecular serial crystallography using synchrotron radiation. IUCrJ, 1(Pt 4), 204–212. https://doi.org/10.1107/S2052252514010070.
Suga, M., Akita, F., Hirata, K., Ueno, G., Murakami, H., Nakajima, Y., et al. (2015). Native structure of photosystem II at 1.95 Å resolution viewed by femtosecond X-ray pulses. Nature, 517(7532), 99–103. https://doi.org/10.1038/nature13991.
Suga, M., Akita, F., Sugahara, M., Kubo, M., Nakajima, Y., Nakane, T., et al. (2017). Light-induced structural changes and the site of O=O bond formation in PSII caught by XFEL. Nature, 543(7643), 131–135. https://doi.org/10.1038/nature21400.
Sugahara, M., Mizohata, E., Nango, E., Suzuki, M., Tanaka, T., Masuda, T., et al. (2015). Grease matrix as a versatile carrier of proteins for serial crystallography. Nature Methods, 12(1), 61–63. https://doi.org/10.1038/nmeth.3172.
Sugahara, M., Nakane, T., Masuda, T., Suzuki, M., Inoue, S., Song, C., et al. (2017). Hydroxyethyl cellulose matrix applied to serial crystallography. Scientific Reports, 7(1), 703. https://doi.org/10.1038/s41598-017-00761-0.
Sugahara, M., Song, C., Suzuki, M., Masuda, T., Inoue, S., Nakane, T., et al. (2016). Oil-free hyaluronic acid matrix for serial femtosecond crystallography. Scientific Reports, 6, 24484. https://doi.org/10.1038/srep24484.
Tenboer, J., Basu, S., Zatsepin, N., Pande, K., Milathianaki, D., Frank, M., et al. (2014). Time-resolved serial crystallography captures high-resolution intermediates of photoactive yellow protein. Science, 346(6214), 1242–1246. https://doi.org/10.1126/science.1259357.
Terwilliger, T. C., & Berendzen, J. (1995). Difference refinement: Obtaining differences between two related structures. Acta Crystallographica. Section D, Biological Crystallography, 51(Pt 5), 609–618.
Tosha, T., Nomura, T., Nishida, T., Saeki, N., Okubayashi, K., Yamagiwa, R., et al. (2017). Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate. Nature Communications, 8(1), 1585. https://doi.org/10.1038/s41467-017-01702-1.
Umena, Y., Kawakami, K., Shen, J.-R., & Kamiya, N. (2011). Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature, 473, 55. https://doi.org/10.1038/nature09913.
Ursby, T., & Bourgeois, D. (1997). Improved estimation of structure-factor difference amplitudes from poorly accurate data. Acta Crystallogr. Sect. A, 53(5), 564–575. https://doi.org/10.1107/S0108767397004522.
Weierstall, U., James, D., Wang, C., White, T. A., Wang, D., Liu, W., et al. (2014). Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nature Communications, 5, 3309. https://doi.org/10.1038/ncomms4309.
Weierstall, U., Spence, J. C., & Doak, R. B. (2012). Injector for scattering measurements on fully solvated biospecies. The Review of Scientific Instruments, 83(3), 035108. https://doi.org/10.1063/1.3693040.
Weinert, T., Olieric, N., Cheng, R., Brunle, S., James, D., Ozerov, D., et al. (2017). Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons. Nature Communications, 8(1), 542. https://doi.org/10.1038/s41467-017-00630-4.
Wickstrand, C., Dods, R., Royant, A., & Neutze, R. (2015). Bacteriorhodopsin: Would the real structural intermediates please stand up? Biochimica et Biophysica Acta, 1850(3), 536–553. https://doi.org/10.1016/j.bbagen.2014.05.021.
Wohri, A. B., Katona, G., Johansson, L. C., Fritz, E., Malmerberg, E., Andersson, M., et al. (2010). Light-induced structural changes in a photosynthetic reaction center caught by Laue diffraction. Science, 328(5978), 630–633.
Yano, J., Kern, J., Irrgang, K. D., Latimer, M. J., Bergmann, U., Glatzel, P., et al. (2005). X-ray damage to the Mn4Ca complex in single crystals of photosystem II: A case study for metalloprotein crystallography. Proceedings of the National Academy of Sciences of the United States of America, 102(34), 12047–12052.
Yeremenko, S., van Stokkum, I. H. M., Moffat, K., & Hellingwerf, K. J. (2006). Influence of the crystalline state on photoinduced dynamics of photoactive yellow protein studied by ultraviolet-visible transient absorption spectroscopy. Biophysical Journal, 90(11), 4224–4235. https://doi.org/10.1529/biophysj.105.074765.
Young, I. D., Ibrahim, M., Chatterjee, R., Gul, S., Fuller, F. D., Koroidov, S., et al. (2016). Structure of photosystem II and substrate binding at room temperature. Nature, 540(7633), 453–457.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Colletier, JP., Schirò, G., Weik, M. (2018). Time-Resolved Serial Femtosecond Crystallography, Towards Molecular Movies of Biomolecules in Action. In: Boutet, S., Fromme, P., Hunter, M. (eds) X-ray Free Electron Lasers. Springer, Cham. https://doi.org/10.1007/978-3-030-00551-1_11
Download citation
DOI: https://doi.org/10.1007/978-3-030-00551-1_11
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00550-4
Online ISBN: 978-3-030-00551-1
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)