Abstract
Plastids evolved from free-living cyanobacteria that were engulfed by a host cell. One critical element for the stable establishment of endosymbiosis was the acquisition of the ability of the endosymbiont to multiply inside the host cell. In all plants, plastids divide by binary fission via the assembly and subsequent constriction of a multiproteic scaffold called the plastid division ring. This highly complex structure associates proteins that have been retained from the bacterial ancestor, such as FtsZ that forms a ring inside the plastid, with proteins brought by the host cell, that form a ring outside the plastid. Over the past 20 years, dramatic progress has been made in our understanding of the mechanisms underlying plastid division, thanks to the combination of various approaches including direct and reverse genetics, electron microscopy and biochemistry. In this review we describe in detail the different steps of plastid division, from the choice of the division site to the sequential assembly of all constituents of the plastid division ring. We also discuss the current knowledge of the regulation of this process, which is still in its infancy but raises fascinating questions for future research.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
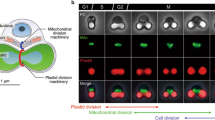
Although the hypothesis that plastid and mitochondria derived from a free living prokaryote was first formulated at the end of the nineteenth century, the question of the biogenesis of plastids in plant cells was still debated in the 1960s: some authors postulated that plastids could arise from small submicroscopic vesicles called proplastid precursors, even though a wealth of microscopic observations provided evidence for plastid division by binary fission [118] . It is now clear that plant cells cannot generate plastids de novo, and that binary fission is the main process allowing the multiplication of these organelles, although budding has been reported in ripening tomato fruits [30]. When observed under the microscope, the chloroplast division process can be described as follows: initially spherical chloroplasts become more ovoid, and constriction becomes visible at chloroplast midpoint. As constriction proceeds chloroplasts become dumbbell shaped and finally deeply constricted with a narrow neck between the two future organelles. At the final stage of division the two chloroplasts can twist around this neck and finally become completely separated. Observation of dividing chloroplasts was reported in the nineteenth century, but only in the last 20 years have we come to understand how plastid division actually takes place.
The study of plastid division has highly benefited from three types of approach: transmission electron microscopy (TEM), forward genetics and reverse genetics. TEM images of dividing Cyanidium caldarium cells revealed the existence of an electron dense structure at the division site of chloroplasts [90]. This observation was subsequently generalized to all plants, demonstrating that a complex structure is responsible for plastid division. Genetic studies were then required to identify the constituents of this structure. In the early 1990s, K. Pyke and R. Leech isolated several mutants specifically affected for plastid division which were called arc for Accumulation and Replication of Chloroplasts [122, 123]. A total of 12 arc mutants have been described, most of which show a reduction of chloroplast number compensated by an increase in chloroplast size (Fig. 6.1). Because positional cloning is a long-term approach, especially when dealing with mutants whose phenotype must be studied under the microscope, more than 10 years passed before the first ARC gene (ARC5) was identified [40]. In the meantime, complete sequencing of the Arabidopsis genome allowed K. Osteryoung and E. Vierling to discover homologues of the FtsZ gene [113]; the bacterial FtsZ protein is distantly related to tubulin and can polymerize in a GTP-dependent manner. Its ability to form contractile rings allows cell division by binary fission. Since then, several studies have demonstrated that plastid division does require homologues of FtsZ and involves several other proteins of prokaryotic origin, but also proteins that were encoded by the genome of the host cell. The resulting machinery is extremely complex, and how this multi-proteic scaffold functions at the molecular level is only beginning to be elucidated.
In this chapter, we will focus mainly on chloroplast division in higher plants, because it is the best described model in many respects, but it is generally assumed that the division mechanisms are common to all plastid types. Over the past 15 years, our knowledge of the mechanisms allowing plastid division has made considerable progress. By contrast, very little is known at present regarding the regulation of plastid division. However, a few recent studies provide insights into this regulation and can now allow new interpretation of older reports regarding the factors that influence plastid division. In the last paragraph of this chapter we will therefore try to summarize these reports, illustrating how the recent discoveries about plastid division can allow a new understanding of early microscopic analyses.
2 Placement of the Plastid Division Site
Our knowledge of bacterial cell division has greatly contributed to the understanding of all aspects of chloroplast division, including division ring placement. To date, the best characterized model is Escherichia coli in which two distinct mechanisms allow the division machinery to assemble at midcell: the nucleoid occlusion system prevents FtsZ ring formation in the close vicinity of chromosomes (i.e. all sites except midcell and cell poles) and the Min system avoids FtsZ polymerization at cell poles [24]. In the absence of Min proteins, division frequently takes place at cell poles, giving rise to minicells. MinC is the inhibitor of FtsZ polymerization, while MinD and MinE regulate its location in the cell and thereby its activity. To affect FtsZ polymerization, MinC needs to be attached to the membrane, and this association requires MinD, an ATPase which forms dimers and engages its C-terminus into the membrane upon ATP binding. After ATP hydrolysis and release of ADP, MinD dissociates from the membrane. MinCD complexes oscillate from pole to pole, owing to the topological factor MinE. MinE forms a ring that appears to cap the Min polar zone and prevent it from extending past midcell: MinE stimulates the ATPase activity of MinD, thus promoting its dissociation from the membrane. The MinE ring then moves towards the pole, until most proteins are released from the membrane and assemble again at the opposite pole [11]. In other rod-shaped bacteria such as Bacillus substilis, the Min system is conserved, but MinCD proteins and their topology factors DivIVA and MinJ do not appear to oscillate from pole to pole. They rather seem to accumulate at the poles at early steps of division and to be dynamically localized to the mature division apparatus, thereby labeling the new cell poles after division is complete [11]. The Min system is also conserved in cyanobacteria, which are the closest relatives of chloroplasts, but their dynamic behavior during the division process has not been elucidated [85].
2.1 Many Proteins are Involved in the Choice of the Division Site
Homologues of MinD and MinE have been identified in Arabidopsis and appear to have antagonistic functions towards FtsZ polymerization . Mesophyll cells of AtMinE over-expressers and atminD/arc11 mutants contain chloroplasts of heterogeneous size, showing aberrant Z-ring positioning and multiple Z-rings in one chloroplast [20, 34, 55, 77]. By contrast, AtMinD overexpressers and atmine/arc12 show inhibition of plastid division [45]. These opposite effects of AtMinD and AtMinE on chloroplast division are due to their impact on FtsZ polymerization: FtsZ proteins form multiple rings in atminD mutants and AtMinE overexpressers whereas they form short filaments in atminE mutants and AtMinD overexpressers, indicating that AtMinD inhibits FtsZ polymerization whereas AtMinE promotes it [37, 45, 147]. Proper balance between AtMinD and AtMinE activities is therefore required for Z-ring formation at midplastid [35]. Molecular data further supports the notion that the Min system operates in chloroplasts in a similar way as in bacterial cells. As observed for their bacterial homologues, AtMinD and AtMinE can form homo or heterodimers [79]. Furthermore, the ATPase activity of AtMinD seems to be important for its proper localization in chloroplasts, and AtMinE, like its bacterial counterpart, stimulates this activity [4]. Finally, neither AtMinD nor AtMinE seem to interact with FtsZ proteins, suggesting that as observed in bacteria, a third component is required to regulate Z-ring positioning.
Although MinC is conserved in cyanobacteria [85] and the chloroplast division machinery is sensitive to overexpression of the E. coli MinC protein [143], plant genomes lack MinC homologues. ARC3 has been suggested to replace MinC functionally. The phenotype of arc3 mutants and over-expressers is similar to arc11 and AtMinD over-expressers respectively [80, 134], providing evidence for an inhibitory function of ARC3 on Z-ring assembly away from the mid-plastid point. ARC3 interacts with AtMinE and AtMinD via its central domain and with AtFtsZ1 via its N-terminal FtsZ-like domain, although this domain lacks the tubulin signature and amino acids crucial for GTP hydrolysis [80]. Taken together with the division site misplacement in the arc3 mutant, the presence of ARC3 in an FtsZ/Min complex suggests that ARC3 plays a central role in division site placement.
Two more Arabidopsis proteins participate in division site placement, bringing further complexity to the system compared with bacterial cell division: MCD1 [100] and PARC6/CDP1 [46, 155]. PARC6 is a paralogue of the ARC6 protein which destabilizes FtsZ ring (see below). PARC6 is conserved from algae to land plants whereas MCD1 is specific to land plants. Both proteins appear to be negative regulators of Z-ring formation, but how they achieve this function is not completely clear. In mcd1 mutants, FtsZ proteins form long filaments, and overexpression of AtMinD has little effect on FtsZ polymerization. In this background the AtMinD protein accumulates at wild-type level, but shows diffuse localization in chloroplasts; taken together, these results suggest that MCD1 may enhance the inhibitory effect of AtMinD on FtsZ polymerization by modulating its intra-plastidial localization [100]. PARC6/CDP1 negatively regulates FtsZ polymerization, possibly by interacting with ARC3 [46, 155]. Interestingly, this interaction appears to be mediated by the C-terminal MORN domain of ARC3 [46]. Mesophyll cells of parc6 mutants contain chloroplasts of heterogeneous sizes suggesting that PARC6 participates in the choice of the division site, but also constricted chloroplasts blocked at a later stage of division, pointing at additional roles of PARC6 ([46], see below).
Finally, mechano-sensitive channel (MSC) proteins appear to be involved in division site positioning [49]. A systematic functional analysis of MSC of the S family (Msc-S Like) led to the observation that MSL2 and MSL3 play redundant roles in chloroplast division: msl2 and msl3 mutants are identical to the wild-type whereas msl2msl3 double mutants are variegated and show abnormal chloroplast size and shape [49]. Further genetic analyses revealed that MSL2 and 3 proteins function as inhibitors of Z-ring formation in a Min/ARC3 dependent manner , although their molecular mechanism of action is not clear [149]. Strikingly, in B. subtilis, the MinE functional equivalent DivIVA can recognize curved membrane regions and has therefore been suggested to have the intrinsic capacity to bind and accumulate at negatively curved membranes [11]. MSL proteins, by regulating ion fluxes to or from the chloroplast may affect membrane curvature and in this way influence the polarity of the organelle.
2.2 How Do Proteins Involved in the Choice of the Division Site Work Together?
Although many players of the Z-ring placement have been identified in the past 10 years, our understanding of the underlying mechanism remains very incomplete and the molecular functions of all the identified players are not fully elucidated yet. Based on interaction data (summarized in Fig. 6.2), and on the effect of these proteins on FtsZ polymerization, AtMinD, AtMinE and ARC3 may be the functional equivalent of the bacterial MinCDE complex. However, it is not clear whether AtMinD engages the chloroplast envelope upon ATP binding and dimerization, like its bacterial counterpart. By contrast, MCD1 is a membrane spanning protein which seems to be required for proper AtMinD localization and function [100]. It is therefore tempting to speculate that MCD1 may be required to attach the MinDE/ARC3 complex to the chloroplast inner envelope. PARC6 may also play a similar role since it is a transmembrane protein and can interact with ARC3 [46]. All these proteins may hence form a complex attached to the chloroplast inner membrane as pictured on Fig. 6.2a, but it is not clear whether a single complex harboring all proteins or several sub-complexes differing by their composition exist in chloroplasts.
Regulation of Z-ring positioning during chloroplast division. a The Z-ring regulatory complex. This picture summarizes the reported interactions between proteins and their membrane association. It is not clear however whether such a complex exists in chloroplasts, or several sub-complexes differing by their composition co-exist. b–d Three alternative models for Z-ring positioning based on localization data. Dark green and light green boxes represent complexes of FtsZ regulatory proteins. Dark green boxes are for a combination of proteins that completely inhibit FtsZ polymerization, whereas light green boxes represent a complex capable to modulate FtsZ dynamics at the division site. b FtsZ regulatory proteins (FRP) operate predominantly at chloroplast poles to inhibit ectopic Z-ring formation. After Z-ring assembly (red ring), a different set of FRP may be recruited at the division site and regulate FtsZ dynamics at the division site. FRP could also be required to label the newly formed poles, thereby maintaining chloroplast polarity. c Alternatively, the polar localization of FRP may mark the site of the future division site. d FRP inhibit Z-ring formation at all sites except midpoint. At the division site, another regulatory complex differing from the previous one by its composition regulates the dynamics of the Z-ring
Even more elusive is how and where in the chloroplast all these proteins function. This gap in our knowledge can be attributed to the lack of live imaging of proteins regulating Z-ring assembly, and to conflicting results regarding their localization in dividing chloroplasts. Depending on the technique used for localization experiments, two types of distributions have been reported for division site placement machinery. According to fluorescent protein (FP) fusions and BiFC assays, AtMinD and AtMinE co-localize into large dots at chloroplast poles [77, 79]. The same localization was reported for MSL2 and MSL3 [49]. PARC6 [46] was also found in pole localized dots, but also at the division site, where it seems to function at later steps of the division process. The distribution of ARC3 seems more complex since it was found to co-localize with FtsZ1 in ring-like structures and to form discrete dots containing MinD [80], or multiple dots in which it interacts with PARC6/CDP1 [155]. By contrast, immuno-fluorescence experiments suggest that MCD1 [100] and AtMinD [37] are localized in ring-like structures at the chloroplast midpoint as well as punctuate structures dispersed throughout the envelope, whereas ARC3 is localized at the division ring during early and middle steps of chloroplast division [134]. Several hypotheses can explain these discrepancies. In some of these studies, localization of FP-fusion proteins was performed in transient expression assays using tobacco leaves in which chloroplast division may have stopped, and the localization of these proteins might therefore be altered. Furthermore, Fujiwara et al reported that the AtMinD-YFP fusion cannot complement the arc11/minD mutant [36], suggesting that the reported localization of the fusion protein may not reflect the proper distribution of AtMinD. Reciprocally, some of the observed immuno-fluorescence signals are very faint, and the inferred distribution of the target protein may not be accurate.
Based on these localization experiments, several models can be proposed for the choice of chloroplast division site (Fig. 6.2b–d). All these models assume that the Z-ring regulatory complex is present at least transiently at the division site, implying that a minimum of two complexes differing by their proteic composition must exist in chloroplasts, one of them being an inhibitor of Z-ring formation (Fig. 6.2b–d; dark green boxes), and the other a regulator of FtsZ dynamics (Fig. 6.2b–d; light green boxes). One possibility would be that inhibitors of FtsZ polymerization act predominantly at the poles, as suggested by FP-fusion experiments (Fig. 6.2b, c). Recognition of the poles by this inhibitory complex may involve membrane curvature and require MSL proteins. In addition, accumulation of proteins involved in the choice of the division site at the poles may reflect their role in the regulation of chloroplast polarity [44]. Indeed, in B. subtilis, Min proteins are associated to the cell poles, and are recruited at the division site to mark the newly formed poles after division (Fig. 6.2b). Another possibility would be that Min-containing dots mark the site of the future division (Fig. 6.2c). Indeed, the cyanobacteria Synechocystis sp divides in alternating perpendicular planes [85]. Finally, as suggested by immuno-fluorescence analysis, inhibitors of FtsZ polymerisation may be distributed both in discrete dots dispersed throughout the inner membrane and at the division site (Fig. 6.2d). If this were the case, the discrete dots would represent inhibitory complexes preventing ectopic Z-ring formation, whereas the inhibitory action of AtMinD, ARC3, MCD1 and PARC6 would somehow be alleviated by AtMinE or another yet unidentified factor at the division site (Fig. 6.2d). Furthermore, the presence of ARC3 and PARC6 at the division site may be required to modulate the dynamics of AtFtsZ polymerization.
Models illustrated by Fig. 6.2c, d have the advantage that they may better account for the choice of the division site. Indeed, unlike bacteria in which the nucleoid occlusion system prevents the formation of the division apparatus everywhere except at midcell and at the poles, chloroplasts contain numerous small nucleoids that are distributed throughout the stroma [67]. Therefore, preventing Z-ring assembly at the poles may not be sufficient to ensure symmetric division of plastids. To clarify the model for Z-ring positioning, time-course analysis of the localization of all the above mentioned proteins during chloroplast division is absolutely indispensable.
3 Assembly and Dynamics of the FtsZ Ring
FtsZ is a stable protein and in bacteria the intracellular concentration changes little during cell division, therefore, rapid responses required for regulation of the Z-ring occur through control over FtsZ assembly. Recent evidence indicates that regulation of Z-ring assembly/maintenance in plastids may be achieved through the opposing effects of ARC6 and PARC6 and also through the action of the plastid chaperone system. Further complexity is added to the potential regulatory mechanisms in plants because at variance with prokaryotes, plant genomes encode several FtsZ isoforms.
3.1 Z-ring Assembly Requires Two Isoforms of FtsZ
In prokaryotes, FtsZ is usually encoded by a single gene even though exceptions have been reported [82]. By contrast, three homologues of FtsZ distributed in two families are present in Arabidopsis (AtFtsZ1, AtFtsZ2-1 and AtFtsZ2-2). Divergence between the two FtsZ families occurred early during evolution, probably before the divergence between Chlorophycean and Charophycean [138], and the number of isoforms in each family seems to vary from one plant to another. For example, only one FtsZ1 isoform can be found in Arabidopsis thaliana versus four in Nicotiana tabacum. AtFtsZ1 and AtFtsZ2 are assumed to play distinct role in chloroplast division because mutants cannot be complemented by overexpression of a member of the other family [132] and AtFtsZ1 and AtFtsZ2 differ by their ability to interact with other plastid division proteins: ARC3 interacts specifically with AtFtsZ1 [80] whereas ARC6 binds only to AtFtsZ2 ([147] see below). In Arabidopsis, ftsz2-2 null mutants show only mild plastid division defects, and this phenotype can be complemented by expression of AtFtsZ2-1. Reciprocally, the more severe phenotype of ftsz2-1 mutants can be rescued by AtFtsZ2-2 expression, suggesting that the two proteins are functionally redundant but differ by their expression level [132]. However, the two FtsZ2 isoforms may differ in their role in thylakoid development [59], see below).
AtFtsZ1 and AtFtsZ2-1 proteins co-localize and form rings at chloroplast midpoint (Fig. 6.3) [79, 86]. FtsZ proteins are structural homologues of tubulins, and can form both longitudinal and lateral interactions. Longitudinal interactions are stimulated by GTP binding and allow the formation of proto-filaments that consist of a head-to-tail linear polymer of FtsZ. However, unlike tubulin, FtsZ proto-filaments do not associate into microtubules but instead form bundles or sheets. Whether FtsZ1 and FtsZ2 proteins form distinct proto-filaments or hetero-multimeric ones is not clear. In chloroplasts, AtFtsZ1 and AtFtsZ2-1 can form curved filaments in the absence of the other protein, even if these structures are often disordered and more numerous than in the wild-type [146]. Purification of FtsZ complexes from Arabidopsis suggests that Z-rings contain both AtFtsZ1 and AtFtsZ2 at a 1:2 ratio [87], and the stoichiometry between the two proteins is probably important to their function because overexpression of either isoform severely inhibits chloroplast division [114]. Interestingly, AtFtsZ1 and AtFtsZ2-1 can form hetero-polymers in vitro and FtsZ1 promotes co-assembly, suggesting it can favor lateral interactions between proto-filaments [109]. Finally, phosphorylation of AtFtsZ2 proteins has recently been reported to somewhat affect their ability to interact with AtFtsZ1 and ARC6 and to form rings in vivo, providing evidence for an additional layer of complexity in the mechanisms governing Z-ring assembly [41].
The stromal and cytoplasmic division machineries of chloroplasts are assembled in a step-wise manner. a In Arabidopsis plastid division is initiated by the assembly of AtFtsZ1-1 (F1), AtFtsZ2-1 and AtFtsZ2-2 (F2) at the centre of chloroplasts to form the Z-ring. Z-ring assembly/maintenance may be achieved through the opposing effects of ARC6 (6) and PARC6 (P6). b After completion of the Z-ring the inner PD ring (iPD) assembles on the inside of the Z-ring. This is followed by the middle PD ring in the inter membrane space, if present (not shown). c Subsequently the outer PD ring (oPD) forms on the cytosolic face of the chloroplast. In C. merolae the oPD is composed of PRD1 and polyglucan filaments but the composition in higher plants is unknown. d Finally dynamin is recruited to the division site. In Arabidopsis this is achieved by the combined action of PDV1 and PDV2. DRP5B is faintly detected on un-constricted but only forms ring-like structures after constriction has begun. The timings of the assembly of the rings are inferred from microscopy studies in both Arabidopsis and C. merolae
3.2 Opposing Roles of ARC6 and PARC6 on Z-Ring Formation
One major difference between AtFtsZ1 and AtFtsZ2 is that only AtFtsZ2 possesses a C-terminal extension harboring a CORE domain allowing its interaction with ARC6 (Fig. 6.3) [79]. arc6 mutants are the most severely affected of all arc mutants, containing only 2 chloroplasts per mesophyll cell on average [129]. ARC6 is an integral membrane protein homologous to the Cyanobacterial protein Ftn2 [64, 147], which appears to affect FtsZ polymerization positively: FtsZ2 forms short filaments in arc6 mutants whereas it forms long polymers in ARC6 over-expressing lines [147]. In this respect, ARC6 seems to function similarly to AtMinE. Nevertheless, chloroplast division is almost completely abolished by ARC6 overexpression or loss of function, and asymmetric division was never observed: ARC6 is therefore a component of the chloroplast division machinery and not involved in the choice of the division site. ARC6 has been reported to form a discontinuous ring at the chloroplast division site [79], and is likely to function as a Z-ring anchor, stabilizing FtsZ polymers and associating them with the inner envelope of the chloroplast, This is similar to the role played by Zip and FtsA in E. coli which interact with FtsZ through the CORE domain and play a role in maintaining/stabilizing the Z-ring [117].
Unlike ARC6, PARC6 lacks a functional J-domain and subsequent associated co-chaperone activity [46]. This observation has led to the suggestion that PARC6 arose in vascular plants as a new functional class of plastid division proteins and evidence suggests that PARC6 carries out at least two, potentially overlapping functions, both playing a role in Z-ring placement (see above) and influencing Z-ring assembly and/or maintenance. PARC6 appears to act antagonistically to ARC6 in its role in Z-ring stability; in arc6 chloroplasts FtsZ filaments are short, indicating that ARC6 acts to stabilize the Z-ring, but in contrast to this in parc6 chloroplasts FtsZ filaments are relatively long and appear as multiple rings or spirals, indicating that PARC6 inhibits Z-ring assembly [46, 147]. These data point to ARC6 and PARC6 having opposing effects on the formation/stability of the Z-ring, despite the high levels of homology between them.
ARC6 is proposed to mediate its effects on the Z-ring by anchoring the ring to the membrane through a direct interaction of the N-terminal conserved domain of ARC6 with AtFtsZ2-1 and AtFtsZ2-2 [79, 132]. Given the high levels of similarity between the N-terminal conserved domains of ARC6 and PARC6, the possibility that PARC6 directly interacted with AtFtsZ1 or AtFtsZ2-1 was investigated by yeast two-hybrid assays, but no interactions were detected [46]. Unlike ARC6, PARC6 is predicted to contain two trans-membrane domains, which would also orientate the C-terminal domain of PARC6 in the stroma and it will be important to test if the C-terminal domain can interact with any of the plastid FtsZ proteins to determine if PARC6 can directly influence the Z-ring.
The N-terminal stromal domain of PARC6 has been found to interact with ARC3. This interaction is dependent on the C-terminal MORN domain of ARC3, which is the same domain of ARC3 which inhibits the interaction of ARC3 with AtFtsZ1 in yeast two-hybrid studies [46, 80]. If PARC6 exerts the destabilizing effects on the Z-ring through ARC3, the availability of the MORN domains could play a vital regulatory role. The interaction of PARC6 with ARC3 could sequester the MORN domain, thus enabling ARC3 to interact with the Z-ring and render the Z-ring unstable. In the absence of the PARC6-ARC3 interaction, the MORN domain could inhibit the interaction of ARC3 with AtFtsZ1, thus rendering the Z-ring more stable, as seen in the parc6 mutant [46]. In an additional level of complexity the availability of ARC3 to interact with PARC6 could also be influenced by the other interacting partners of ARC3, including MinD, MinE, and MCD1 [80, 100], and much work will be required to unravel the role of PARC6 on the placement and assembly of the Z-ring.
3.3 Chaperone-Regulation of Z-Ring Assembly/Maintenance
Recent evidence has shed light on the possible role of molecular chaperones in the assembly and/or maintenance of the Z-ring. Chaperones assist in the folding or unfolding of proteins and the assembly or disassembly of protein complexes. The first chaperones shown to play a direct role in the process of plastid division were members of the chaperonin 60 (cpn60, hsp60) family in Arabidopsis [142]. The Cpn60 family of chaperonins is highly conserved and found in bacteria, mitochondria and plastids [51]. The best characterized members include the GroEL complex in E. coli, which together with the cochaperonin GroES forms a large (~ 1 MDa) chaperone complex termed GroE. In E. coli GroE function is required for the formation of the cell division machinery and cells with impaired GroE activity exhibit filamentous cell morphology, characteristic of cell division inhibition [33, 104]. The filamentous phenotypes are also observed in GroE-depleted Streptococcus mutans and Caulobacter crescentus, suggesting that GroE plays a universal role in cell division in bacteria [72, 141].
In Arabidopsis at least two members of the Cpn60 family, ptCpn60α1 and ptCpn60β1, are required for chloroplast division [142]. A null mutation in ptCpn60α1 or the double ptcpn60β1-1 ptcpn60β2 mutant results in albino and dwarf seedlings harboring small, colorless plastids, indicating Cpn60 function is vital for plant development [5, 142]. However a moderate reduction in the levels of activity of ptCpn60βB (caused by a premature stop codon in the ptCpn60β1) or of ptCpn60α1 (caused by a missense mutation, Ala-342-Val), results in the modest impairment of plastid division and mesophyll cells that harbor fewer but larger chloroplasts than those in wild type cells [142]. In E. coli, treatment of cells with penicillin has been shown to trigger the localization of GroE to possible division sites in an FtsZ dependent manner [104], although insults with other stresses did not replicate these results [16]. ptCpn60β is localized as speckles throughout the chloroplasts in Arabidopsis [142] and there is no evidence to date of localization to the division machinery. However In both the ptCpn60α1 and ptCpn60β1 mutants FtsZ forms long, disorganized filaments, similar to those observed in Arabidopsis plants in which ARC6 is over-expressed, suggesting that reduced ptCpn60 levels result in excessively stable FtsZ filaments.
How changes in the levels of Cpn60 function might alter FtsZ filament stability in plastids is not yet clear. In bacteria proteome-wide analysis of GroE substrates have revealed several cell division proteins as putative targets of GroE [15, 52, 62]. To date GroE has been shown to assist in the folding of the FtsE [33]. FtsE interacts with FtsZ and works together with FtsX to support septal ring assembly and most likely ring constriction [6, 21]. FtsE is absent in plant and algal genomes and in Arabidopsis FtsZ is properly imported and processed in the ptCpn60β1-1 mutant, suggesting that the plastid Cpn60 system targets a different plastid division substrate(s) [142]. However, it is attractive to speculate that akin to bacterial system, the impaired chloroplast division phenotype and excessively stable FtsZ filament phenotype observed in the cpn60 mutants are as a result of changes in the stability of a regulator of Z-ring formation and identification of possible targets of Cpn60 chaperonin activity will be important.
Given the striking similarity in the structure of the long, disorganized FtsZ filaments observed the cpn60 mutants and in Arabidopsis plants overexpressing ARC6, it is interesting to observe that ARC6 might also play a role in the chloroplast chaperone systems. ARC6 harbors a J domain characteristic of co-chaperones that interact with the Hsp70 family of chaperone proteins [17, 147, 148]. J domains can associate with unfolded polypeptide chains and deliver them to Hsp70 chaperones for processing and also regulate the activity of the Hsp70 chaperones [148]. Most J domains harbor a central His-Pro-Asp motif, but in ARC6 and its orthologues only the central Pro is conserved uniformly. The His-Pro-Asp motif is commonly crucial for interaction with the Hsp70 chaperone partners, although exceptions exist [50]. Z-ring formation in E. coli is known to be influenced by Hsp70 [145] and it is possible that in addition to the proposed role in anchoring the Z-ring, ARC6 may have additional functions.
Whilst it remains to be shown whether ARC6 can interact with a chaperone through its atypical J-domain, recently it was shown that ARC6 can interact with a second chloroplast protein harboring a J domain, Chloroplast J-like Domain 1 (CJD1) [3]. Like ARC6, CJD1 is an inner envelope protein and harbors an N-terminal J-like domain that resides in the stroma. The J-domain of CJD1 interacts with the ARC6 J-like domain and the adjacent conserved region. Interestingly, during mitochondrial protein import in yeast the dynamic interaction of two J-domain containing proteins (the J co-chaperone Tim14 and the J-like protein Tim16) has been shown to regulate the co-chaperone activity of Tim14 on mtHsp70, which plays an important regulatory role [98] and it is possible that the ARC6-CJD1 interaction has a similar regulatory role.
CJD1 plants were identified as containing altered levels of the leaf galactolipids monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG) [3]. These two galactolipids commonly constitute up to 80 % of the total chloroplast lipids and play an essential role in chloroplast membrane biogenesis and are indispensable for correct protein import into chloroplasts [31]. Mutant analysis failed to reveal evidence for a direct link between the function of CJD1 and ARC6, however in the tgd-1-1, fad6-1 and tgd-4-3, fad6-1 double mutants, a strong reduction in the levels of MGDG and DGDG is coupled with a reduction in the number of chloroplasts per cell [29]. The authors suggest that an inadequate lipid supply for membrane proliferation during chloroplast division is responsible. This hypothesis is supported by the recent finding that impairment of very-long-chain fatty acids biosynthesis via genetic of pharmacologic approaches affects plastid division [103]. Whilst the picture is complicated by the lack of strong linkage between lipid mutants and chloroplast division mutants phenotypes, it will be interesting to determine the functional significance of interaction between CJD1 and ARC6.
4 Assembly of the Plastid Division Machinery
In bacteria the Z-ring is known to act as a scaffold protein, playing an essential role in the sequential and coordinated recruitment of up to nine additional cell division proteins to the division site (FtsX, FtsE, FtsK, FtsQ, FtsL, FtsB, FtsI, FtsW and FtsN) to form the divisome or septal ring (reviewed in [24]). Studies in Arabidopsis and algae have also shown that assembly of the chloroplast division machinery appears to occur in a linear order. After the formation of the Z-ring the timed addition of division machinery components results in assembly of up to four additional ring-like structures at the division site (Fig. 6.3).
4.1 The Plastid Dividing Rings
Many algae have just one chloroplast per cell and have been used extensively for the study of the coordinated assembly of the chloroplast division machinery because, unlike in Arabidopsis in which each cell can contain as many as 100 chloroplasts that divide asynchronously, algal cell and organelle division can be highly synchronized by light/dark cycles . The completion of Z-ring formation is followed by the appearance of ring like structures known as plastid dividing (PD) rings. PD rings were first identified as fuzzy plaques in electron microscopy images in young wheat leaves [71]. This fuzzy plaque was subsequently shown to be an electron-dense ring-like structure in the alga C. caldarium [90]. Studies in both Avena sativa and Cyanidioschyzon merolae further resolved this ring to be composed of two or three electron-dense rings: an inner ring on the stromal face of the inner envelope, an outer ring on the cytosolic face of the outer envelope, and occasionally a middle ring in the intermembrane space. PD rings have subsequently been detected in many plant and algae species and are thought to be a universal feature of plastid division machineries [47, 68, 90–92, 94].
The first PD ring to assemble after the completion of the Z-ring is the inner PD ring, which forms between the inner envelope membrane and the Z-ring [93, 94]. This is followed by the middle PD ring in the inter membrane space (IMS), if present [91]. The components of the inner and middle PD ring are as yet unidentified. Finally the outer PD ring, which is composed of a bundle of 5–7 nm filaments, forms on the outer chloroplast envelope (Fig. 6.3) [95] .
The identity of the components of the PD rings remained elusive for many years. It was found that isolated PD machineries from C. merolae could not be fully decomposed with proteases , indicating a non-proteic component, and electron dense deposits appeared on the PD rings after staining with periodic acid–horseradish peroxidase, indicating that it is saccharic in nature [153]. Proteomic analysis of the isolated PD machinery identified more than 20 putative components, one of which was a glycogenin-like protein and was designated Plastid Dividing Ring 1 (PDR1) [153]. PDR1 forms a ring that is recruited to the division site after the formation of the Z-ring and down-regulation of C. merolae PRD1 by antisense suppression prevents the formation of the PDR1 ring, but does not affect the assembly of the Z-ring. PDR1 harbors a glycosyltransferase domain in the C-terminal region, putatively associated with the transfer of a sugar moiety from an activated donor sugar onto an acceptor molecule. Immunoblot analysis of synchronized cell cultures revealed that during chloroplast division the band corresponding to PDR1 shifted downwards before decreasing in intensity. The authors propose that PDR1 synthesizes the sugars that constitute the outer PD ring and is glycosylated during chloroplast division before decomposing [153] .
The precise role of PRD1 and the polyglucan filaments is unclear, but down-regulation of PRD1 leads to a decrease in the frequency of chloroplast division in C. merolae compared with control cells [153], indicating that the correct formation of the PD ring is essential for chloroplast division. PDR1 is widely conserved from red alga to land plants, and in Arabidopsis there is a family of 6 PDR1 proteins (PGSIP1-6). PGSIP1 and PGSIP3 are located in the Golgi [105], but the remaining proteins are yet to be investigated. It will be interesting to determine if any members of the family are involved in chloroplast division and if the outer PD ring is also a bundle of PDR1-mediated polyglucan filaments in higher plants .
4.2 PDV1, PDV2 and Dynamin Recruitment to the Division Site
Although the outer PD ring is not well characterized in higher plants, microscopy studies have revealed that the formation of the Z-ring is followed by the recruitment of two novel proteins, Plastid Division 1 and Plastid Division 2 (PDV1 and PDV2) to the division site on the outer envelope [97]. PDV1 and PDV2 are integral outer envelope proteins that show no significant sequence similarity to known proteins. PDV1 and PDV2 have similar domain structure and are composed of a large N-terminal domain residing in the cytosol, a single transmembrane domain and a small C-terminal domain residing in the in the IMS [44, 97]. The cytosolic domains harbor two short conserved regions and a coiled-coil domain, whilst the IMS domains both terminate in a conserved C-terminal glycine. The PDV1 family of proteins is found in only higher plants, whilst the PDV2 family is found in higher plants and in moss [97]. The primary defining feature between the two PDV families is the presence of a 28 amino acid extension in the C-terminal domain of the PDV2 proteins, that is absent in all PDV1 proteins [44].
Both PDV1 and PDV2 localize to patches on the outer membrane of the chloroplasts and are recruited to ring-like structures at the division site after Z-ring formation. The analysis of the localization of PDV1 and PDV2 in the pdv2 and pdv1 mutants, respectively, reveals that both proteins are recruited to the division site independently of each other [46]. PDV2 is recruited to the division site by a direct interaction of the IMS domain of PDV2 with the IMS domain of ARC6 [44]. Disruption of this interaction, by deletion of the conserved C-terminal glycine of PDV2 (PDV2G307D) destroys PDV function in vivo, as demonstrated by the inability of PDV2G307D to complement the pdv2 mutant [44]. The IMS of PDV1 does not interact with ARC6 and it is possible that the unique 28 amino acid extension present in the PDV2 family of proteins mediates this interaction [44]. The recruitment of PDV1 to the division site is dependent on PARC6, although no direct interaction has been detected between the IMS domains of PDV1 and PARC6 [46]. Consequently the mechanism by which PDV1 is recruited to the division site is not clear.
A mutation of the C-terminal conserved glycine residue of PDV1 (PDV1G272D) does not disrupt the insertion of PDV1 into the outer chloroplast membrane but, PDV1G272D is localized dispersed throughout the membrane and is unable to be recruited to the division site [97]. It will be important to determine the protein interacting partner that mediates the ability of PDV1 to identify and/or localize to the division site through the C-terminal Gly. Interestingly, analysis of a truncated form of ARC6 lacking the IMS domain (ARC6ΔIMS) found that ARC6ΔIMS is able to insert into the inner chloroplast envelope and to correctly localize to the division site [44]. Furthermore, whilst the ARC6 ring is detected in both the pdv1 and pdv2 mutant background, neither PDV1 nor PDV2 are recruited to the division site in arc6 mutant [44]. These data indicate that ARC6 acts upstream of PDV1 and PDV2 and is necessary for the membrane recruitment of both proteins. The link between the PDV proteins and ARC6 (and potentially PARC6) is the first clue as to how the cytosolic and stromal division machineries can be coordinated across the inner and outer chloroplast envelope. In addition to conveying topological information regarding the location of the division site, it may also act to convey signals concerning the status of the stromal to the cytosolic division machineries.
Disruption or deletion of either PDV1 or PDV2 results in mesophyll cells containing chloroplasts that are fewer in number and larger than those in the wild type, and which frequently show prevalent constrictions [97]. This phenotype suggests that plants lacking functional PDV proteins are able to initiate constriction of the division sites but are unable to complete division. A strikingly similar phenotype is observed in Arabidopsis plants expressing a truncated form of DRP5B (Dynamin-related protein 5B; ARC5) [40], the last chloroplast division protein known to be recruited to the division site, and recent work has shown these three proteins work together as part of the cytosolic chloroplast division machinery.
DRP5B was first identified through cloning of the arc5 mutant and around the same time a dynamin-like protein in C. merolae (CmDnm2) was also identified as a late stage chloroplast division protein [40, 96]. The dynamin and dynamin-like protein superfamily of self-assembling GTPases are multidomain proteins and are well documented to participate in fission and fusion events of intracellular membrane structures (such as endocytosis, intracellular vesicle trafficking, cytokinesis, mitochondrial fusion and fission and peroxisome division) reviewed in [22]. How dynamin proteins function to bring about such a diverse range of events has been the focus of much study.
There are 16 dynamin-like proteins in Arabidopsis and only two in C. merolae. CmDnm2 and AtDRP5B, along with AtDRP5A which is involved in cytokinesis, represent a novel plant-specific group of dynamin-related proteins. This group harbor the three domains central to dynamin proteins: N-terminal GTPase domain, a middle domain (MD) and a C-terminal GTPase effector domain (GED), thought to interact directly with the GTPase domain and to mediate self-assembly [22]. Both AtDRP5B and CmDnm2 also have a pleckstrin homology (PH) domain, which have been shown to mediate lipid binding of other dynamin-like proteins. DRP5B is expressed as two alternatively spliced forms in Arabidopsis, with the shorter protein (714 amino acids) harboring a shorter PH domain than the longer variant (777 amino acids), although the functional significance of this is not yet apparent [40].
Both CmDnm2 and DRP5B relocate from the cytosol, where they localize as patches, to the division site where they form a ring like structure on the surface of the chloroplast [40, 96]. In Arabidopsis this dynamin ring is faintly detected in un-constricted chloroplasts but in C. merolae immunoblot analysis has shown that the dynamin ring only associates with chloroplasts during the division phase [96]. DRP5B is recruited to the division site by the combined action of PDV1 and PDV2. DRP5B is observed to localize to the division site in both the single pdv1 and pdv2 mutants, but not in the double pdv1,pdv2 mutant, indicating that PDV1 and PDV2 function independently to recruit DRP5B to the division site [97]. The precise mechanism of recruitment is unclear and no interaction has been detected between DRP5B and PDV1 or PDV2 [97, 107]. However, both PDV proteins harbor cytoplasmic coiled-coil domains and there are several examples of proteins that recruit dynamin proteins to the mitochondrial division site via a direct interaction with their coiled-coil domain, for example Mdv1p recruits Dnm1p during mitochondrial division [144].
In C. merolae the PDV families of proteins have not been identified, indicating that a novel mechanism may exist to recruit CmDnm2 to the chloroplasts division site in algae. Immunofluorescence microscopy demonstrated that PDR1 appears earlier than CmDnm2 in the cytoplasm and forms a ring from the beginning to the end of chloroplast division. CmDnm2 forms a ring between the outer most rings of the PD1/polyglucan filaments, leading to the hypothesis that PD ring plays a role in organizing the dynamin ring [153].
Recently it was found that DRP5B is also required for peroxisome division in Arabidopsis [154]. Akin to the phenotype observed in the chloroplasts of the arc5 mutant, these plants exhibit enlarged peroxisomes and peroxisomes that have undergone membrane constriction but failed to complete fission and therefore are unable to separate from each other [154]. Two other dynamin-like proteins, two dynamin-related proteins (DRP3A and DRP3B) are also implicated in peroxisome division [32] and it will be interesting to see both how these they are coordinated with DRP5B and also how peroxisome division is coordinated with chloroplast division.
To date the dynamin-like proteins are the final component known to be recruited to the chloroplast division machinery, however it is doubtful that we have identified all of the components of the stromal and cytosolic division machineries. In the future the collection of arc mutants may provide novel insights into chloroplast division, as the mutations in at least four of the original arc mutants are yet to be identified. Furthermore, disruptions of several Arabidopsis genes have been demonstrated to lead to defects in chloroplast division, for example GC1/AtSulA, CRL (crumpled leaf) and ARTEMIS, although a direct link to the division machinery has yet to be demonstrated [8, 39, 78, 126, 140]. Likewise, the negative regulator of chloroplast division ARC1 has recently been identified as FtsHi, but does not affect accumulation of chloroplast division proteins or Z-ring formation [58]. Further characterization of these proteins will be important to identify the full complement of chloroplast division machinery proteins.
5 Constriction and Separation of Chloroplasts
After formation of the division machinery on the inner and outer envelope, microscopic observations reveal that constriction of the plastid is initiated and the septum progressively tightens to eventually separate the two new daughter plastids. Whilst events mediating the placement and the temporal assembly of the machinery are beginning to be elucidated, less clear is how the chloroplast division machinery is initiated to begin constriction, how the force for constriction is generated and which molecules/mechanisms mediate the final scission event.
The analysis of plants in which the mechanism of chloroplast division is disrupted, either through the absence of a functional form of a division protein or through overexpression of a division protein, has revealed distinct classes of chloroplast division phenotypes. Most striking are the presence of either (i) enlarged chloroplasts which are globular/round in shape or (ii) enlarged chloroplasts which are frequently constricted and have a dumbbell-shaped appearance. The first category are commonly caused by diminution of functional stromal ring division components, such as ARC6 or FtsZ [86, 125, 139], whilst the second class are only caused by diminution of the cytosolic division components [40, 97] (Fig. 6.1). These phenotypic differences suggest that the early and late stages of plastid constriction are governed by different forces, generated by either the stromal or the cytosolic division machineries.
5.1 Force Generation by the Stromal Plastid Division Machinery
It is thought that FtsZ is necessary and sufficient to generate the force for constriction of the septum during bacterial cell division [9, 76]. In support of this in some bacteria the sole cell division protein is FtsZ, suggesting that FtsZ alone is sufficient to bring about cell division, furthermore FtsZ protofilaments are observed to undergo a conformational change from straight to curved that would be capable of generating a force [75, 81]. This hypothesis was strengthened when membrane tethered FtsZ successfully assembled into Z-rings on the inside of liposomes [110, 111]. The Z-rings formed were dynamic and capable of constricting the liposomes in a GTP dependent manner, proving that FtsZ alone is capable of self-assembly and force generation. How FtsZ might generate the force for constriction in bacteria is still controversial but years of genetic, biochemical and structural studies have revealed many properties of the protein.
FtsZ has the ability to bind GTP and FtsZ proteins from at least seven bacteria, as well as both AtFtsZ1 and AtFtsZ2-1 from Arabidopsis, have been shown to be functional GTPase enzymes, although both AtFtsZ1 and AtFtsZ2-1 hydrolyze GTP at a slower rate than their bacterial counterparts [25, 74, 109, 137]. Monomeric FtsZ hydrolyses GTP very slowly but can self-assemble into filaments. When assembled into a protofilaments the bottom interface of one FtsZ makes contact with the GTP pocket of the subunit below [131].
FtsZ filaments have three preferred conformations; straight, intermediate curved and highly curved [28]. Whilst the structure of the Z-ring in cell or chloroplast division is not yet clear, it is thought to be formed from small, overlapping subunits of curved FtsZ filaments that are tethered to the membrane via interaction with FtsA, ZipA or ARC6 [79, 117]. GTP hydrolysis is not necessary for FtsZ self-assembly and FtsZ polymers from bacteria have been shown to contain a substantial amount of GTP, suggesting that hydrolysis takes place with some lag after polymer formation [28, 108]. Despite many years of research it is not known what happens following GTP hydrolysis in a protofilament. It is known that GDP substantially destabilizes the protofilaments and one suggestion is that after hydrolysis the protofilament fragments, however it has been possible to detect FtsZ protofilaments that contain a 1:1 ratio of GTP:GDP and many believe that the polymers fragment only at the ends [18, 130].
Several mathematical and physical models have been proposed to explain how the physical properties of FtsZ can generate the force necessary to constrict a membrane. It is proposed that the membrane tethered Z-ring generates force by the protofilaments exerting a bending force on the membrane as they are induced into a curved conformation. In support of this elegant experiments were performed in which Z-rings were reconstituted in liposomes by either targeting the C-terminal end of FtsZ (FtsZ-c) to the membrane (to mimic the in vivo orientation of FtsZ tethered to the membrane) or by targeting the N-terminal end of FtsZ (FtsZ-n) to the membrane (to reverse the orientation). As described above, the Z-rings reconstituted from FtsZ-c were capable in inducing concave constrictions of the liposomes. In stark contrast, the Z-rings formed from FtsZ-n caused convex bulges to form on the liposomes, indicating that the bending force was generated in the opposing direction [111, 112]. It will be extremely interesting to determine if each of the three Arabidopsis FtsZ proteins is capable of generating such bending forces alone and whether they act cooperatively.
If FtsZ is not (solely) responsible for generating the force necessary to initiate constriction of the chloroplast division site, what other proteins could play a role? To date the best candidates are the components of the PD rings. During constriction the inner PD ring has been observed to remain a constant thickness and loses components as constriction proceeds, disassembling late in constriction, indicating that any role played in force generation is not related to final constriction/scission [93]. Identification of the components of these rings and analysis of the phenotype of mutants lacking functional components will be vital in determining possible roles.
5.2 Force Generation by the Cytosolic Plastid Division Machinery
The principal candidates to generate force for constriction on the outer envelope are the chloroplast division dynamin-like proteins. Dynamin-like proteins are well established to be essential for constriction during the division of organelles in many species. For example mitochondrial division is known to employ dynamin-like proteins in mammals (Drp1/Dlp1), yeast (Dnm1), higher plants (ADL1 and ADL2) and C. merolae (CmDnm1) (reviewed in [69]), and the involvement of dynamin-related proteins in peroxisome division has been demonstrated in Arabidopsis (DRP5B, DRP3A, DRP3B), humans (DLP1,Fis1) and yeast (Vps1, Dnm1) (reviewed in [61]). Best characterized is the role of Dnm1p/Drp1 in mitochondrial division and current models leave no doubt that dynamin is the key player mediating membrane scission.
The study of how the dynamin chloroplast division proteins function during division is in its relative infancy but many parallels can be drawn between the mechanisms of chloroplast division and mitochondrial division. Like the chloroplast dynamin division proteins, Dnm1p/Drp1 interacts with other proteins to form the mitochondrial division machinery (reviewed in [116]). Furthermore, Dnm1p/Drp1 are believed to be involved in late stages of division: C. elegans with mutant drp-1 have constrictions in the outer mitochondrial membranes but fail to complete division, a phenotype reminiscent of the chloroplasts in the arc5 mutant, which are frequently constricted and dumbbell shaped [70]. Also, the majority of Dnm1p/Drp1 is present in the cytosol and is recruited to the mitochondrial outer membrane during the late stage of division [136]. Similarly, CmDnm2 and DRP5B also relocate from cytosolic patches to associate with the chloroplast division site [40, 101].
Drp1 exists as small oligomers (dimers/tetramers) that can assemble into larger multimeric structures at the mitochondrial outer membrane [38, 135]. DRP5B can interact with itself in vivo [154], suggesting DRP5B can form higher order structures on chloroplasts. Drp1 can polymerize in vivo into spirals with a diameter of approximately 100 nm, which is considerably smaller than the diameter of mitochondria (400 nm) [54] and it is thought that as higher order structures of dynamin are only observed at later stages of mitochondrial division, they may only form once the membrane has undergone some constriction. In C. merolae small dynamin patches are discontinuously localized at the division site at the onset of constriction and only form a continuous ring at later stages, suggesting that the same mechanism may be in place in chloroplast division [96].
Once assembled, dynamin can undergo GTPase activity, which is stimulated by interactions between the GTPase domain, the MD and the GED [14, 54]. The GTPase activity is stimulated once a ‘critical mass’ is reached and the resulting activity resembles a chain reaction. When dynamin hydrolyses GTP there is a conformation change in the protein which results in considerable bending of the filament, sufficient to bring about constriction of the membrane [88]. Although the identified dynamin-like proteins from Arabidopsis and C. merolae are yet to be proven to be GTPases, given the high levels of conservation in the domain architecture it is likely that they are functional enzymes. Additionally the membrane-free PD machineries isolated from C. merolae formed super-twisted rings, circular rings and spirals which is attributed to the motive force generated for contraction by CmDnm2 [152].
What is the role of the outer PD ring? PDR1 is only glycosylated during division and after constriction has been initiated the outer PD ring is observed to widen and thicken, and remains attached until division has been completed [94, 153]. It is possible that the PDR1 and sugar ring is required to organize the dynamin ring throughout the division process, but it could also play a role in force generation.
The mechanism behind the final scission of chloroplasts remains unknown. It is possible that dynamin is necessary and sufficient to bring about this stage, but other factors may also be required. After constriction is completed, plastids are separated and can move inside the cell via the actin cytoskeleton [66, 133]. Very recently, the CLUMPED CHLOROPLAST 1 protein (CLMP1) has been identified as an important factor for chloroplast separation [151]. In clmp1 mutants, chloroplasts remain interconnected and form clumps instead of being distributed throughout the cells, resulting in the formation of some aplastidic cells. This phenotype is transient and almost completely disappears as leaves age. Interestingly, both early and late steps of chloroplast division appear to occur normally since FtsZ and DRP5B localization is normal in this mutant. Ultrastructure analysis revealed that some chloroplasts within the clumps are held together by thin membranous-connections, similar in structure to the isthmus, suggesting that only the final scission of chloroplasts is impaired in this mutant [151]. CLMP1 does not localize at the division site, and further analysis will be required to fully understand possible roles in scission and how its activity affects chloroplast separation.
6 Regulation of Plastid Division
It is now clear that all plastids originate from the division of a pre-existing organelle, and fusion between plastids appears to occur very rarely if at all [7]. Because plastid number is maintained constant depending on the species and the cell type, plastid division must be tightly regulated to achieve the correct number of organelles in a given cell. Exogenous cues such as light, temperature or nutrition have also been reported to affect chloroplast division (reviewed in [118]), but the underlying mechanisms remain largely unknown.
6.1 Regulation During Cell Proliferation
Regulation of chloroplast division during the cell cycle has been extensively studied in unicellular algae. In such model organisms cells contain one or a few chloroplasts, and the requirement for a strict regulation of chloroplast division during the cell cycle is obvious: if chloroplast division did not occur prior to cell division, aplastidic daughter cells would be generated. This can be observed in the case of Hatena sp, a protist that hosts an endosymbiotic green algal partner but inherits it unevenly because the algae does not divide inside the host cell [106]. The ability of the host cell to promote and control the division of the endosymbiont has hence clearly been a major step during the evolution of chloroplasts. In the red algae C. merolae, accumulation of chloroplast division proteins CmFtsZ1 and CmFtsZ2 precedes accumulation of tubulin during mitosis [102]. In the brown algae Seminavis robusta, chloroplast division has been shown to occur during the S/G2 phase of the cell cycle, and this regulation may involve transcriptional regulation of FtsZ genes expression [43]. In Chlamydomonas reinhardtii, expression of FtsZ and Min genes is also cell cycle regulated [2, 53].
In higher plants, the relationship between cell and plastid division is far more complex because (i) cells contain numerous plastids, and a strict coordination between cell cycle and plastid division is therefore no longer required to avoid the formation of aplastidic cells, and (ii) plastid division can occur in two contrasting cellular contexts: in proliferating cells or in differentiating cells. Indeed, mature Arabidopsis mesophyll cells contain on average 120 chloroplasts in the L-er background, whereas meristematic cells contain about 10 proplastids [83]. Hence, proplastid division is required to keep pace with cell division, and in developing leaves, differentiated chloroplasts can divide both in dividing and in expanding cells.
The existence of mechanisms regulating plastid division during the cell cycle in higher plants is debated. Early observations of dividing chloroplasts in various plant species do not provide evidence for synchrony of chloroplast division in a given cell. Furthermore, chloroplast division can occur independently of the cell cycle in wheat leaves where the maximum rate of chloroplast division is observed in a region of the leaf where cell division has ceased [10]. However, plastid division might be regulated depending on nuclear DNA (nDNA) content (or synthesis) because there is a correlation between chloroplast number and cell ploidy: chloroplast number is increased in a polyploid plant compared to a diploid plant of the same species, and chloroplast number in a given cell type has also been proposed to vary according to endoreduplication (a particular type of cell cycle during which the cell undergoes subsequent phases of DNA replication without mitosis) [12]. In agreement with this hypothesis, in tomato fruit, in which endoreduplication is particularly high, chromoplast number can be up to 1000 per cell [121].
To date, data is lacking to firmly establish that chloroplast division is cell cycle regulated in higher plants, or to provide a molecular mechanism underlying this regulation, but some studies have provided insights into this process. In algae, cell cycle-dependent regulation of FtsZ seems to be the rule, but is not so clear in higher plants: it was observed in BY-2 cells [27], but not in micro-arrays on synchronized Arabidopsis cells [89]. Furthermore, expression of genes encoding proteins involved at early steps of plastid division such as FtsZ or ARC6 does not seem to vary much in developing leaves [107]. In Arabidopsis, silencing of the AtCDT1a gene, encoding a protein involved in nuclear DNA replication licensing, resulted in an inhibition of cell cycle progression and chloroplast division. AtCDT1a was also found to interact with ARC6, suggesting that this factor may not only be a key factor for nuclear DNA replication but also play a direct role in plastid division, and providing evidence for a common regulatory pathway to both processes at the onset of S-phase [127]. In tomato, Caspi et al (2008) reported that the high pigment 1 (hp1) mutation, affecting the DDB1 protein, results in increased chloroplast number per cell [13]. Interestingly, DDB1 is a sub-unit of an ubiquitin-E3 ligase complex and CDT1 is one of its targets; this result may hence support the view of a CDT1-mediated coordination of cell cycle and chloroplast division at the onset of S-phase.
6.2 Plastid Division and Plastid DNA Replication
To date, it is not clear whether plastid DNA replication and plastid division are strictly coupled processes. Plastids contain multiple copies of their genome, and replication of plastid DNA is therefore not a pre-requisite to plastid division. In some instances, chloroplast division can occur independently of cpDNA replication since in the latest stages of leaf development DNA copy number per chloroplast decreases [48]. However, in dividing chloroplasts, nucleoid replication usually occurs before chloroplast division [121]. The phenotype of the crinkled leaves 8 mutant (cls-8) may provide further evidence for a link between cpDNA replication and chloroplast division. These plants are deficient for the large sub-unit of ribonucleotide reductase (RNR), but display severe inhibition of chloroplast division. Despite a clear reduction in dTTP and dATP pools, little effect was observed on nDNA replication whereas chloroplast DNA (CpDNA) copy number was drastically reduced [42]. Finally, the YlmG protein was recently identified in cyanobacteria and the red alga C. merolae for its putative role in nucleoid partitioning [57]. Its overexpression inhibits chloroplast division whereas loss of function only affects nucleoid localization, suggesting that partitioning of chloroplast DNA may somehow be related to the chloroplast division process.
As illustrated by the above paragraphs, several lines of evidence suggest that plastid division may be influenced by nuclear and plastidial DNA replication, although further work is needed to further support this hypothesis and to decipher the molecular mechanisms involved.
6.3 Plastid Division Is Regulated by Cell Differentiation, and Cell Expansion
Plastid number strikingly differs depending on the cell type: for example in Arabidopsis, meristematic cells contain about 10 proplastids [83] whereas stomata guard cells contain only 4 [129], petal cells 15–20 [124], and mesophyll cells more than 100 [83]. How plastid division is regulated according to cell type remains largely unknown, but cytokinins are probably responsible for the stimulation of chloroplast division in young developing leaves. Early reports supporting this hypothesis came from the isolation of the PC22 mutant in the moss Physcomitrella patens. In this mutant, chloroplast division is severely impaired, but can be partially restored by exogenously applied cytokinins [1, 60]. A positive effect of cytokinins on chloroplast division was also reported in bean leaves [99] and Brassica rapa leaf disks [150], but not in spinach leaf disk [119]. The molecular basis for this regulation is beginning to be elucidated and is mediated at least partly by PDV proteins. Indeed, unlike FtsZ or ARC6, accumulation of PDV1 and PDV2 is higher in young leaves, where chloroplast division is most active, than in older leaves, and their overexpression is sufficient to stimulate chloroplast division, like exogenous cytokinin application. Furthermore, the cytokinin response factor CRF2 was shown to stimulate PDV genes expression and chloroplast division [107]. More recently, two cytokinin responsive transcription factors, GATA Nitrate-inducible Carbon metabolism involved (GNC) and Cytokinin-Responsive GATA1 (CGA1) have also been involved in the cytokinin-dependent stimulation of chloroplast division downstream of B-type ARRs [19]. Whether these factors regulate chloroplast division via PDV proteins or via an independent mechanism remains unclear, but PDV proteins nevertheless appear to be key regulators of chloroplast division in developing leaves. The mechanisms regulating plastid division in other organs such as roots, flowers, tuber or seeds have not been investigated, but it is tempting to speculate that other phytohormones such as gibberellins in the case of flowers [23] may play a role in regulating plastid division in non-green tissues.
Additional regulatory factors probably affect chloroplast division in leaves. Indeed in mesophyll cells, chloroplast number clearly depends on cell area: in Arabidopsis, a very nice correlation is observed between cell size and chloroplast content in the mesophyll cells [123]. This correlation is retained between different species: for example cocoa mesophyll cells contain on average 3 chloroplasts but are very small [121]. Isolation of arc mutants led to the conclusion that total chloroplast area, rather than chloroplast number is under tight control in mesophyll cells. Diminution of chloroplast number due to impaired chloroplast division is compensated by an increase in chloroplast size, so that the total chloroplast plan area remains almost constant [123]. Reciprocally, in the few arc mutants characterized by an increased chloroplast number, chloroplast size is reduced [120]. Finally, comparison of mesophyll cell size, chloroplast number and chloroplast size in different species demonstrated that total chloroplast area in cells is strictly related to cell size over a tenfold range of cell sizes [121]. Because cell size is generally related to cell ploidy [65], some authors postulate that chloroplast division is regulated directly by cell size and independently of DNA replication. In agreement with this hypothesis, chloroplast number still correlates with cell size in transgenic plants over-expressing an inhibitor of cell cycle regulation, resulting in increased cell size and reduced cell ploidy [56].
As shown by the above summary, several studies have tackled the issue of the regulation of plastid division during development , but little is known yet regarding the exogenous control of this cellular process. Due to their sessile nature, external conditions affect many aspects of plant growth and development, and in this respect are also likely to affect chloroplast division, but evidence for such regulatory pathways is scarce.
6.4 Chloroplast Division Is Regulated by Exogenous Cues
The most obvious external signal that could affect chloroplast division is probably light. Evidence for light regulation of chloroplast division has been provided long ago by Hashimoto and Possingham [48], in spinach leaf disks they found chloroplast generation time to be more than 2 times longer in the dark. Interestingly, the late phases of chloroplast division are most affected: chloroplasts remain constricted (at the dumbbell stage) for 22 h instead of 3.5 h [48]. Recently DRP5B expression has been shown to be regulated by FHY3, a transcription factor involved in phytochrome A signaling [115]. This result fits nicely with early microscopic observations since chloroplasts in arc5 mutants are arrested at the dumbbell stage of division [83]. FHY3 is specifically involved in far-red signaling and appears to stimulate DRP5B expression in far-red light. In agreement with this observation, mutants deficient for FHY3 or its homolog are affected in chloroplast division, and accumulation of DRP5B at the division site is no longer detected in fhy3. Chloroplast division is even more severely impaired in fhy3 far1 double mutants, demonstrating that FHY3 and FAR1 act partially redundantly to promote chloroplast division in response to far-red light. Nevertheless, other light dependent signaling pathways regulating chloroplast division may exist. Indeed, the chloroplast division phenotype of the PC22 mutant in P. patens can be reverted by blue light treatment [128], suggesting that other photoreceptors such as cryptochromes may play a role in regulating chloroplast division.
Capture of light energy by chloroplasts and active photosynthesis can modify the redox status of the plastoquinone pool in the chloroplast, and the redox state of PQ is likely to regulate nuclear gene expression [63]. In addition, chloroplasts generate high amounts of reactive oxygen species, especially when submitted to excess light, which can result in severe photo-oxidative stress. Because the chloroplast redox balance is most important for the survival and function of plant cells, redox regulation of chloroplast division may well occur. In agreement with such a hypothesis, ntrc mutants deficient for the chloroplast NADPH-thioredoxin reductase appear to accumulate fewer chloroplasts than the wild-type during their first month of growth [73], but further work is needed to confirm this observation and determine how this regulation may function.
Finally, many other factors have been reported to affect chloroplast division including temperature [119] or mineral nutrition [118], but it is not clear whether these effects are direct or indirect, and the underlying molecular mechanisms have yet to be investigated.
Now that the mechanism for chloroplast division is better understood, an increasing number of studies have investigated its regulation, and are providing molecular basis accounting for early cytological observations. Regulation of plastid division by external cues is only beginning to be unraveled but the role of internal cues is more thoroughly described. Two contrasting situations can be distinguished: in unicellular algae containing one or a few chloroplasts, cell cycle regulation of chloroplast division appears to be the rule, and seems to involve transcriptional regulation of genes involved at early steps of the process such as FtsZ or Min . By contrast in higher plants, cell cycle regulation of plastid division is still debated, an although several lines of evidence suggest that plastid division may be modulated by nuclear DNA replication, the only consensus is that total chloroplast area is modulated according to cell area, but the molecular mechanisms for this regulation have yet to be described. One striking feature of plastid division in higher plants is that expression of FtsZ proteins or ARC6 does not seem to vary a lot and proteins involved at later stages of division such as ARC5 or PDV seem to be targets for regulatory pathways. This may point to additional functions for FtsZ proteins unrelated to plastid division. Indeed, in Arabidopsis, FtsZ proteins are associated with thylakoid membranes [26, 59], and fsZ2-2 mutants show defects in chloroplast shape and thylakoid development [59]. Likewise, in P. patens, the five FtsZ isoforms appear to play distinctive roles in cell patterning, chloroplast shaping, plant development and gravity sensing [84] suggesting that in multi-cellular plants, the functions of FtsZ proteins may have diversified.
Abbreviations
- CGA1:
-
Cytokinin-responsive GATA1
- CJD1:
-
Chloroplast J-like domain 1
- CLMP1:
-
Clumped chloroplast 1
- CLS-8:
-
Crinkled leaves 8
- CpDNA:
-
Chloroplast DNA
- Cpn60:
-
Chaperonin 60
- CRL:
-
Crumpled leaf
- DGDG:
-
Digalactosyldiacylglycerol
- DRP5B:
-
Dynamin-related protein 5B
- FP:
-
Fluorescent protein
- FRP:
-
FtsZ regulatory proteins
- FtsZ:
-
Filamenting temperature-sensitive mutant Z
- GED:
-
GTPase effector domain
- HP1:
-
High pigment 1
- IMS:
-
Inter membrane space
- MCD1:
-
Multiple chloroplast division site 1
- MD:
-
Middle domain
- MGDG:
-
Monogalactosyldiacylglycerol
- Min:
-
Membrane Occupation and Recognition Nexus
- MSC:
-
Mechano-sensitive channel
- nDNA:
-
Nuclear DNA
- PARC6:
-
Paralog of ARC6
- PD:
-
Plastid dividing
- PDR1:
-
Plastid dividing ring 1
- PH:
-
Pleckstrin homology
- RNR:
-
Ribonucleotide reductase
- TEM:
-
Transmission electron microscopy
References
Abel W, Knebel W, Koop H, Marienfeld J, Quader H, Reski R, Schnepf E, Spörlein B (1989) A cytokinin-sensitive mutant of the mos, Physcomitrella patens, defective in chloroplast division. Protoplasma 152:1–13
Adams S, Maple J, Moller SG (2008) Functional conservation of the MIN plastid division homologues of Chlamydomonas reinhardtii. Planta 227:1199–1211
Ajjawi I, Coku A, Froehlich JE, Yang Y, Osteryoung KW, Benning C, Last RL (2011) A j-like protein influences fatty acid composition of chloroplast lipids in Arabidopsis. PLoS ONE 6:e25368
Aldridge C, Moller SG (2005) The plastid division protein AtMinD1 is a Ca2+-ATPase stimulated by AtMinE1. J Biol Chem 280:31673–31678
Apuya NR, Yadegari R, Fischer RL, Harada JJ, Zimmerman JL, Goldberg RB (2001) The Arabidopsis embryo mutant schlepperless has a defect in the chaperonin-60alpha gene. Plant Physiol 126:717–730
Arends SJ, Kustusch RJ, Weiss DS (2009) ATP-binding site lesions in FtsE impair cell division. J Bacteriol 191:3772–3784
Arimura S, Yamamoto J, Aida GP, Nakazono M, Tsutsumi N (2004) Frequent fusion and fission of plant mitochondria with unequal nucleoid distribution. Proc Natl Acad Sci U S A 101:7805–7808
Asano T, Yoshioka Y, Kurei S, Sakamoto W, Machida Y (2004) A mutation of the CRUMPLED LEAF gene that encodes a protein localized in the outer envelope membrane of plastids affects the pattern of cell division, cell differentiation, and plastid division in Arabidopsis. Plant J 38:448–459
Bi EF, Lutkenhaus J (1991) FtsZ ring structure associated with division in Escherichia coli. Nature 354:161–164
Boffey SA, Ellis JR, Sellden G, Leech RM (1979) Chloroplast division and DNA synthesis in light-grown wheat leaves. Plant Physiol 64:502–505
Bramkamp M, van Baarle S (2009) Division site selection in rod-shaped bacteria. Curr Opin Microbiol 12:683–688
Butterfass T (1973) Control of plastid division by means of nuclear DNA amount. Protoplasma 76:167–195
Caspi N, Levin I, Chamovitz DA, Reuveni M (2008) A mutation in the tomato DDB1 gene affects cell and chloroplast compartment size and CDT1 transcript. Plant Signal Behav 3:641–649
Chang CR, Blackstone C (2010) Dynamic regulation of mitochondrial fission through modification of the dynamin-related protein Drp1. Ann N Y Acad Sci 1201:34–39
Chapman E et al (2006) Global aggregation of newly translated proteins in an Escherichia coli strain deficient of the chaperonin GroEL. Proc Natl Acad Sci U S A 103:15800–15805
Charbon G, Wang J, Brustad E, Schultz PG, Horwich AL, Jacobs-Wagner C, Chapman E (2011) Localization of GroEL determined by in vivo incorporation of a fluorescent amino acid. Bioorg Med Chem Lett 21:6067–6070
Cheetham ME, Caplan AJ (1998) Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones 3:28–36
Chen Y, Erickson HP (2009) FtsZ filament dynamics at steady state: subunit exchange with and without nucleotide hydrolysis. Biochemistry 48:6664–6673
Chiang YH, Zubo YO, Tapken W, Kim HJ, Lavanway AM, Howard L, Pilon M, Kieber JJ, Schaller GE (2012) Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant Physiol 160:332–348
Colletti KS, Tattersall EA, Pyke KA, Froelich JE, Stokes KD, Osteryoung KW (2000) A homologue of the bacterial cell division site-determining factor MinD mediates placement of the chloroplast division apparatus. Curr Biol 10:507–516
Corbin BD, Wang Y, Beuria TK, Margolin W (2007) Interaction between cell division proteins FtsE and FtsZ. J Bacteriol 189:3026–3035
Danino D, Hinshaw JE (2001) Dynamin family of mechanoenzymes. Curr Opin Cell Biol 13:454–460
Davis SJ (2009) Integrating hormones into the floral-transition pathway of Arabidopsis thaliana. Plant Cell Environ 32:1201–1210
de Boer PA (2010) Advances in understanding E. coli cell fission. Curr Opin Microbiol 13:730–737
de Boer P, Crossley R, Rothfield L (1992) The essential bacterial cell-division protein FtsZ is a GTPase. Nature 359:254–256
El-Kafafi el S, Mukherjee S, El-Shami M, Putaux JL, Block MA, Pignot-Paintrand I, Lerbs-Mache S, Falconet D (2005) The plastid division proteins, FtsZ1 and FtsZ2, differ in their biochemical properties and sub-plastidial localization. Biochem J 387:669–676
El-Shami M, El-Kafafi S, Falconet D, Lerbs-Mache S (2002) Cell cycle-dependent modulation of FtsZ expression in synchronized tobacco BY2 cells. Mol Genet Genomics 267:254–261
Erickson HP, Anderson DE, Osawa M (2010) FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol Mol Biol Rev 74:504–528
Fan J, Xu C (2011) Genetic analysis of Arabidopsis mutants impaired in plastid lipid import reveals a role of membrane lipids in chloroplast division. Plant Signal Behav 6:458–460
Forth D, Pyke KA (2006) The suffulta mutation in tomato reveals a novel method of plastid replication during fruit ripening. J Exp Bot 57:1971–1979
Froehlich JE, Benning C, Dormann P (2001) The digalactosyldiacylglycerol (DGDG) synthase DGD1 is inserted into the outer envelope membrane of chloroplasts in a manner independent of the general import pathway and does not depend on direct interaction with monogalactosyldiacylglycerol synthase for DGDG biosynthesis. J Biol Chem 276:31806–31812
Fujimoto M et al (2009) Arabidopsis dynamin-related proteins DRP3A and DRP3B are functionally redundant in mitochondrial fission, but have distinct roles in peroxisomal fission. Plant J 58:388–400
Fujiwara K, Taguchi H (2007) Filamentous morphology in GroE-depleted Escherichia coli induced by impaired folding of FtsE. J Bacteriol 189:5860–5866
Fujiwara MT, Nakamura A, Itoh R, Shimada Y, Yoshida S, Moller SG (2004) Chloroplast division site placement requires dimerization of the ARC11/AtMinD1 protein in Arabidopsis. J Cell Sci 117:2399–2410
Fujiwara MT, Hashimoto H, Kazama Y, Abe T, Yoshida S, Sato N, Itoh RD (2008) The assembly of the FtsZ ring at the mid-chloroplast division site depends on a balance between the activities of AtMinE1 and ARC11/AtMinD1. Plant Cell Physiol 49:345–361
Fujiwara MT, Li D, Kazama Y, Abe T, Uno T, Yamagata H, Kanamaru K, Itoh RD (2009) Further evaluation of the localization and functionality of hemagglutinin epitope- and fluorescent protein-tagged AtMinD1 in Arabidopsis thaliana. Biosci Biotechnol Biochem 73:1693–1697
Fujiwara MT, Sekine K, Yamamoto YY, Abe T, Sato N, Itoh RD (2009) Live imaging of chloroplast FtsZ1 filaments, rings, spirals, and motile dot structures in the AtMinE1 mutant and overexpressor of Arabidopsis thaliana. Plant Cell Physiol 50:1116–1126
Fukushima NH, Brisch E, Keegan BR, Bleazard W, Shaw JM (2001) The GTPase effector domain sequence of the Dnm1p GTPase regulates self-assembly and controls a rate-limiting step in mitochondrial fission. Mol Biol Cell 12:2756–2766
Fulgosi H, Gerdes L, Westphal S, Glockmann C, Soll J (2002) Cell and chloroplast division requires ARTEMIS. Proc Natl Acad Sci USA 99:11501–11506
Gao H, Kadirjan-Kalbach D, Froehlich JE, Osteryoung KW (2003) ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc Natl Acad Sci U S A 100:4328–4333
Gargano D, Maple-Grodem J, Moller SG (2012) In vivo phosphorylation of FtsZ2 in Arabidopsis thaliana. Biochem J 446:517–521
Garton S, Knight H, Knight MR, Thorlby GJ (2007) Nucleotide depletion and chloroplast division. Plant Signal Behav 2:197–198
Gillard J et al (2008) Physiological and transcriptomic evidence for a close coupling between chloroplast ontogeny and cell cycle progression in the pennate diatom Seminavis robusta. Plant Physiol 148:1394–1411
Glynn JM, Froehlich JE, Osteryoung KW (2008) Arabidopsis ARC6 coordinates the division machineries of the inner and outer chloroplast membranes through interaction with PDV2 in the intermembrane space. Plant Cell 20:2460–2470
Glynn JM, Miyagishima SY, Yoder DW, Osteryoung KW, Vitha S (2007) Chloroplast division. Traffic 8:451–461
Glynn JM, Yang Y, Vitha S, Schmitz AJ, Hemmes M, Miyagishima SY, Osteryoung KW (2009) PARC6, a novel chloroplast division factor, influences FtsZ assembly and is required for recruitment of PDV1 during chloroplast division in Arabidopsis. Plant J 59:700–711
Hashimoto H (1986) Double ring structure around the constricting neck of dividing plastids of Avena sativa. Protoplasma 135:166–172
Hashimoto H, Possingham JV (1989) Effect of light on the chloroplast division cycle and DNA synthesis in cultured leaf discs of spinach. Plant Physiol 89:1178–1183
Haswell ES, Meyerowitz EM (2006) MscS-like proteins control plastid size and shape in Arabidopsis thaliana. Curr Biol 16:1–11
Hennessy F, Cheetham ME, Dirr HW, Blatch GL (2000) Analysis of the levels of conservation of the J domain among the various types of DnaJ-like proteins. Cell Stress Chaperones 5:347–358
Hill JE, Hemmingsen SM (2001) Arabidopsis thaliana type I and II chaperonins. Cell Stress Chaperones 6:190–200
Houry WA, Frishman D, Eckerskorn C, Lottspeich F, Hartl FU (1999) Identification of in vivo substrates of the chaperonin GroEL. Nature 402:147–154
Hu Y, Chen Z-W, Liu W-Z, Liu X-L, He Y-K (2008) Chloroplast division is regulated by the circadian expression of FTSZ and MIN genes in Chlamydomonas reinhardtii. Eur J Phycol 43:207–215
Ingerman E, Perkins EM, Marino M, Mears JA, McCaffery JM, Hinshaw JE, Nunnari J (2005) Dnm1 forms spirals that are structurally tailored to fit mitochondria. J Cell Biol 170:1021–1027
Itoh R, Fujiwara M, Nagata N, Yoshida S (2001) A chloroplast protein homologous to the eubacterial topological specificity factor minE plays a role in chloroplast division. Plant Physiol 127:1644–1655
Jasinski S, Saraiva Leite C, Domenichini S, Stevens R, Raynaud C, Perennes C, Bergounioux C, Glab N (2003) NtKIS2, a novel tobacco cyclin-dependent kinase inhibitor is differentially expressed during the cell cycle and plant development. Plant Physiol Biochem 41:667–676
Kabeya Y, Nakanishi H, Suzuki K, Ichikawa T, Kondou Y, Matsui M, Miyagishima SY (2010) The YlmG protein has a conserved function related to the distribution of nucleoids in chloroplasts and cyanobacteria. BMC Plant Biol 10:57
Kadirjan-Kalbach DK, Yoder DW, Ruckle ME, Larkin RM, Osteryoung KW (2012) FtsHi1/ARC1 is an essential gene in Arabidopsis that links chloroplast biogenesis and division. Plant J 72(5):856–867
Karamoko M, El-Kafafi el S, Mandaron P, Lerbs-Mache S, Falconet D (2011) Multiple FtsZ2 isoforms involved in chloroplast division and biogenesis are developmentally associated with thylakoid membranes in Arabidopsis. FEBS Lett 585:1203–1208
Kasten B, Buck F, Nuske J, Reski R (1997) Cytokinin affects nuclear- and plastome-encoded energy-converting plastid enzymes. Planta 201:261–272
Kaur N, Hu J (2009) Dynamics of peroxisome abundance: a tale of division and proliferation. Curr Opin Plant Biol 12:781–788
Kerner MJ et al (2005) Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell 122:209–220
Kleine T, Voigt C, Leister D (2009) Plastid signalling to the nucleus: messengers still lost in the mists? Trends Genet 25:185–192
Koksharova OA, Wolk CP (2002) A novel gene that bears a DnaJ motif influences cyanobacterial cell division. J Bacteriol 184:5524–5528
Kondorosi E, Roudier F, Gendreau E (2000) Plant cell-size control: growing by ploidy? Curr Opin Plant Biol 3:488–492
Krzeszowiec W, Rajwa B, Dobrucki J, Gabrys H (2007) Actin cytoskeleton in Arabidopsis thaliana under blue and red light. Biol Cell 99:251–260
Kuroiwa T (1991) The replication, differentiation, and inheritance of plastids with emphasis on the concept of organelle nuclei. Int Rev Cytol 128:1–62
Kuroiwa H, Mori T, Takahara M, Miyagishima SY, Kuroiwa T (2002) Chloroplast division machinery as revealed by immunofluorescence and electron microscopy. Planta 215:185–190
Kuroiwa T, Nishida K, Yoshida Y, Fujiwara T, Mori T, Kuroiwa H, Misumi O (2006) Structure, function and evolution of the mitochondrial division apparatus. Biochim Biophys Acta 1763:510–521
Labrousse AM, Zappaterra MD, Rube DA, van der Bliek AM (1999) C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol Cell 4:815–826
Leech RM, Thomson WW, Platt-Aloika KA (1981) Observations on the mechanism of chloroplast division in higher plants. New Phytologist 87:1–9
Lemos JA, Luzardo Y, Burne RA (2007) Physiologic effects of forced down-regulation of dnaK and groEL expression in Streptococcus mutans. J Bacteriol 189:1582–1588
Lepisto A, Kangasjarvi S, Luomala EM, Brader G, Sipari N, Keranen M, Keinanen M, Rintamaki E (2009) Chloroplast NADPH-thioredoxin reductase interacts with photoperiodic development in Arabidopsis. Plant Physiol 149:1261–1276
Lu CL, Stricker J, Erickson HP (1998) FtsZ from Escherichia coli, Azotobacter vinelandii, and Thermotoga maritima-Quantitation, GTP hydrolysis, and assembly. Cell Motil Cytoskeleton 40:71–86
Lu C, Reedy M, Erickson HP (2000) Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J Bacteriol 182:164–170
Lutkenhaus J (1993) FtsZ ring in bacterial cytokinesis. Mol Microbiol 9:403–409
Maple J, Chua NH, Moller SG (2002) The topological specificity factor AtMinE1 is essential for correct plastid division site placement in Arabidopsis. Plant J 31:269–277
Maple J, Fujiwara MT, Kitahata N, Lawson T, Baker NR, Yoshida S, Moller SG (2004) GIANT CHLOROPLAST 1 is essential for correct plastid division in Arabidopsis. Curr Biol 14:776–781
Maple J, Aldridge C, Moller SG (2005) Plastid division is mediated by combinatorial assembly of plastid division proteins. Plant J 43:811–823
Maple J, Vojta L, Soll J, Moller SG (2007) ARC3 is a stromal Z-ring accessory protein essential for plastid division. EMBO Rep 8:293–299
Margolin W (2001) Spatial regulation of cytokinesis in bacteria. Curr Opin Microbiol 4:647–652
Margolin W, Long SR (1994) Rhizobium meliloti contains a novel second homolog of the cell division gene ftsZ. J Bacteriol 176:2033–2043
Marrison JL, Rutherford SM, Robertson EJ, Lister C, Dean C, Leech RM (1999) The distinctive roles of five different ARC genes in the chloroplast division process in Arabidopsis. Plant J 18:651–662
Martin A, Lang D, Hanke ST, Mueller SJ, Sarnighausen E, Vervliet-Scheebaum M, Reski R (2009) Targeted gene knockouts reveal overlapping functions of the five Physcomitrella patens FtsZ isoforms in chloroplast division, chloroplast shaping, cell patterning, plant development, and gravity sensing. Mol Plant 2:1359–1372
Mazouni K, Domain F, Cassier-Chauvat C, Chauvat F (2004) Molecular analysis of the key cytokinetic components of cyanobacteria: FtsZ, ZipN and MinCDE. Mol Microbiol 52:1145–1158
McAndrew RS, Froehlich JE, Vitha S, Stokes KD, Osteryoung KW (2001) Colocalization of plastid division proteins in the chloroplast stromal compartment establishes a new functional relationship between FtsZ1 and FtsZ2 in higher plants. Plant Physiol 127:1656–1666
McAndrew RS, Olson BJ, Kadirjan-Kalbach DK, Chi-Ham CL, Vitha S, Froehlich JE, Osteryoung KW (2008) In vivo quantitative relationship between plastid division proteins FtsZ1 and FtsZ2 and identification of ARC6 and ARC3 in a native FtsZ complex. Biochem J 412:367–378
Mears JA, Lackner LL, Fang S, Ingerman E, Nunnari J, Hinshaw JE (2011) Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat Struct Mol Biol 18:20–26
Menges M, Murray JA (2002) Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J 30:203–212
Mita T, Kanbe T, Tanaka K, Kuroiwa T (1986) A ring structure around the dividing plane of the Cyanidium caldarium chloroplast. Protoplasma 130:211–213
Miyagishima S-y, Itoh R, Toda K, Takahashi H, Kuroiwa H, Kuroiwa T (1998) Identification of a triple ring structure involved in plastid division in the primitive red alga Cyanidioschyzon merolae. J Electron Microsc 47:269–272
Miyagishima S, Itoh R, Aita S, Kuroiwa H, Kuroiwa T (1999) Isolation of dividing chloroplasts with intact plastid-dividing rings from a synchronous culture of the unicellular red alga cyanidioschyzon merolae. Planta 209:371–375
Miyagishima SY, Takahara M, Mori T, Kuroiwa H, Higashiyama T, Kuroiwa T (2001) Plastid division is driven by a complex mechanism that involves differential transition of the bacterial and eukaryotic division rings. Plant Cell 13:2257–2268
Miyagishima S, Kuroiwa H, Kuroiwa T (2001) The timing and manner of disassembly of the apparatuses for chloroplast and mitochondrial division in the red alga Cyanidioschyzon merolae. Planta 212:517–528
Miyagishima S, Takahara M, Kuroiwa T (2001) Novel filaments 5 nm in diameter constitute the cytosolic ring of the plastid division apparatus. Plant Cell 13:707–721
Miyagishima SY, Nishida K, Mori T, Matsuzaki M, Higashiyama T, Kuroiwa H, Kuroiwa T (2003) A plant-specific dynamin-related protein forms a ring at the chloroplast division site. Plant Cell 15:655–665
Miyagishima SY, Froehlich JE, Osteryoung KW (2006) PDV1 and PDV2 mediate recruitment of the dynamin-related protein ARC5 to the plastid division site. Plant Cell 18:2517–2530
Mokranjac D, Bourenkov G, Hell K, Neupert W, Groll M (2006) Structure and function of Tim14 and Tim16, the J and J-like components of the mitochondrial protein import motor. EMBO J 25:4675–4685
Momotani E, Aoki K, Tsuji H (1991) Effect of benzyladenine on diurnal changes in DNA content per chloroplast and chloroplast replication in intact bean leaves. J Exp Bot 42:1287–1293
Nakanishi H, Suzuki K, Kabeya Y, Miyagishima SY (2009) Plant-specific protein MCD1 determines the site of chloroplast division in concert with bacteria-derived MinD. Curr Biol 19:151–156
Nishida K, Takahara M, Miyagishima SY, Kuroiwa H, Matsuzaki M, Kuroiwa T (2003) Dynamic recruitment of dynamin for final mitochondrial severance in a primitive red alga. Proc Natl Acad Sci U S A 100:2146–2151
Nishida K, Yagisawa F, Kuroiwa H, Nagata T, Kuroiwa T (2005) Cell cycle-regulated, microtubule-independent organelle division in Cyanidioschyzon merolae. Mol Biol Cell 16:2493–2502
Nobusawa T, Umeda M (2012) Very-long-chain fatty acids have an essential role in plastid division by controlling Z-ring formation in Arabidopsis thaliana. Genes Cells 17:709–719
Ogino H, Wachi M, Ishii A, Iwai N, Nishida T, Yamada S, Nagai K, Sugai M (2004) FtsZ-dependent localization of GroEL protein at possible division sites. Genes Cells 9:765–771
Oikawa A, Joshi HJ, Rennie EA, Ebert B, Manisseri C, Heazlewood JL, Scheller HV (2010) An integrative approach to the identification of Arabidopsis and rice genes involved in xylan and secondary wall development. PLoS ONE 5:e15481
Okamoto N, Inouye I (2005) A secondary symbiosis in progress? Science 310:287
Okazaki K, Kabeya Y, Suzuki K, Mori T, Ichikawa T, Matsui M, Nakanishi H, Miyagishima SY (2009) The PLASTID DIVISION1 and 2 components of the chloroplast division machinery determine the rate of chloroplast division in land plant cell differentiation. Plant Cell 21:1769–1780
Oliva MA, Huecas S, Palacios JM, Martin-Benito J, Valpuesta JM, Andreu JM (2003) Assembly of archaeal cell division protein FtsZ and a GTPase-inactive mutant into double-stranded filaments. J Biol Chem 278:33562–33570
Olson BJ, Wang Q, Osteryoung KW (2010) GTP-dependent heteropolymer formation and bundling of chloroplast FtsZ1 and FtsZ2. J Biol Chem 285:20634–20643
Osawa M, Anderson DE, Erickson HP (2008) Reconstitution of contractile FtsZ rings in liposomes. Science 320:792–794
Osawa M, Anderson DE, Erickson HP (2009) Curved FtsZ protofilaments generate bending forces on liposome membranes. EMBO J 28:3476–3484
Osawa M, Erickson HP (2011) Inside-out Z rings-constriction with and without GTP hydrolysis. Mol Microbiol 81:571–579
Osteryoung KW, Vierling E (1995) Conserved cell and organelle division. Nature 376:473–474
Osteryoung KW, Stokes KD, Rutherford SM, Percival AL, Lee WY (1998) Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell 10:1991–2004
Ouyang X et al (2011) Genome-wide binding site analysis of FAR-RED ELONGATED HYPOCOTYL3 reveals its novel function in Arabidopsis development. Plant Cell 23:2514–2535
Palmer CS, Osellame LD, Stojanovski D, Ryan MT (2011) The regulation of mitochondrial morphology: intricate mechanisms and dynamic machinery. Cell Signal 23:1534–1545
Pichoff S, Lutkenhaus J (2002) Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J 21:685–693
Possingham JV (1980) Plastid replication and development in the life cycle of higher plants. Annu Rev Plant Physiol 31:113–129
Possingham J, Smith J (1972) Factors affecting chloroplast replication in spinach J Exp Bot 23:1050–1059
Pyke K (1997) The genetic control of plastid division in higher plants. Am J Bot 84:1017
Pyke KA (1999) Plastid division and development. Plant Cell 11:549–556
Pyke KA, Leech RM (1991) Rapid image analysis screening procedure for identifying chloroplast number mutants in mesophyll cells of Arabidopsis thaliana (L.) Heynh. Plant Physiol 96:1193–1195
Pyke KA, Leech RM (1992) Chloroplast division and expansion is radically altered by nuclear mutations in Arabidopsis thaliana. Plant Physiol 99:1005–1008
Pyke KA, Page AM (1998) Plastid ontogeny during petal development in Arabidopsis. Plant Physiol 116:797–803
Pyke KA, Rutherford SM, Robertson EJ, Leech RM (1994) arc6, A fertile Arabidopsis mutant with only two mesophyll cell chloroplasts. Plant Physiol 106:1169–1177
Raynaud C, Cassier-Chauvat C, Perennes C, Bergounioux C (2004) An Arabidopsis homolog of the bacterial cell division inhibitor SulA is involved in plastid division. Plant Cell 16:1801–1811
Raynaud C, Perennes C, Reuzeau C, Catrice O, Brown S, Bergounioux C (2005) Cell and plastid division are coordinated through the prereplication factor AtCDT1. Proc Natl Acad Sci U S A 102:8216–8221
Reski R, Wehe M, Hadeler B, Marienfeld J, Abel WO (1991) Cytokinin and light quality interact at the molecular level in the chloroplast-mutant PC22 of the moss Physcomitrella patens. J Plant Physiol 138: 236–243
Robertson EJ, Pyke KA, Leech RM (1995) arc6, an extreme chloroplast division mutant of Arabidopsis also alters proplastid proliferation and morphology in shoot and root apices. J Cell Sci 108(Pt 9):2937–2944
Scheffers DJ, den Blaauwen T, Driessen AJ (2000) Non-hydrolysable GTP-gamma-S stabilizes the FtsZ polymer in a GDP-bound state. Mol Microbiol 35:1211–1219
Scheffers DJ, de Wit JG, den Blaauwen T, Driessen AJ (2002) GTP hydrolysis of cell division protein FtsZ: evidence that the active site is formed by the association of monomers. Biochemistry 41:521–529
Schmitz AJ, Glynn JM, Olson BJ, Stokes KD, Osteryoung KW (2009) Arabidopsis FtsZ2-1 and FtsZ2-2 are functionally redundant, but FtsZ-based plastid division is not essential for chloroplast partitioning or plant growth and development. Mol Plant 2:1211–1222
Sheahan MB, Rose RJ, McCurdy DW (2004) Organelle inheritance in plant cell division: the actin cytoskeleton is required for unbiased inheritance of chloroplasts, mitochondria and endoplasmic reticulum in dividing protoplasts. Plant J 37:379–390
Shimada H, Koizumi M, Kuroki K, Mochizuki M, Fujimoto H, Ohta H, Masuda T, Takamiya K (2004) ARC3, a chloroplast division factor, is a chimera of prokaryotic FtsZ and part of eukaryotic phosphatidylinositol-4-phosphate 5-kinase. Plant Cell Physiol 45:960–967
Shin HW, Takatsu H, Mukai H, Munekata E, Murakami K, Nakayama K (1999) Intermolecular and interdomain interactions of a dynamin-related GTP-binding protein, Dnm1p/Vps1p-like protein. J Biol Chem 274:2780–2785
Smirnova E, Griparic L, Shurland DL, van der Bliek AM (2001) Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell 12:2245–2256
Smith AG, Johnson CB, Vitha S, Holzenburg A (2010) Plant FtsZ1 and FtsZ2 expressed in a eukaryotic host: GTPase activity and self-assembly. FEBS Lett 584:166–172
Stokes KD, Osteryoung KW (2003) Early divergence of the FtsZ1 and FtsZ2 plastid division gene families in photosynthetic eukaryotes. Gene 320:97–108
Stokes KD, McAndrew RS, Figueroa R, Vitha S, Osteryoung KW (2000) Chloroplast division and morphology are differentially affected by overexpression of FtsZ1 and FtsZ2 genes in Arabidopsis. Plant Physiol 124:1668–1677
Sugita C, Kato Y, Yoshioka Y, Tsurumi N, Iida Y, Machida Y, Sugita M (2012) CRUMPLED LEAF (CRL) homologs of Physcomitrella patens are involved in the complete separation of dividing plastids. Plant Cell Physiol 53:1124–1133
Susin MF, Baldini RL, Gueiros-Filho F, Gomes SL (2006) GroES/GroEL and DnaK/DnaJ have distinct roles in stress responses and during cell cycle progression in Caulobacter crescentus. J Bacteriol 188:8044–8053
Suzuki K, Nakanishi H, Bower J, Yoder DW, Osteryoung KW, Miyagishima SY (2009) Plastid chaperonin proteins Cpn60 alpha and Cpn60 beta are required for plastid division in Arabidopsis thaliana. BMC Plant Biol 9:38
Tavva VS, Collins GB, Dinkins RD (2006) Targeted overexpression of the Escherichia coli MinC protein in higher plants results in abnormal chloroplasts. Plant Cell Rep 25:341–348
Tieu Q, Nunnari J (2000) Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J Cell Biol 151:353–366
Uehara T, Matsuzawa H, Nishimura A (2001) HscA is involved in the dynamics of FtsZ-ring formation in Escherichia coli K12. Genes Cells 6:803–814
Vitha S, McAndrew RS, Osteryoung KW (2001) FtsZ ring formation at the chloroplast division site in plants. J Cell Biol 153:111–120
Vitha S, Froehlich JE, Koksharova O, Pyke KA, van Erp H, Osteryoung KW (2003) ARC6 is a J-domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. Plant Cell 15:1918–1933
Walter S, Buchner J (2002) Molecular chaperones-cellular machines for protein folding. Angew Chem Int Ed Engl 41:1098–1113
Wilson ME, Jensen GS, Haswell ES (2011) Two mechanosensitive channel homologs influence division ring placement in Arabidopsis chloroplasts. Plant Cell 23:2939–2949
Yagisawa F, Mori T, Higashiyama T, Kuroiwa H, Kuroiwa T (2003) Regulation of Brassica rapa chloroplast proliferation in vivo and in cultured leaf disks. Protoplasma 222:139–148
Yang Y, Sage TL, Liu Y, Ahmad TR, Marshall WF, Shiu SH, Froehlich JE, Imre KM, Osteryoung KW (2011) CLUMPED CHLOROPLASTS 1 is required for plastid separation in Arabidopsis. Proc Natl Acad Sci U S A 108:18530–18535
Yoshida Y, Kuroiwa H, Misumi O, Nishida K, Yagisawa F, Fujiwara T, Nanamiya H, Kawamura F, Kuroiwa T (2006) Isolated chloroplast division machinery can actively constrict after stretching. Science 313:1435–1438
Yoshida Y et al (2010) Chloroplasts divide by contraction of a bundle of nanofilaments consisting of polyglucan. Science 329:949–953
Zhang X, Hu J (2010) The Arabidopsis chloroplast division protein DYNAMIN-RELATED PROTEIN5B also mediates peroxisome division. Plant Cell 22:431–442
Zhang M, Hu Y, Jia J, Li D, Zhang R, Gao H, He Y (2009) CDP1, a novel component of chloroplast division site positioning system in Arabidopsis. Cell Res 19:877–886
Acknowledgements
Jodi Maple was supported by a grant from the Norwegian Research Council (Grant no. 204761/F20), and Cécile Raynaud by a grant from the French National Research Agency (ANR 2010 JCJC1207 01). We thank Daniela Gargano for critical reading of the manuscript and Ingrid Holtsmark for help with microscopy.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Science+Business Media New York
About this chapter
Cite this chapter
Maple-Grødem, J., Raynaud, C. (2014). Plastid Division. In: Theg, S., Wollman, FA. (eds) Plastid Biology. Advances in Plant Biology, vol 5. Springer, New York, NY. https://doi.org/10.1007/978-1-4939-1136-3_6
Download citation
DOI: https://doi.org/10.1007/978-1-4939-1136-3_6
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4939-1135-6
Online ISBN: 978-1-4939-1136-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)