Abstract
The essential molecular chaperone heat shock protein 90 (Hsp90) is a highly conserved protein hub, which modulates the genotype to phenotype translation in animals, plants, and fungi. In doing so, Hsp90 interacts with up to 10 % of the eukaryotic cell’s proteome. In the leading fungal pathogen of humans, Candida albicans, Hsp90 governs virulence, morphogenesis, and drug resistance. While specific Hsp90 clients have been identified and described, a global overview of Hsp90 interactions in this pathogen remained elusive until recently.
Here, we discuss recent advancements in mapping the C. albicans Hsp90 chaperone network. We describe the first Hsp90 genetic interaction network in C. albicans, discuss its divergence from that of its relative Saccharomyces cerevisiae and illustrate how the network informs our understanding of fungal biology, stress responses and virulence. Deciphering the Hsp90 chaperone network holds great promise for the development of suitable measures to combat fungal drug resistance and counter the ever-increasing number of Candida infections.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Candida albicans
- Saccharomyces cerevisiae
- Hsp90
- Drug resistance
- Virulence
- Morphogenesis
- Evolution
- CK2
- Mkc1
- Calcineurin
- Cdc37
- Hog1
1 The Commensal Yeast Candida albicans Is the Leading Fungal Pathogen of Humans
Candida albicans is a member of the human microbiome, colonizing mucosal surfaces, such as the oral cavity and the gastrointestinal tract in 30–70 % of healthy adults [1]. While fungal carriage does not present a problem in most immunocompetent individuals, it can cause serious complications in individuals with compromised immune function. The impact on human health and quality of life is immense. Every year up to 11,200 patients in the USA alone die from C. albicans infections of the bloodstream or the organs (candidemia) [2]. Recent estimates suggest that ~ 400,000 patients suffer from life-threatening C. albicans infections worldwide [3]. C. albicans is the fourth most common cause of hospital-acquired bloodstream infections [4], with mortality rates of up to 50 % and an attributable economic burden of US$ 40,000 per episode and patient due to extended stays at the hospital [1] .
Patients at risk of contracting candidemia include those who received solid organ transplants, are cared for at neonatal intensive care units, underwent abdominal surgery, or were diagnosed with solid tumors [4]. As this patient population is growing due to recent advances in medical care, so is the incidence rate of candidemia, which increased between 1979 and 2000 by 207 % [5]. This already dire situation is exacerbated by the two main challenges associated with the treatment of C. albicans infections. First, due to the close association between C. albicans and its human host, the patient acts as a reservoir for endogenous infections. Second, therapeutic strategies are quickly compromised by the rapid emergence of antifungal drug resistance , the occurrence of intrinsically resistant C. albicans strains, or severe side effects caused by antifungal drugs. To make matters worse, development of more effective therapeutic strategies is hampered by the paucity of fungal-specific drug targets and C. albicans’ status as commensal. This emphasizes a pressing need for more efficacious treatment strategies .
Currently, the two most widely utilized drug classes for the treatment of life-threatening candidemia are the azoles and the echinocandins. They affect the fungal cell’s outer layers, the cell membrane and the cell wall. The azoles block the production of ergosterol, the fungal analog of human cholesterol, by inhibiting C14α-demethylase, thereby causing a toxic sterol intermediate to accumulate in the cell membrane [6]. This results in increased vulnerability to further membrane damage. The echinocandins are the only new class to enter the clinic in decades and inhibit 1,3-β-d-glucan synthase, which is essential for cell wall production [7]. Importantly, both basal tolerance and resistance to these drugs acquired by diverse mechanisms depend upon cellular stress responses, such as those regulated by the molecular chaperone Hsp90. The combination of antifungal drug treatment with Hsp90 compromise abrogates drug resistance in vitro,emphasizing the role of Hsp90 as the emerging fungal Achilles’ heel [8] .
2 Hsp90 Stabilizes Stress-Responsive Signal Transducers and Governs the Genotype-to-Phenotype Transition
Hsp90 was originally discovered as essential for high temperature growth in the eukaryotic model organism Saccharomyces cerevisiae. Unlike other heat shock proteins, Hsp90 is not required for de novo protein folding but keeps metastable client proteins poised for activation [9]. It does so through a chaperone cycle, during which adenosine triphosphate (ATP) hydrolysis and associated co-chaperones assist in opening of the Hsp90 homodimer and client release [10]. Protein folding is facilitated by Hsp90’s molecular structure [11–13]. The tri-partite composition of an ATP-binding N-terminal domain, a client-binding middle segment, and a C-terminal dimerization domain promote chaperoning [13]. Hsp90 client proteins important for human health and biology include the steroid hormone receptors and oncoproteins, such as p53 [14]. The intricate liaison between Hsp90 and cancer proteins is being exploited for the development of anticancer therapeutics, and there are currently 13 Hsp90 inhibitors in clinical trials for the treatment of diverse cancers [15]. Hsp90 inhibitors commonly used in laboratory settings for the dissection of Hsp90 function include the antibiotics geldanamycin (GdA) and radicicol, which are structurally unrelated and competitively bind Hsp90’s ATP-binding pocket, thereby inhibiting Hsp90 function [16] .
Hsp90 serves as a central hub in the cellular circuitry by chaperoning diverse regulators of cellular signalling in a manner that is contingent upon environmental cues. Although Hsp90 is one of the most abundant proteins in the cell, environmental perturbation can exhaust the Hsp90 system by increasing the cellular burden of misfolded proteins. Modulation of Hsp90 function by environmental stress can have profound impacts on the genotype-to-phenotype translation on a global level [17]. Hsp90 can function as a capacitor by buffering genetic or epigenetic variation, such that it remains in a phenotypically silent state until Hsp90 function is compromised and new traits are revealed [18]. Perturbations of the Hsp90 chaperone system have been found to reveal new traits in animals, plants, and fungi [17, 19, 20]. Hsp90 can also function as a potentiator of genetic variation by stabilizing mutant regulators and enabling new genetic variants to have immediate phenotypic effects. In this context, compromise of the Hsp90 chaperone system has been found to mask new traits in fungi as well as mammalian cancer cells [14, 17, 21] .
Hsp90 does not operate solitarily. For proper function, it requires more than 20 different co-chaperones that contribute to distinct tasks, such as regulating ATPase activity, targeting specific clients, and assisting with client maturation [12]. The composition of the Hsp90 co-chaperone machinery differs considerably across the tree of life [22] while Hsp90 itself is highly conserved. For example, Hsp90 amino acid sequence identity between humans and C. albicans is 62 %, and between C. albicans and S. cerevisiae, it is 84 % [23] .
3 C. albicans Hsp90 Modulates Antifungal Drug Resistance, Morphogenesis, and Virulence in Concert With its Co-chaperone Sgt1 (suppressor of G2 allele of skp1), and its Client Proteins Mkc1 and Calcineurin
Hsp90 operates in concert with its co-chaperone machinery, which comprises 10 co-chaperones in both C. albicans and S. cerevisiae [24] . In S. cerevisiae, the co-chaperone Sgt1 has been demonstrated to function as an adaptor protein [25], as it connects Hsp90 with several of its clients, one of which is the adenylyl cyclase Cyr1 [26]. Cyr1 generates 3′-5′-cyclic adenosine monophosphate (cAMP), whose signalling properties are a key factor in C. albicans’ yeast-to-filament transition [27]. The morphogenetic transition between yeast and filamentous growth is a temperature-dependent developmental program, and a key virulence trait [28]. The basis of temperature dependence of C. albicans morphogenesis remained enigmatic for decades, until it was discovered that Hsp90 represses filamentation, such that inhibition of Hsp90 induces filamentation under conditions that normally favor yeast-form growth [29]. Consistent with the importance of Sgt1 for cAMP-protein kinase A (PKA) signalling in S. cerevisiae [30], C. albicans Sgt1 was found to be a key regulator of cAMP-PKA signalling and morphogenesis [31]. Genetic depletion of Sgt1 phenocopies Hsp90 compromise and induces filamentation in the absence of other cues, such as elevated temperature, serum, or CO2 [31]. Co-immunoprecipitation experiments showed that Hsp90 and Sgt1 interact with each other, and that Sgt1 interacts with Cyr1. Consistent with the impact of Hsp90 on antifungal drug resistance , Sgt1 was also found to be a key regulator of basal tolerance and resistance to both the azoles and echinocandins [31].
Hsp90 governs the emergence and maintenance of antifungal drug resistance [8, 32]. Initial studies with S. cerevisiae revealed that genetic reduction of Hsp90 levels blocks the rapid evolution of resistance to azoles [21]. Pharmacological inhibition of Hsp90 with GdA or radicicol, structurally distinct Hsp90 inhibitors that competitively bind to the ATP-binding pocket [16], blocks the rapid evolution of azole resistance in C. albicans, and abrogates resistance acquired by different mechanisms [21]. Hsp90 also has a profound impact on azole resistance of C. albicans that evolved in a human host. Analysis of a series of 17 C. albicans isolates from a human immunodeficiency virus (HIV) patient undergoing therapy with the azole fluconazole for two years [33] revealed that inhibition of Hsp90 reduced resistance of all of the isolates, with the largest impact on those isolates recovered earlier during treatment [21]. Targeting Hsp90 also transforms azoles from fungistatic to fungicidal [34] and overcomes the intrinsic resistance of C. albicans biofilms to azole treatment [35]. Inhibition of Hsp90 function in C. albicans abrogates resistance not only to azoles but also to echinocandins. Genetic or pharmacological compromise of Hsp90 function reduces resistance of laboratory strains and clinical isolates that evolved resistance in the human host [36, 37].
One of the key downstream effectors through which Hsp90 governs resistance to both azoles and echinocandins is calcineurin . Calcineurin is a protein phosphatase that was known to regulate cellular responses to various environmental stressors, including the azoles [38]. Work in S. cerevisiae demonstrated that calcineurin physically interacts with Hsp90 [39] . Consistent with an important role for Hsp90 in regulating calcineurin-dependent stress responses essential for azole resistance, inhibition of calcineurin with the structurally distinct molecules cyclosporin A or FK506, phenocopies Hsp90 inhibition and reduces azole resistance [21]. Deletion of the catalytic or regulatory subunits of calcineurin also phenocopies genetic depletion of Hsp90, abrogating resistance to both azoles and echinocandins [21, 36]. Calcineurin was the first Hsp90 client protein identified in C. albicans based on the physical interaction between Hsp90 and the catalytic subunit of calcineurin, Cna1, and the destabilization of Cna1 upon depletion of Hsp90 [36] .
Although calcineurin is a key mediator of Hsp90-dependent drug resistance , additional targets have recently been implicated in C. albicans. Another client protein that is important for resistance to both azoles and echinocandins is Mkc1 , the terminal mitogen-activated protein kinase (MAPK) of the Pkc1 cell wall integrity pathway. Depletion of Hsp90 leads to destabilization of Mkc1 [40], consistent with the interaction of Hsp90 with the homologous MAPK, Slt2, in S. cerevisiae [41]. Beyond Mkc1, targeting Pkc1 with a specific inhibitor phenocopies inhibition of Hsp90 or calcineurin , suggesting that Pkc1 may be an additional client protein in C. albicans, as is the case in mammalian cells [42].
Given Hsp90’s key role in fungal drug resistance, morphogenesis, and virulence, targeting Hsp90 with one of the inhibitors currently in clinical trials as an anticancer agent in combination with an antifungal has emerged as a promising approach for treating candidemia. This prospect has been explored in multiple models of infection. In larvae of the Greater Wax Moth Galleria mellonella , combination therapy with an Hsp90 inhibitor structurally related to GdA, 17-N-allylamino-17-demethoxygeldanamycin (17-AAG), rescues infections with C. albicans that are otherwise lethal and recalcitrant to treatment with azoles alone [34]. In a rat central venous catheter model of C. albicans biofilm infection, 17-AAG transforms azoles from ineffective to highly efficacious, such that the combination sterilizes an infected catheter [35]. While there was no toxicity in the biofilm model where the drug delivery and fungal infection are localized, systemic inhibition of Hsp90 with 17-AAG in the context of a disseminated infection in a murine model of candidemia proved toxic, likely due to inhibition of host Hsp90. This emphasizes the need for Hsp90 inhibitors that can selectively target the pathogen or for alternate strategies to inhibit the Hsp90 chaperone machinery. Recent work has established a powerful approach to map the C. albicans Hsp90 chaperone network, revealing novel effectors upstream and downstream of Hsp90 and new candidate therapeutic targets for combatting fungal infectious disease.
4 The C. albicans Hsp90 Genetic Interaction Network Reveals Environmental Contingency and Evolutionary Rewiring
Genetic interaction networks are important illustrations of which genes are involved in similar biological processes or pathways, permitting gene function annotation, and providing functional insights into protein-protein complexes [43]. These interaction profiles typically emerge from synthetic lethality screens. Synthetic lethality describes the situation in which single mutants are viable while concurrent perturbation of two genes causes lethality, which is indicative of a genetic interaction [44]. To obtain genome-wide genetic interaction data, genomic tools such as gene deletion libraries [45], synthetic genetic array technology (SGA) [43], or ribonucleic acid (RNA) interference [46], are an absolute necessity. Until recently, such tools were restricted to model eukaryotic systems such as S. cerevisiae and the nematode Caenorhabditis elegans.
Proteomic and genomic analyses in the model yeast S. cerevisiae revealed that Hsp90 interacts with approximately 10 % of the proteome. Many interactors are signal transducers such as kinases and transcription factors [47] that are involved in different stress response pathways, such as those governing high osmolarity growth (Hog1) and cell wall disturbances (Mkc1/Slt2) [41]. Detection of genetic and physical interactions was facilitated by the extensive genomic and proteomic tool set available for S. cerevisiae. Hsp90 genetic interactions have been identified by an SGA screen for mutants with a synthetic lethal or synthetic sick phenotype when combined with a temperature-sensitive hsp82 allele as the only source of Hsp90, as well as by screens of gene deletion mutant libraries for hypersensitivity to Hsp90 inhibitors [47–49]. Physical interactions have been uncovered by MALDI-ToF (Matrix-assisted laser desorption/ionization Time of Flight) mass spectrometric analyses following tandem affinity purification of Hsp82-tandem affinity purification (TAP), as well as by yeast two-hybrid assays employing Hsp82 as bait [41, 47]. These studies provide a foundation for analyses of the Hsp90 chaperone network in C. albicans, contingent on the availability of genomic resources such as mutant libraries.
In 2002, the first large-scale set of C. albicans mutants was made available by Prof. Aaron Mitchell [50], and is referred to here as the Mitchell Library. This library of homozygous transposon insertion mutants was created by random insertions of a transposon followed by selection for homozygosity. It quickly became a vital tool for the C. albicans community and expedited mapping of the C. albicans Hsp90 genetic interaction network. Over the years, additional mutant strains have been generated and made publically available. The library now comprises ~ 1,200 mutant strains representing 704 genes, which cover about 10 % of the C. albicans genome. More than half the genes are represented by two mutants, about a third by one mutant, and very few genes are represented by 3−6 mutants. A genome-scale library with 5,400 bar-coded heterozygous gene deletion mutants and 1,152 mutants with promoter replacement of the intact allele with the tetracycline-repressible promoter have only very recently been made publically available [51, 52], providing unprecedented opportunities to expand the Hsp90 genetic interaction network.
We employed the most recent version of the Mitchell Library to map the C. albicans Hsp90 genetic interaction network and to explore the impact of environmental stress on network dynamics [53]. To do so, we optimized six environmental conditions identified as general or specific stresses, which were selected to exemplify conditions similar to those the pathogen may encounter during the course of infection. As a general stressor, we chose high temperature growth, which mimics a febrile host situation (41 °C). To determine if stressors that affect a wide array of cellular processes elicit similar Hsp90 genetic interaction profiles, we compared high temperature growth to osmotic stress. This is a particularly well-characterized stressor, here exerted by NaCl. To test specific stressors, we turned to drugs that target cellular compartments in a well-defined manner. The nucleoside inhibitor tunicamycin exerts stress on the endoplasmatic reticulum by inhibiting the first step of glycoprotein synthesis, thus inducing the unfolded protein response. The azole fluconazole inhibits ergosterol biosynthesis and exerts stress on the cell membrane while the echinocandin caspofungin targets cell wall synthesis and exerts cell wall stress. All assays were carried out in RPMI (Roswell Park Memorial Institute), a cell culture medium that is formulated to approximate human physiological conditions and is the standard used for clinical analysis of antifungal drug susceptibility.
We screened the Mitchell Library, which is arrayed in 96-well format, for growth inhibition in the presence of GdA in six different environmental conditions (standard growth at 37 °C, two general stresses, and three specific stresses [53]). Hypersensitivity to Hsp90 inhibition is indicative of an Hsp90 genetic interactor. The screen revealed a total of 226 Hsp90 genetic interactors (Table 8.1), including the only two Hsp90 interactors that were previously described in C. albicans. Identification of the two reported client proteins, Mkc1 [40] and Cna1 [36], lent support to the validity of our approach. This C. albicans Hsp90 network also includes known S. cerevisiae Hsp90 interactors, such as Hog1 and Mkc1/Slt2 [41].
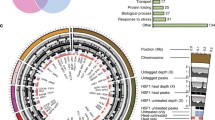
A subsequent network analysis using Cytoscape [54] revealed relationships between interactors and the environmental conditions they were identified in [53]. The network structure was found to be highly dependent on environmental conditions (Fig. 8.1) , such that the vast majority of the interactors were relevant for growth under only one or two conditions. Only four interactors were identified in five or six screens. The number of interactors differed dramatically between screens (Table 8.1). The cell wall stressor caspofungin elicited a total of 73 interactions, only 21 of which were shared with other conditions. The dependence of cell wall integrity on Hsp90 function suggests a broader role for the chaperone in cell wall regulation than previously appreciated.
Global C. albicans Hsp90 genetic interaction network. The Cytoscape network analysis of 226 genetic interactors (filled circles) revealed an environmentally contingent network structure. Interactors were identified in six different environmental conditions (black squares), including standard growth (37 °C), general stress conditions (41 °C, NaCl), and the specific stressors tunicamycin (T), caspofungin (C), and fluconazole (F). Each interactor is color-coded depending on its degree of connectivity (frequency of detection in six screens), which ranges from 1 (grey) to 6 (red). The edges connecting interactors indicate the screen in which the interactor had originally been identified. Thus, an interactor that has been identified more than once is connected by multiple edges to different conditions. (Figure reproduced from [53])
Next, we screened the set of Hsp90 genetic interactors for enrichment of gene ontology (GO) gene function categories relative to the set represented by the Mitchell Library [53]. Hsp90 genetic interactors were enriched for macromolecular complexes, protein complexes, protein modification processes, biopolymer modification, and posttranslation protein modifications. We then analyzed GO category enrichment for each environmental condition separately (Table 8.1). Significant enrichment could be detected in three environmental conditions, namely both general stresses and tunicamycin. Interestingly, patterns of GO category enrichment differed between the general stresses. Phosphate-based protein modifications were more relevant during high temperature growth while various complexes were identified as important for growth during osmotic stress. Protein phosphorylation and phosphate/phosorphorus metabolic processes were particularly important for growth at 41 °C. During osmotic stress, the SAGA (Spt-Ada-Gcn5-acetyltransferase) complex, which regulates transcription by RNA polymerase II [55], the CK2 complex, a promiscuous kinase complex with some 300 targets in the cell [56], and the UTP-C (U three proteins complex) complex, which comprises the four CK2 subunits as well as two more proteins (Rrp7 and Utp22) that form a subcomplex of the 90S preribosome [57], were relevant for growth during osmotic stress. This conforms with previous insights gleaned from Hsp90 clients in S. cerevisiae, many of which are signal transducers [47], and the finding of distinct interactors to be relevant during different conditions [48].
We found that analysis of Hsp90 genetic interactions in multiple conditions is crucial for a comprehensive appreciation of client proteins. For example, we identified Hog1 as a C. albicans Hsp90 interactor only in our 41 °C screen [53]. Depletion of Hsp90 reduced Hog1 transcript and protein levels, and blocked Hog1 activation, confirming its position as a conserved Hsp90 client protein. S. cerevisiae Hog1 [41] and the mammalian Hog1 homolog p38 [58] are both established Hsp90 clients. Thus, analysis of genetic interactors in multiple environmental conditions was instrumental for the identification of client proteins in C. albicans.
The extensive degree of environmental contingency of the C. albicans Hsp90 chaperone network revealed important properties that enabled prediction of whether interactors function upstream or downstream of Hsp90 [53]. Our finding that only four of the 226 interactors were important for growth in five or six conditions led to the model that interactors important for growth in many conditions (defined here as high-connectivity), operate upstream of Hsp90 affecting its expression or function. In contrast, those interactors that were only important for growth in one or two conditions (low-connectivity) function downstream of Hsp90 to regulate a specific subset of Hsp90-dependent cellular processes. The simple expectation is that for downstream effectors, Hsp90 would impact on their stability or activation while Hsp90 would have little impact on upstream effectors. We tested this model by analyzing the effect of Hsp90 depletion on the transcript and protein levels of three high-connectivity interactors (Hos2, Ckb1, and Ckb2) and six low-connectivity interactors (Hog1, Cka1, Cka2, Mkk2, Cdr1, and Cmk1). As predicted, Hsp90 depletion had a significantly greater effect on levels of the low-connectivity interactors than the high-connectivity interactors (P < 0.0430). Thus, our chemical genomic approach to identify Hsp90 interactors in multiple environmental conditions provided a powerful strategy to predict functional relationships within the Hsp90 chaperone network.
We aimed to further investigate how the four high-connectivity interactors might affect Hsp90 function. First, we focused on Ckb1 and Ckb2, the regulatory subunits of the protein kinase CK2 complex. We found this complex enriched during osmotic stress, one of the general stressors (Table 8.1). Previous research in S. cerevisiae demonstrated that CK2 phosphorylates Hsp90 residue T22, which regulates high temperature growth [59]. Thus, we monitored levels of phosphorylation of Hsp90 and its kinase-targeting co-chaperone Cdc37 in CK2 subunit deletion mutants [53]. As expected, deletion of CKA1 or CKA2, encoding the low-connectivity catalytic subunits, had negligible impact on phosphorylation of Hsp90 or Cdc37. Strikingly, deletion of CKB1 reduced phosphorylation of Hsp90 and Cdc37 while deletion of CKB2 abolished Cdc37 phosphorylation. This provides a compelling example of high connectivity Hsp90 interactors functioning upstream of Hsp90.
To determine the phenotypic consequences of aberrant Hsp90 and Cdc37 phosphorylation, we measured Hog1 protein levels and activation during standard growth and oxidative stress, and further assessed the ability of CK2 deletion mutants to grow in sorbitol, which exerts osmotic stress [53]. During oxidative stress, Hog1 protein levels are reduced in ckb1Δ/Δ and ckb2Δ/Δ mutants but Hog1 activation, as measured by phosphorylation, remains unaffected. Given that Hsp90 is required for Hog1 stability and phosphorylation, this suggests that blocking CK2-mediated phosphorylation of Hsp90 and/or Cdc37 reduces but does not abrogate the function of chaperoning Hog1. The phenotypic assay revealed that the CK2 regulatory subunit deletion mutants were hypersensitive to sorbitol, as is the case with the hog1Δ/Δ mutant, confirming the importance of Ckb1 and Ckb2 for Hog1-mediated growth in osmotic stress. This provides the first example of regulation of Hsp90 function by posttranslational modification in C. albicans, and implicates protein kinase CK2 as an evolutionarily conserved regulator of Hsp90 function.
Next, we sought to explore the function of another high-connectivity interactor that was poised to modulate HSP90 expression, the zinc finger transcription factor Ahr1. This Candida-specific transcription factor has been found to influence morphology [60] and high-temperature growth [61], such that deletion of AHR1 causes aberrant morphologies and reduced biofilm formation [60], as well as reduces the ability to growth at 37 °C and 42 °C [61]. Here, we predicted that Ahr1 regulates HSP90 expression and that the morphological alterations of the ahr1Δ/Δ mutant might reflect reduced HSP90 levels, given that reduction of Hsp90 function or levels induces filamentation [29]. These predictions were confirmed by qRT-PCR and microscopic analyses of the ahr1Δ/Δmutant grown at 30 °C in rich medium that favors yeast growth [53]. Hsp90 levels are significantly reduced in the ahr1Δ/Δmutant, and the strain is filamentous, consistent with reduced Hsp90 function. Thus, Ahr1 is a novel regulator of Hsp90 expression, emphasizing the power of the Hsp90 genetic network for identifying upstream effectors of this global cellular regulator.
Next, we focussed on the lysine deacetylase Hos2, a high connectivity Hsp90 interactor important for growth in all six environmental conditions. Hos2 is part of the Set3C lysine deacetylase (KDAC) complex. Together with Set3, Snt1, and Sif2, Hos2 forms the core of this complex [62] and is required for lysine deacetylation of H3 and H4 histones, thereby ensuring efficient transcription [63]. Hos2 protein levels are not reduced upon Hsp90 depletion, suggesting that Hos2 may function upstream of Hsp90, as with other high-connectivity interactors [53]. Deletion of HOS2 causes colony morphology to appear wrinkly at 37 °C due to filamentous growth [64], consistent with a modest reduction of Hsp90 function. Further, the broad spectrum lysine deacetylase inhibitor trichostatin A has recently been demonstrated to phenocopy Hsp90 inhibitors, blocking the emergence of azole resistance and abrogating resistance that evolved in a human host [65]. In S. cerevisiae, Hsp90 function is regulated by acetylation, such that inhibition of KDACs compromises Hsp90 function, blocks Hsp90 interaction with key client proteins for drug resistance , and alters client protein stability and activation [65]. The key KDACs in S. cerevisiae are Rpd3 and Hda1 while the relevant players in C. albicans remain to be identified. Our network analysis suggests that Hos2 may well be a key regulator of Hsp90 function in this pathogen.
In addition to illuminating novel functional insights into C. albicans Hsp90 and its targets, this network provided the first opportunity for a comparative analysis of a chaperone network. Until recently, comparative analyses were limited by the lack of large-scale interaction data in species other than S. cerevisiae [47, 48] or by the extensive protein interaction network divergence from other eukaryotes, such as with Plasmodium falciparum [66]. Thus, we seized this opportunity and compared C. albicans Hsp90 genetic interactors to those identified in S. cerevisiae [47].
These yeast species diverged from their most recent common ancestor approximately 200 million years ago [67]. During this time, they adapted to diverse environmental niches. C. albicans is a commensal of warm-blooded animals, mostly humans [68] while S. cerevisiae is a saprophyte living on fruit and in the soil. These environments exert distinct selective pressures, such as temperature and glucose content. Given that genetic variation between strains of S. cerevisiae can alter the dependence of diverse traits on Hsp90 [17], it is thus conceivable that the Hsp90 network has diverged considerably between S. cerevisiae and C. albicans. Although a small but significant (P = 0.004) overlap in Hsp90 genetic interactors remained between the species, the network underwent extensive rewiring over evolutionary time [53]. Less than 20 % of the interactors are conserved, despite very similar Hsp90 chaperone machineries in the two species [22].
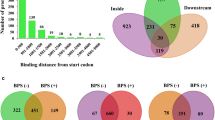
A total of 428 C. albicans genes present in the Mitchell Library have homologs in S. cerevisiae [53], 59 of those are Hsp90 genetic interactors in S. cerevisiae as well (Fig. 8.2). Of these 59, 29 were shared between both species and 30 were unique to S. cerevisiae, indicating extensive rewiring. Conserved interactors are distributed throughout the network. While there is no detectable pattern regarding conservation of high- or low-connectivity interactors, there is some degree of conservation detectable within different environmental conditions. For example, a quarter of Hsp90 interactors relevant for growth during exposure to the cell membrane stressor fluconazole are conserved, but less than 10 % of those important for growth during cell wall stress exerted by caspofungin are conserved. While the extent of interactor conservation may be low, response profiles are conserved. Both species maintain a large number of their interactors when temperatures shifts between standard growth (30 °C for S. cerevisiae [48] and 37 °C for C. albicans) and high-temperature growth (37 °C for S. cerevisiae [48] and 41 °C for C. albicans; Fig. 8.2 inset). Even though the specific interactions may change, similar proportions of the genome remain associated during high-temperature growth. The mechanisms underpinning Hsp90 chaperone network dynamics and evolution remains largely unexplored.
The C. albicans Hsp90 genetic interaction network has been substantially rewired. The C. albicans Hsp90 genetic interactor set was compared to that previously identified for S. cerevisiae, revealing that less than 20 % of Hsp90 genetic interactions are conserved. Interactors with a homolog in S. cerevisiae (blue) and those that are also Hsp90 genetic interactors in the model yeast (red) are mapped onto the global network. The Hsp90 genetic interaction network has been largely rewired since both yeasts diverged from their most common ancestor. (Figure reproduced from [53])
Taken together, the initial work towards elucidating the C. albicans Hsp90 genetic interaction network has imparted a suite of critical insights regarding Hsp90 biology in this leading fungal pathogen of humans: (i) High connectivity Hsp90 genetic interactors are poised to regulate Hsp90 function or expression; (ii) Hsp90 genetic interaction profiles are highly contingent on the environment; and (iii) The network has been substantially rewired over the last 200 million years in response to niche-specific selective pressures. This work has also revealed connections of a multitude of Hsp90 genetic interactors with virulence phenotypes; 52 of the 226 genetic interactors have GO annotations related to traits important for C. albicans virulence, including morphogenesis , biofilm formation, iron metabolism, and drug resistance (Fig. 8.3).
C. albicans Hsp90 genetic interactors relevant for virulence phenotypes. Fifty-two of the 226 genetic interactors have GO annotations for phenotypes important in C. albicans virulence. This subset of interactors is grouped here by phenotype (morphogenesis, biofilm formation, iron metabolism, and drug resistance) and connected by edges to the environmental condition they were identified in. Environmental condition box size is relative to the number of edges connected to this condition. All interactors highlighted here are low-connectivity, except for FCR3, which is connected to four conditions. This emphasizes the high degree of specificity through which Hsp90 fine-tunes complex processes such as biofilm formation or drug resistance
5 Beyond the Current State of the C. albicans Hsp90 Chaperone Network
To date, 10 % of the C. albicans genome have been screened for Hsp90 genetic interactors. Since the first report of the Mitchell Library in 2002 and its extension over the next few years, other labs have created C. albicans mutant libraries and Merck has released its genome-scale mutant libraries. Currently, there are four additional collections available. Two of the libraries were generated by Prof. Alexander Johnson’s laboratory: One library consists of 143 homozygous transcription factor gene deletion mutants [61], and the other consists of ~ 3,000 homozygous gene deletion mutants representing a total of 674 genes [69]. Screens of these libraries have revealed novel insights into C. albicans regulatory circuitries, and identified interesting instances of uncoupling of the important connections between morphology and virulence [61, 69]. Notably, the mutants in the Noble Library are marked with a set of distinct bar codes that enables pooled assays and increases speed of discovery dramatically [69]. Most recently, Merck has made available its gene replacement and conditional expression (GRACETM) [51] and double bar-coded (DBC) [52] heterozygous mutant libraries to the academic community. These resources provide unprecedented opportunity to expand the C. albicans Hsp90 chaperone network.
Despite the creation of five libraries in different strain backgrounds using different strategies and selectable markers, comprehensive screens have the potential to provide important insights and hold a number of advantages. Screening for genetic interactions in different genetic backgrounds could elucidate epistatic regulation of Hsp90 interactions. One gene may interact with Hsp90 only if there are other genes present in a specific background that facilitate interaction. Furthermore, comparing transposon insertions, which only affect a specific region of a gene, with a clean gene deletion may yield insights as to which protein domain or gene region are relevant for the interaction. Additional insight may be leveraged by comparison of homozygous deletion mutants with heterozygotes, and by analysis of essential genes using a gene depletion system.
In addition to developing mutant libraries that could be exploited to study Hsp90 interactions, the C. albicans community has created a suite of tools that facilitate genetic analyses and the detection of protein-protein interactions (PPIs) in this finicky yeast. There are three main challenges associated with conducting molecular biological work in C. albicans. First, C. albicans employs an alternative codon usage. This yeast translates the CUG (cysteine-uracil-guanine) codon into serine instead of leucine [70], which necessitates codon-by-codon alterations of constructs that were previously used successfully in S. cerevisiae. Second, C. albicans is almost exclusively found in the diploid state in nature despite recent advances in the construction of stable haploids that are competent for mating [71]. The lack of a complete meiotic cycle hampers construction of double mutants for SGA and monitoring segregation patterns following sporulation, as can be done with ease in S. cerevisiae. Third, C. albicans does not maintain plasmids, which can render the development of some proteomic tools and the study of essential genes more challenging. Despite these hurdles, significant progress has been made in adapting the yeast two-hybrid assay [72], tandem-affinity purification [73], and the usage of an expanded genetic code for C. albicans [74].
The yeast two-hybrid assay uncovers PPIs via expression of reporter genes. To do so, the protein of interest, such as Hsp90, can be fused to the DNA-binding domain of a transcription factor and any other gene is fused to the activation domain of that transcription factor. Consequently, an interaction between the two test proteins reconstitutes a functional transcription factor that in turn activates expression of a reporter gene. Stynen et al. [72] constructed a yeast two-hybrid system that is compatible for use in C. albicans. This includes selection of a functional DNA-binding domain and activation domain, as well as an effective reporter gene. Following optimization of this system, it was used to verify previously identified PPIs and uncovered novel PPIs in the MAPK pathway regulating filamentous growth [72].
An alternative strategy for identifying PPIs is based on purification of an epitope-tagged version of the protein of interest followed by mass spectrometry of the proteins that co-purify. TAP has been used extensively in S. cerevisiae [75], including for the identification of Hsp90 physical interactions [47]. TAP was optimized for C. albicans with two different epitope tags. First, Kaneko et al. [73] applied this method to study members of the C. albicans septin complex by constructing a tagging vector and 6xHis-FLAG tagged strains. Second, Blackwell and Brown [76] optimized the most widely used TAP tag, which comprises a proximal calmodulin-binding peptide, a cleavage site for the tobacco etch virus protease, and a distal repeated protein A IgG-binding domain. Proteomic approaches for identifying Hsp90 interactors can be challenging due to the transient nature of many Hsp90-client interactions, but remain a promising strategy to expand the chaperone network in C. albicans.
The expanded genetic code is a standard tool for the study of PPIs in model organisms, such as Escherichia coli, S. cerevisiae, and C. elegans. Palzer et al. [74] optimized this approach for use in C. albicans. This required codon optimization of the E. coli orthogonal transfer RNA (tRNA) and tRNA synthetase for the unnatural amino acid p-azido-l-phenylalanine. This amino acid can be photoactivated by ultraviolet (UV) light, which leads to the establishment of covalent bonds between molecules in close vicinity. This system is particularly amenable to investigations of weak or transient interactions in living cells. Covalent bonds are subsequently stabilized by UV cross-linking and interaction partners can be identified using mass spectrometry [74].
With these new tools in place, it should soon be possible to map the Hsp90 physical interaction network and expand the Hsp90 genetic interaction network. Further resolution of the C. albicans Hsp90 chaperone network is particularly relevant in the context of identifying urgently needed regulators of Hsp90 function that are more divergent between pathogen and host than Hsp90, facilitating selective targeting of the pathogen. Furthermore, understanding the broad spectrum of genetic and physical interactors will reveal the mechanisms by which Hsp90 governs fungal morphogenesis , drug resistance, and virulence .
Abbreviations
- Hsp90:
-
Heat shock protein 90
- GdA:
-
Geldanamycin (Hsp90 inhibitor)
- GO:
-
Gene ontology
- cAMP:
-
3’-5’-Cyclic adenosine monophosphate
- PKA:
-
Protein kinase A
- MAPK:
-
Mitogen-activated protein kinase
- 17-AAG:
-
17-N-allylamino-17-demethoxygeldanamycin
- SGA:
-
Synthetic genetic array
- KDAC:
-
Lysine deacetylase
- PPIs:
-
Protein-protein interactions
- TAP:
-
Tandem affinity purification
- CO2:
-
Carbon dioxide
- RPMI:
-
Roswell Park Memorial Institute medium
- UV:
-
ultraviolet
- Sgt1:
-
Suppressor of G2 allele of skp1
- MALDI ToF:
-
Matrix-assisted laser desorption/ionization Time of Flight
- SAGA:
-
Spt-Ada-Gcn5-acetyltransferase
- UTP-C:
-
U three proteins complex
- CUG:
-
Cysteine Uracil Guanine
References
Zaoutis TE, Argon J, Chu J, Berlin JA, Walsh TJ, Feudtner C (2005) The epidemiology and attributable outcomes of candidemia in adults and children hospitalized in the United States: a propensity analysis. Clin Infect Dis 41:1232–1239
Zilberberg MD, Shorr AF, Kollef MH (2008) Secular trends in candidemia-related hospitalization in the United States, 2000–2005. Infect Control Hosp Epidemiol 29:978–980
Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, White TC (2012) Hidden killers: human fungal infections. Sci Transl Med 4:165rv13
Pfaller MA, Diekema DJ (2010) Epidemiology of invasive mycoses in North America. Crit Rev Microbiol 36:1–53
Martin GS, Mannino DM, Eaton S, Moss M (2003) The epidemiology of sepsis in the United States from 1979 through 2000. N Engl J Med 348:1546–1554
Lupetti A, Danesi R, Campa M, Del Tacca M, Kelly S (2002) Molecular basis of resistance to azole antifungals. Trends Mol Med 8:76–81
Turner MS, Drew RH, Perfect JR (2006) Emerging echinocandins for treatment of invasive fungal infections. Expert Opin Emerg Drugs 11:231–250
Cowen LE (2013) The fungal Achilles’ heel: targeting Hsp90 to cripple fungal pathogens. Curr Opin Microbiol 16:377–384
Nathan DF, Vos MH, Lindquist S (1997) In vivo functions of the Saccharomyces cerevisiae Hsp90 chaperone. PNAS 94:12949–12956
Young JC, Moarefi I, Hartl FU (2001) Hsp90: a specialized but essential protein-folding tool. J Cell Biol 154:267–273
Ali MMU, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW et al (2006) Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature 440:1013–1017
Li J, Buchner J (2013) Structure, function and regulation of the Hsp90 machinery. Biomed J 36:106–117
Pearl LH, Prodromou C (2006) Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem 75:271–294
Whitesell L, Lindquist SL (2005) HSP90 and the chaperoning of cancer. Nat Rev Cancer 5:761–772
Sidera K, Patsavoudi E (20134) Hsp90 inhibitors: current development and potential in cancer therapy. Recent Pat Anticancer Drug Discov; 9(1):1–20
Roe SM, Prodromou C, O’Brien R, Ladbury JE, Piper PW, Pearl LH (1999) Structural basis for inhibition of the Hsp90 molecular chaperone by the antitumor antibiotics radicicol and geldanamycin. J Med Chem 42:260–266
Jarosz DF, Lindquist S (2010) Hsp90 and environmental stress transform the adaptive value of natural genetic variation. Science 330:1820–1824
Wong KSK, Houry WA (2006) Hsp90 at the crossroads of genetics and epigenetics. Cell Res 16:742–749
Queitsch C, Sangster TA, Lindquist S (2002) Hsp90 as a capacitor of phenotypic variation. Nature 417:618–624
Rutherford SL, Lindquist S (1998) Hsp90 as a capacitor for morphological evolution. Nature 396:336–342
Cowen LE, Lindquist S (2005) Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science 309:2185–2189
Johnson JL, Brown C (2009) Plasticity of the Hsp90 chaperone machine in divergent eukaryotic organisms. Cell Stress Chaperon 14:83–94
Swoboda RK, Bertram G, Budge S, Gooday GW, Gow NA, Brown AJ (1995) Structure and regulation of the HSP90 gene from the pathogenic fungus Candida albicans. Infect Immun 63:4506–4514
Leach MD, Klipp E, Cowen LE, Brown AJP (2012) Fungal Hsp90: a biological transistor that tunes cellular outputs to thermal inputs. Nat Rev Micro 10:693–704
Bansal PK, Abdulle R, Kitagawa K (2004) Sgt1 associates with Hsp90: an initial step of assembly of the core kinetochore complex. Mol Cell Biol 24:8069–8079.
Dubacq C, Guerois R, Courbeyrette R, Kitagawa K, Mann C (2002) Sgt1p contributes to cyclic AMP pathway activity and physically interacts with the adenylyl cyclase Cyr1p/Cdc35p in budding yeast. Eukaryot Cell 1:568–582
Rocha CR, Schröppel K, Harcus D, Marcil A, Dignard D, Taylor BN et al (2001) Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol Biol Cell 12:3631–3643
Shapiro RS, Robbins N, Cowen LE (2011) Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiol Mol Biol Rev 75:213–267
Shapiro RS, Uppuluri P, Zaas AK, Collins C, Senn H, Perfect JR et al (2009) Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA signaling. Curr Biol 19:621–629
Flom GA, Langner E, Johnson JL (2012) Identification of an Hsp90 mutation that selectively disrupts cAMP/PKA signaling in Saccharomyces cerevisiae. Curr Biol 58:149–163
Shapiro RS, Zaas AK, Betancourt-Quiroz M, Perfect JR, Cowen LE (2012) The Hsp90 co-chaperone Sgt1 governs Candida albicans morphogenesis and drug resistance. PLoS ONE 7:e44734
Cowen LE (2008) The evolution of fungal drug resistance: modulating the trajectory from genotype to phenotype. Nat Rev Micro 6:187–198
White TC (1997) The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14alpha demethylase in Candida albicans. Antimicrob Agents Chemother 1997;41:1488 −14 94
Cowen LE, Singh SD, Köhler JR, Collins C, Zaas AK, Schell WA et al (2009) Harnessing Hsp90 function as a powerful, broadly effective therapeutic strategy for fungal infectious disease. PNAS 106:2818–2823
Robbins N, Uppuluri P, Nett J, Rajendran R, Ramage G, Lopez-Ribot JL et al (2011) Hsp90 governs dispersion and drug resistance of fungal biofilms. PLoS Pathog 7:e1002257
Singh SD, Robbins N, Zaas AK, Schell WA, Perfect JR, Cowen LE (2009) Hsp90 governs echinocandin resistance in the pathogenic yeast Candida albicans via calcineurin. PLoS Pathog 5:e1000532
Singh-Babak SD, Babak T, Diezmann S, Hill JA, Xie JL, Chen Y-L et al (2012) Global analysis of the evolution and mechanism of echinocandin resistance in Candida glabrata. PLoS Pathog 8:e1002718
Cruz MC, Goldstein AL, Blankenship JR, Del Poeta M, Davis D, Cardenas ME et al (2002) Calcineurin is essential for survival during membrane stress in Candida albicans. EMBO J 21:546–559
Imai J, Yahara I (2000) Role of HSP90 in salt stress tolerance via stabilization and regulation of calcineurin. Mol Cell Biol 20:9262–9270
LaFayette SL, Collins C, Zaas AK, Schell WA, Betancourt-Quiroz M, Gunatilaka AAL et al (2010) PKC signaling regulates drug resistance of the fungal pathogen Candida albicans via circuitry comprised of Mkc1, calcineurin, and Hsp90. PLoS Pathog 6:e1001069
Millson SH, Truman AW, King V, Prodromou C, Pearl LH, Piper PW (2005) A two-hybrid screen of the yeast proteome for Hsp90 interactors uncovers a novel Hsp90 chaperone requirement in the activity of a stress-activated mitogen-activated protein kinase, Slt2p (Mpk1p). Eukaryot Cell 4:849–860
Gould CM, Kannan N, Taylor SS, Newton AC (2008) The chaperones Hsp90 and Cdc37 mediate the maturation and stabilization of protein kinase c through a conserved PXXP motif in the c-terminal tail. J Biol Chem 284:4921–4935
Tong AHY, Lesage G, Bader GD, Ding H, Xu H, Xin X et al (2004) Global mapping of the yeast genetic interaction network. Science 303:808–813
Dobzhansky T. (1946) Genetics of natural populations. xiii. Recombination and variability in populations of Drosophila pseudoobscura. Genetics 31:269–290
Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S et al (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391
Tischler J, Lehner B, Fraser AG (2008) Evolutionary plasticity of genetic interaction networks. Nat Genet 40:390–391
Zhao R, Davey M, Hsu Y-C, Kaplanek P, Tong A, Parsons AB et al (2005) Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the Hsp90 chaperone. Cell 120:715–727
McClellan AJ, Xia Y, Deutschbauer AM, Davis RW, Gerstein M, Frydman J (2007) Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell 131:121–135
Franzosa EA, Albanèse V, Frydman J, Xia Y, McClellan AJ (2011) Heterozygous yeast deletion collection screens reveal essential targets of Hsp90. PLoS ONE 6:e28211
Davis DA, Bruno VM, Loza L, Filler SG (2002) Mitchell AP. Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics 162:1573–1581
Roemer T, Jiang B, Davison J, Ketela T, Veillette K, Breton A et al (2003) Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol Microbiol 50:167–181
Xu D, Jiang B, Ketela T, Lemieux S, Veillette K, Martel N et al (2007) Genome-wide fitness test and mechanism-of-action studies of inhibitory compounds in Candida albicans. PLoS Pathog 3:e92
Diezmann S, Michaut M, Shapiro RS, Bader GD, Cowen LE (2012) Mapping the Hsp90 genetic interaction network in Candida albicans reveals environmental contingency and rewired circuitry. PLoS Genet 8:e1002562
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504
Wu P-YJ, Ruhlmann C, Winston F, Schultz P (2004) Molecular architecture of the S. cerevisiae SAGA complex. Mol Cell 15:199–208
Pinna LA (2002) Protein kinase CK2: a challenge to canons. J Cell Sci 115:3873–3878
Perez-Fernandez J, Roman A, Las RDeJ, Bustelo XR, Dosil M (2007) The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol Cell Biol 27:5414–5429
Ota A, Zhang J, Ping P, Han J, Wang Y (2010) Specific regulation of noncanonical p38 activation by Hsp90-Cdc37 chaperone complex in cardiomyocyte. Circ Res 106:1404–1412
Mollapour M, Tsutsumi S, Truman AW, Xu W, Vaughan CK, Beebe K et al (2011) Threonine 22 phosphorylation attenuates Hsp90 interaction with cochaperones and affects its chaperone activity. Mol Cell 41:672–681
Askew C, Sellam A, Epp E, Mallick J, Hogues H, Mullick A et al (2010) The zinc cluster transcription factor Ahr1p directs Mcm1p regulation of Candida albicans adhesion. Mol Microbiol 79:940–953
Homann OR, Dea J, Noble SM, Johnson AD (2009) A phenotypic profile of the Candida albicans regulatory network. PLoS Genet 5:e1000783
Pijnappel WW, Schaft D, Roguev A, Shevchenko A, Tekotte H, Wilm M et al (2001) The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Gene Dev 15:2991–3004
Wang A, Kurdistani SK, Grunstein M (2002) Requirement of Hos2 histone deacetylase for gene activity in yeast. Science 298:1412–1414
Hnisz D, Majer O, Frohner IE, Komnenovic V, Kuchler K (2010) The Set3/Hos2 histone deacetylase complex attenuates cAMP/PKA signaling to regulate morphogenesis and virulence of Candida albicans. PLoS Pathog 6:e1000889
Robbins N, Leach MD, Cowen LE (2012) Lysine deacetylases Hda1 and Rpd3 regulate Hsp90 function thereby governing fungal drug resistance. Cell Reports 2:878–888
Suthram S, Sittler T, Ideker T (2005) The plasmodium protein network diverges from those of other eukaryotes. Nature 438:108–112
Taylor JW, Berbee ML (2006) Dating divergences in the fungal tree of life: review and new analyses. Mycologia 98:838–849
Odds FC. (1988) Candida and Candidosis. Elsevier Science Health Science Division, 468 p.
Noble SM, French S, Kohn LA, Chen V, Johnson AD (2010) Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet 42:590–598
Santos MA, Tuite MF (1995) The CUG codon is decoded in vivo as serine and not leucine in Candida albicans. Nucleic Acids Res 23:1481–1486
Hickman MA, Zeng G, Forche A, Hirakawa MP, Abbey D, Harrison BD et al (2013) The “obligate diploid” Candida albicans forms mating-competent haploids. Nature 494:55–59
Stynen B, Van Dijck P, Tournu HA (2010) CUG codon adapted two-hybrid system for the pathogenic fungus Candida albicans. Nucleic Acids Res 38:e184–e184
Kaneko A, Umeyama T, Hanaoka N, Monk BC, Uehara Y, Niimi M (2004) Tandem affinity purification of the Candida albicans septin protein complex. Yeast 21:1025–1033
Palzer S, Bantel Y, Kazenwadel F, Berg M, Rupp S, Sohn K (2013) An expanded genetic code in Candida albicans to study protein-protein interactions in vivo. Eukaryot Cell 12:816–827.
Gavin A-C, Bösche M, Krause R, Grandi P, Marzioch M, Bauer A et al (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141–147
Blackwell C, Brown JD (2009) The application of tandem-affinity purification to Candida albicans. Methods Mol Biol 499:133–148
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Science+Business Media New York
About this chapter
Cite this chapter
Diezmann, S., Cowen, L. (2014). The Candida albicans Hsp90 Chaperone Network Is Environmentally Flexible and Evolutionarily Divergent. In: Houry, W. (eds) The Molecular Chaperones Interaction Networks in Protein Folding and Degradation. Interactomics and Systems Biology, vol 1. Springer, New York, NY. https://doi.org/10.1007/978-1-4939-1130-1_8
Download citation
DOI: https://doi.org/10.1007/978-1-4939-1130-1_8
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4939-1129-5
Online ISBN: 978-1-4939-1130-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)