Abstract
Yearly epidemics and occasional pandemic of influenza are responsible for a substantial global disease burden related to both the acute effects of infection and the high rates of complications particularly in individuals at the extremes of age and with chronic medical conditions. This chapter provides a review of the basic biology of influenza, the epidemiologic patterns of disease and estimates of disease burden, and the pathogenesis and immune response. Potential methods of control and prevention, including vaccines and antiviral agents, are also reviewed.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
More than 80 years after the recognition of influenza virus as the cause of the syndrome of influenza, or grippe, influenza continues to be a major cause of acute respiratory illnesses. These illnesses rival acute gastroenteritis as causes of morbidity and mortality throughout the world. Although the majority of cases of influenza are not identified by laboratory methods, a variety of direct and indirect methods have attributed a substantial proportion of the overall burden of acute respiratory illness directly to influenza. This burden includes both deaths and hospitalizations, which occur most commonly at the extremes of age and in persons with other underlying heart or lung diseases and in pregnancy, as well as an enormous burden of transiently disabling illness in all ages that result in substantial economic and productivity losses.
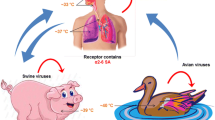
Infection with influenza virus stimulates a coordinated response of the innate, and cellular and humoral adaptive, immune response that leads to effective and long-lived resistance to reinfection with the same strain of virus. However, influenza viruses uniquely subvert this immune response through rapid evolution of the viral surface glycoproteins resulting in antigenic changes that allow infection in the presence of immunity to prior strains. Relatively minor antigenic changes, traditionally referred to as “antigenic drift,” typically result in new viruses that cause the familiar seasonal epidemics of acute influenza. In addition, influenza A viruses occasionally and unpredictably undergo major changes in antigenicity referred to as “antigenic shift” that lead to worldwide, more severe epidemics, or pandemics of influenza. In many cases, it appears that pandemics viruses emerge from the enormous gene pool of influenza A viruses in migratory waterfowl. Surveillance for these viruses in birds, and their transmission to domestic poultry, swine, other mammals, and ultimately man, has therefore become an object of intense interest in recent years.
Two specific forms of influenza control approaches have been developed in the 80 years since the discovery of these viruses, vaccines, and antiviral agents. In theory, developing effective vaccines for influenza should be straightforward. However, the same antigenic variation that circumvents natural immunity also has confounded efforts to develop effective influenza vaccines, and currently available live or inactivated vaccines must be reformulated and readministered annually to keep pace with these antigenic changes. Despite intense effort over many decades, a truly universal influenza vaccine has never been successfully developed, although recent progress in influenza immunology has increased optimism along these lines. Nevertheless, the current vaccines do provide a substantial measure of protection and are important tools for reducing the overall disease burden of influenza.
The same high rate of evolution of influenza viruses has also complicated efforts to develop antiviral agents. Two viral proteins, the influenza A virus M2 protein and the neuraminidase (NA) protein of influenza A and B viruses, are the targets of the current classes of antiviral agents, the M2-inhibitors (amantadine and rimantadine) and NA-inhibitors (zanamivir and oseltamivir), respectively. Both classes of drug have been shown to reduce the duration of illness and viral shedding in acutely infected individuals and to reduce the rate of hospitalization and death when used relatively early in the course of illness. However, the rapid development of antiviral resistance has significantly impacted the utility of these agents. At the moment, essentially all current influenza A viruses are completely resistant to the M2 inhibitors, and there has been frequent development of resistance to the NAI oseltamivir, particularly in the N1 neuraminidases. The use of multiple antiviral agents with synergistic activities may represent a strategy to reduce the emergence of resistance.
In this chapter, we will briefly review the history of influenza throughout. We will then describe the classification and the basic virology of these agents and consider the data used to estimate their yearly impact on human health. We will review the basics of the immune response to influenza and the characteristics of illness induced by these viruses and then discuss the current approaches to prevention and control.
2 Historical Background
The characteristics of influenza epidemics, such as the high attack rates, explosive spread of disease, and characteristic cough and fever, have allowed identification of past influenza epidemic throughout history. Older studies identified probable influenza epidemics occurring at an average interval of 2.4 years between 1173 and 1875 [1]. The greatest pandemic in recorded history occurred in 1918–1919 when, during three “waves” of influenza, 21 million deaths were recorded worldwide, among them 549,000 in the United States [2].
William Farr introduced the concept of “excess mortality” in his vivid description of the London epidemic of 1847 [3]. Frost [4] first used this concept in the United States to describe the 1918 influenza epidemic, but it was Selwyn Collins who systematically used excess mortality as an index for recognition of influenza epidemics [5, 6]. Collins estimated baseline mortality by calculating weekly arithmetic means, and Serfling refined the baseline estimate by deriving a regressin function to describe seasonal variation in baseline mortality [7], the basic methodology which is still in use today for assessing excess mortality.
The first influenza virus was isolated from chickens with fowl plague in 1901, but it was not recognized that this was an influenza A virus until 1955. Shope isolated the swine influenza virus in 1931 [8, 9]. Influenza A virus was first transmitted from humans to ferrets and recognized as the cause of influenza in 1933 [10]. Influenza B virus was isolated by Francis in 1939 [11] and influenza C virus by Taylor in 1950 [12]. The discovery by Burnet in 1936 that influenza virus could be grown in embryonated hens’ eggs allowed extensive study of the properties of the virus and the development of inactivated vaccines [13]. Animal cell culture systems for the growth of influenza viruses were developed in the 1950s [14]. The phenomenon of hemagglutination, which was discovered by Hirst in 1941, led to simple and inexpensive methods for the measurement of virus and specific antibody [15].
Evidence of the protective efficacy of inactivated vaccines was developed in the 1940s [16]. The use of live vaccines for influenza was first suggested shortly after the virus was discovered [17], but the first live vaccine was not licensed in the United States until 2003, approximately 70 years later. Finally, four antiviral agents in two classes have been approved for prevention and treatment of influenza. These include the so-called M2 inhibitors, amantadine in the mid-1960s, rimantadine in 1993, and the neuraminidase inhibitors zanamivir and oseltamivir in 2000.
3 Biological Characteristics
Influenza viruses are members of the Orthomyxoviridae family of viruses and are enveloped, single-stranded, negative-sense RNA viruses. The viruses are further divided into types A, B, and C based on substantial differences in proteins and genomic structure. Types A and B influenza are causes of seasonal epidemics of acute respiratory disease in children, adults, and elders. Type C influenza is primarily a minor cause of mild respiratory illness in children.
Type A influenza viruses are further subdivided into subtypes based on antigenic and sequence differences in the two major surface glycoproteins: the hemagglutinin (HA) and neuraminidase (NA). To date, a total of 16 distinct HA subtypes have been identified, designated H1 to H16, and nine distinct NA subtypes, N1 to N9. Recently, a new, unique influenza virus was detected in yellow-shouldered bats in Guatemala [18]. Preliminary characterization of this virus suggests that its HA and NA should be designated H17 and N10, although crystal structure suggests that the N10 neuraminidase does not actually have neuraminidase activity [19].
While influenza A viruses of the H1N1, H2N2, and H3N2 subtypes have caused substantial disease in humans, the natural hosts of influenza A viruses are probably migratory waterfowl, in which a genetically diverse ecology of all known HA and NA subtypes have been found. These avian species thus are thought to act as a reservoir of genetic diversity of influenza A viruses and as the source of emerging pandemic influenza viruses (discussed below). Avian influenza viruses are frequently transmitted from waterfowl to domestic poultry, where they can cause widespread epidemics of severe or fatal disease. Transmission of influenza A viruses also commonly occurs to pigs, horses, mink and ferrets, and marine mammals, in which the viruses can become established and maintained for years. Influenza in each of these populations is typically limited to certain subtypes, with H1 and H3 viruses in pigs, H7 and H3 viruses in horses, and H7 in marine mammals. Recently, influenza A has also been described in felines and dogs.
Influenza B viruses do not exhibit the same type of antigenic and genetic variation in the HA and NA, and therefore do not have subtypes. However, since 2001, two antigenically lineages of influenza B viruses, termed the “Victoria” lineage and the “Yamagata” lineage, have cocirculated in humans [20]. In contrast to influenza A viruses, influenza B viruses appear to be limited to humans, although isolation of influenza B from seals has been described [21]. Possibly for this reason, influenza B viruses have not been responsible for pandemics of influenza.
An important feature of influenza viruses is that the genome is segmented. Each gene segment is responsible for the synthesis of one or more viral proteins. Both influenza A and B viruses contain eight gene segments, although the specific proteins assigned to each gene segment differ between these two types. The consequences of this are that when a cell is infected with two different influenza viruses, the resulting progeny virus can in theory contain any combination of gene segments from the two parent viruses, meaning that 256 possible combinations of genes can be derived from reassortment event between two influenza viruses. However, some combinations may not be compatible [22], and all combinations are probably not equally likely. Genetic reassortment has been demonstrated to play an important role in the generation of pandemic influenza A viruses and has also been taken advantage of for the construction of attenuated live influenza vaccines.
Influenza viruses enter the cell by attachment of the viral hemagglutinin to sialic-acid-containing receptors on the cell membrane, followed by internalization of the virus into an acidic endosome. In the acidic environment of the endosome, the HA undergoes a conformational change that liberates a fusion peptide and results in fusion of the viral envelope with the endosomal membrane. At the same time, the third envelope protein, the M2 protein, acts as an ion channel allowing H + ions to enter the virion from the endosome. This in turn allows the viral gene segments to leave the virion and enter the cytoplasm, a process known as uncoating.
Viral gene segments are transported to the nucleus, where the viral polymerase complex, comprised of the proteins PB1, PB2, and PA, directs the synthesis of the plus sense messenger RNA as well as synthesis of negative-sense copies that will serve as progeny genomic RNA. The polymerase proteins also may play a role in disruption of host cell protein synthesis. Because replication takes place in the nucleus, some mRNA are spliced, giving rise in the case of influenza A viruses to the M2 protein from the M gene segment, and the NS2, or NEP (nuclear export protein) from the NS gene segment. Alternative start codons also give rise to a number of additional proteins from the polymerase genes of influenza A viruses including PB1-F2 [23] and PA-X [24], which may play roles in pathogenesis.
Negative-sense daughter virion RNAs are encased in nucleoprotein, associated with one copy of the polymerase complex and transported to the cytoplasm for assembly at the cell surface. Envelope proteins are glycosylated and transported to the cell surface. Virions bud from selected lipid rafts at the cell surface, acquiring an envelope derived from the cell membrane and decorated with HA, NA, and small amounts of M2 protein. Finally, the NA removes sialic acid from receptors on the cell surface or on the viral envelope, allowing the progeny viruses to leave the infected cell.
4 Descriptive Epidemiology
The epidemiology of influenza is characterized by seasonal epidemics of acute respiratory disease, punctuated, in the case of influenza A, at random intervals by worldwide epidemics of varying levels of severity, referred to as pandemics. Both of these events are felt to be driven by antigenic variation. In the case of seasonal epidemics of influenza, it is felt that population immunity drives the selection of mutations in the immunologically critical HA and NA proteins that result in sufficient antigenic differences from previously circulating viruses to allow the selected antigenic variant to efficiently replicate and infect individuals who have developed immunity to previous viruses of the same subtype. Because these antigenic changes are relatively minor and are generally the result of a few mutations in critical antibody epitopes, this process is frequently referred to as antigenic drift.
In contrast, radical changes in the HA and NA result in the emergence of viruses that are able to spread rapidly in populations with little or no effective immunity, resulting in pandemics with high attack rates and, usually, high levels of morbidity and mortality (Fig. 19.1).
Schematic diagram of the subtype circulation of influenza A viruses in man. The pandemic of 1918 was caused by a virus of the 1918 subtype, which may have been introduced from an unknown animal reservoir. Pandemics of 1957 and 1968 were reassortment events between previous human and avian viruses (see text). In 1977 and 2009, novel H1N1 viruses which were antigenically related to previous H1N1 viruses (dotted lines) were introduced into the population
Classically, pandemics involve the replacement of influenza A viruses of one subtype with influenza A viruses of a different subtype. For example, the most severe pandemic of influenza in recently recorded history was the so-called Spanish flu pandemic of 1918, due to a virus of the H1N1 subtype (the pandemic occurred before influenza virus was discovered and before the recognition of subtypes but was classified as H1N1 retrospectively). Subsequent seasonal epidemics of influenza were due to H1N1 viruses with various degrees of antigenic change, most remarkably in 1947 when a pseudopandemic was experienced due to an H1N1 virus (A/Fort Monmouth/47) with enough antigenic change from previous viruses that it was initially characterized as a new influenza A virus, so-called A’ influenza [25]. H1N1 viruses continued to cause seasonal epidemics in man until 1957, when an influenza virus with an entirely new H and N subtype, A/Japan/57 (H2N2), emerged to cause a second pandemic of the twentieth century, the Asian flu. Initially, these viruses were referred to as strain A2, until the modern typing system was developed and they were categorized as H2N2 viruses. For reasons that are unclear, H1N1 viruses ceased to circulate in man after the emergence of H2N2 viruses.
H2N2 viruses then underwent antigenic drift resulting in seasonal influenza until 1968 when these viruses were replaced by the new subtype H3N2 viruses or the so-called Hong Kong flu. This virus represented a slightly different circumstance in that while the HA was a new subtype, the NA was retained from the previous H2N2 virus. Several studies have suggested that residual immunity to the retained N2 component of the H3N2 viruses substantially ameliorated the severity of this pandemic worldwide [26]. Again, for unclear reasons, the emergence of H3N2 viruses coincided with the disappearance of the previous H2N2 viruses, which have not circulated in man since then. Studies of the serologic reactivity of banked sera (so-called seroarcheology) have suggested that an H3 virus, possibly of the H3N8 subtype, may have also caused a pandemic in 1889–1891 [27].
H1N1 viruses reemerged in 1977 and resulted in a relatively more mild pandemic referred to as the Russian flu (A/USSR/77). This virus was genetically and antigenically identical to influenza A viruses that had circulated in man in 1950, and the mechanism that led to the preservation of this virus over the subsequent 27 years has never been fully explained. As expected, disease was largely restricted to younger persons born after 1957 who had not been previously exposed to H1N1 viruses, and the overall impact of this pandemic was much less severe than previous pandemics. In addition, the emergence of the H1N1 viruses did not result in the disappearance of the previous H3N2 virus. These H1N1 viruses cocirculated with the H3N2 viruses until 2009, when a new variant of H1N1 virus emerged from swine and replaced the previous H1N1, but not H3N2 viruses.
4.1 Emergence of Pandemic Viruses from Birds
Extensive surveillance studies have identified influenza A viruses of all 16 HA subtypes and all 9 NA subtypes in migratory waterfowl. In these birds influenza A causes mild illness or may be shed asymptomatically at high levels and for long duration in the feces. These birds may transmit influenza to other animals, including domestic poultry, horses, swine, and marine mammals, which may in turn transmit these viruses to man. Comparisons of sequence data from animal and human influenza viruses isolates have suggested that the 1918 virus was introduced into humans from such an animal population. In contrast, the 1957 and 1968 pandemic influenza viruses were reassortant viruses that derived some genes from previously circulating human viruses, while deriving the HA and sometimes NA genes from an avian influenza virus [28].
Because of the likely role of avian influenza viruses in the generation of emerging pandemics, there has been intense interest in recent outbreaks involving transmission of avian influenza viruses to man, with resulting disease. Most of these transmission events have been quite limited, with small numbers of persons affected, relatively mild disease, and little or no evidence of person-to-person transmission. In most cases, virus has been transmitted to humans from infected domestic poultry, but cases have also occurred in association with marine mammals and possibly wild birds.
Subtype H7 viruses have been responsible for several small outbreaks. Human infections with H7 AI viruses have generally been sporadic and mild in nature, with infected individuals presenting with mild flu-like illness and/or conjunctivitis [29, 30]. Human cases have typically been associated with outbreaks in birds, although one case of human H7 infection was reported in a laboratory worker who was sneezed upon by a seal that was infected with an H7N7 influenza virus [31]. The largest known cluster of H7N7 infections of humans occurred in 2003 in association with an outbreak of highly pathogenic avian influenza in commercial poultry farms in the Netherlands [32, 33]. While almost all cases in this outbreak were mild or subclinical, there was one confirmed fatal case in a 57-year-old otherwise healthy veterinarian, who developed pneumonia [33].
A new outbreak of influenza illness due to viruses of the H7N9 subtype has been recognized in western China since the spring of 2013 [34]. In contrast to previous H7 disease which has been predominantly mild respiratory illness with conjunctivitis, cases in this outbreak have been more severe, with hospitalizations and an approximately 20% case fatality rate [35]. Cases have been mostly recognized in older adults, for unknown reasons, and fatalities have largely occurred in individuals with underlying heart of lung disease , somewhat similar to seasonal influenza [36]. Almost all cases have direct contact with poultry, mostly in live bird markets. Because H7N9 viruses are not highly lethal in poultry, outbreaks in markets are much harder to recognize and control, which may be contributing to the persistence of this outbreak in affected areas.
Subtype H9 viruses have rarely caused human disease. H9N2 virus was isolated from two children in Hong Kong with mild febrile pharyngitis in 1999 [37]. Retrospective serologic cohort studies of individuals exposed to these two H9N2-infected children did not suggest person-to-person transmission [38]. Subsequently, H9N2 infection has been detected from five individuals with typical influenza in China [39] and from a child with a relatively severe influenza in Hong Kong.
The greatest concern has been for H5N1 viruses, which were first recognized in humans in 1997 [40] and which have continued to cause substantial numbers of human cases since that time. From 2003 to October 1, 2012, a total of 608 laboratory-confirmed human cases of H5N1 infection had been reported to the WHO, of which 359 cases were fatal. Cases have ranged in age from 3 months to 75 years with the median age being 20 years. Half of all cases have been in people aged less than 20 years and 90 % of cases have been in those less than 40 years of age. The median duration from onset of illness to hospitalization has been 4 days (range of 0–18 days). The case fatality rates have been the highest for those in the 10–19-year age group, lowest for people 50 or older, and in between for children aged <10 years [41].
Most cases have had close contact with ill poultry in the week before the onset of illness. Activities like plucking and preparing diseased birds, playing with birds, especially asymptomatically infected ducks, and handling fighting cocks are risk factors for infection [42]. Other apparent modes of acquisition have included eating undercooked poultry or drinking raw duck blood or exposure to contaminated water [43]. However, instances of person-to-person transmission have been rare. Fifteen family clusters of infection involving ≥2 family members were documented between January 2004 and July of 2005 [44], with the largest cluster identified thus far involving seven confirmed cases in family members of a woman who died of an acute respiratory illness [45]. In addition, there is one well-documented transmission of virus from an ill child in Thailand to her mother [46].
4.2 Emergence of Pandemic Viruses from Swine
Domestic swine have also been recognized as a potential source of pandemic influenza viruses in man. Genomic data have suggested that influenza A (H1N1) viruses were introduced into swine populations at around the same time that H1N1 viruses emerged in man in 1918 [47]. Since that time, these H1N1 viruses, or classic swine viruses, continued to be maintained in domestic swine where they caused minor illnesses and underwent relatively little antigenic evolution. During this time, swine were also occasionally infected with influenza A viruses from humans and from birds and have always been considered to represent a potential “mixing vessel” in which reassortment between human and avian influenza viruses could occur. This concept was strengthened by the recognition that the swine respiratory tract contains abundant receptors of both the α2-3 and α2-6 types favored by avian and human viruses, respectively [48].
In the late 1990s, one such reassortment event has been recognized leading to a unique virus containing polymerase genes of both swine and avian influenza virus origin. This unique combination of genes, referred to as the triple reassortant cassette, apparently increased the frequency with which these viruses underwent reassortment with other variants in swine populations. These viruses were also occasionally transmitted to humans, typically in the context of state agricultural fairs. While most of the resulting disease was relatively mild, occasional severe disease and deaths were reported, primarily in pregnant women. However, person-to-person transmission was not observed.
This situation changed in early 2009, when cases of swine-origin influenza A H1N1 viruses were first recognized in the United States and rapidly spread throughout the world [49]. The virus responsible for this pandemic was determined to be a quadruple reassortant virus derived from the triple reassortant virus by the addition of M and NA genes from a swine virus of Eurasian lineage (Fig. 19.2). The exact circumstances that led to this event remain mysterious, but it appears that the M1 protein derived from the Eurasian M gene conferred on these viruses an enhanced ability to transmit from person to person [50].
The age distribution of cases in the resulting pandemic displayed the relative sparing of older adults that has been observed in previous pandemics and might be explained by the exposure of older adults to antigenically similar viruses in their childhood [51]. Thus, the bulk of the disease occurred in adolescents and young adults. As a result, the estimated number of excess deaths due to the pandemic in 2009 was estimated to be only 12,000 in the United States, which, while substantial, was considerably less than often experienced during seasonal influenza, especially in years predominated by H3N2 viruses. However, as many of these deaths occurred in young people, the impact on years of life lost was much greater and overall more representative of the impact of the pandemic.
Since the emergence of the pH1N1 viruses, influenza surveillance activities have increased their focus on domestic swine, and a new potential pandemic threat in the form of quadruple reassortant viruses with the same internal genes as the pH1N1 but containing variant H3 HA genes has been recognized [52]. Antigenically, these viruses resemble human H3 viruses that circulated in the early 1970s [53]. Approximately 300 cases of human disease have been identified, almost all as the result of transmission from swine to humans during agricultural fairs. As expected, almost all of these cases have occurred in children. There has been little evidence of person-to-person spread, but there remains a need for continued vigilance regarding this possibility.
4.3 Seasonal Influenza
Influenza epidemics during the interpandemic period generally display a marked seasonal periodicity in regions with temperate climates, with the majority of disease activity occurring between November and April in the Northern Hemisphere and between May and September in the Southern Hemisphere (Fig. 19.3).
Seasonal periodicity is also observed in tropical climates, with increased activity during periods of low absolute humidity, although influenza can occur throughout the year and seasonal fluctuations are not as marked [54]. The reasons for these seasonal changes are not entirely clear but might be the result of more favorable environmental conditions for virus survival [55]. Studies in a model of transmission of influenza in guinea pigs have also supported a role for conditions of cold temperature and dry humidity in facilitating transmission [56]. Colder temperatures are also associated with behavioral changes that may increase transmission, such as indoor crowding or school attendance.
4.3.1 Disease Impact
Influenza epidemics are regularly associated with excess morbidity and mortality, usually expressed in the form of excess rates of pneumonia- and influenza-associated (P&I) hospitalizations and deaths during epidemics. In order to estimate the disease burden of influenza, observed P&I events during periods of influenza epidemic activity are compared with an expected seasonal baseline derived from a time-series regression model, and the excess event rate attributable to influenza is calculated. Because not all influenza-related deaths are manifested as pneumonia, P&I mortality statistics may underestimate the true impact of influenza on the population [57]. Although less precise, seasonal excess all-cause mortality is probably a more accurate reflection of the total burden of influenza.
From 1979 to 1991, these methodologies have led to estimated rates of influenza-associated hospitalizations ranging from 55,000 to 431,000 annually, with an overall average of 226,000 hospitalizations attributable to influenza [58]. Estimates of influenza-associated deaths have increased in recent decades, possibly because of the increasing numbers of older, at-risk members of the population. Recent estimates suggest an average of approximately 8,000 excess P&I-associated deaths annually, but much higher numbers of all-cause mortality associated with influenza, with an average of 36,000 deaths annually with a range as high as 51,000 deaths in the United States [59]. Generally, the level of excess mortality is highest in years when influenza A (H3N2) viruses predominate [60] and lower in years with predominant H1N1 activity, possibly reflecting age-related susceptibility to these two subtypes.
Excess morbidity and mortality are particularly high in those with certain high-risk medical conditions, including adults and children with cardiovascular and pulmonary conditions such as asthma, or those requiring regular medical care because of chronic metabolic disease, renal dysfunction, hemoglobinopathies, or immunodeficiency, and in individuals with neurologic conditions that compromise handling respiratory secretions [61]. Influenza also results in more severe disease and significant mortality in individuals with human immunodeficiency virus (HIV) infection [62, 63].
The increased risk of influenza during pregnancy was dramatically demonstrated during the 2009 pandemic [64]. Previous studies had identified an increased risk of hospitalization associated with influenza epidemics during pregnancy, especially in the second and third trimester and in the immediate postpartum period [65]. Several groups at increased risk for severe influenza-related disease and deaths were recognized during the 2009 H1N1 pandemic. Women in each stage of pregnancy, or in the immediate post partum period, were clearly over represented among those admitted to hospitals and ICUs. This observation was perhaps not surprising as pregnancy has long been recognized as a risk factor for influenza mortality during previous pandemics and to a lesser extent during seasonal influenza as well. The mechanism(s) by which pregnancy enhances the risk of influenza is not clear but might include the increased cardiovascular demands of pregnancy as well as hormonally mediated changes to the innate and adaptive immune response.
Obesity also emerged as a risk factor for influenza morbidity and mortality that had not been recognized in previous seasonal epidemics or pandemics. While compromise to respiratory mechanics as a direct result of extreme obesity undoubtedly plays a role, there is also evidence to support a detrimental role of adipose tissue in the inflammatory response that might also enhance the influenza disease process. As a result of these observations, obesity is not recognized as an important factor for targeting influenza vaccine.
Influenza is usually associated with a U-shaped epidemic curve (Fig. 19.4). Attack rates are generally highest in the young, whereas mortality is generally highest among older adults [66, 67], in part because the prevalence of high-risk conditions is greater in this group. Influenza is also recognized as an important health problem in young children. Rates of influenza-related hospitalizations are particularly high in healthy children under 2 years of age, where rates approach those of older children with high-risk conditions [68–70]. In addition, a high rate of secondary complications, particularly otitis media and pneumonia, occurs in children with influenza infection [71]. Outpatient clinic visit rates for laboratory-documented influenza have been observed at 50–95 and emergency room visit rates of 6–27 per thousand person years in children under five [72]. While rare, influenza-related deaths occur each year in previously healthy children [61]. Notably, many of these deaths occur in children who were not recognized to have high-risk conditions prior to their illness.
Age-related annual incidence of acute illness visits, hospitalizations, and deaths during the interpandemic era in Houston, Texas (Data from Couch et al. [66])
Much of the impact of influenza is related to the malaise and consequent disability that it produces, even in young, healthy individuals. It has been estimated that a typical case of influenza, on average, is associated with 5–6 days of restricted activity, 3–4 days of bed disability, and about 3 days lost from work or school [73]. The average number of medical visits for cases in which medical attention was sought was from 1.1 to 3.6 per year, depending on severity of the outbreak and age of the patient. In children, outpatient visits are 10–250 times more common than hospitalizations [72]. It is worth noting that direct medical costs of illness account for only about 20 % of the total expenses of a case of influenza, with a major proportion (30–50 %) of the economic impact due to loss of productivity [74]. In one study, influenza in schoolchildren resulted in 37 missed school days by children and 20 days of missed work by parents, per 100 children [75]. Influenza is also associated with decreased job performance in working adults [76] and reduced levels of independent functioning in older adults [77]. Data from the Tecumseh Community Health Study have been used to estimate that influenza is responsible for 13.8–16.0 million excess respiratory illnesses per year in the United States among individuals less than 20 years of age and for 4.1–4.5 million excess illnesses in older individuals [78].
5 Mechanisms and Routes of Transmission
Influenza viruses are transmitted from person to person via the respiratory route. Three potential modes of transmission have been suggested [79]. Coughing and sneezing could generate small particle aerosols (<10 um mass diameter) which can remain suspended in air for many hours and could transmit infection to individuals at a substantial difference. Larger particles or droplets will typically fall to the ground within 3 m of the infected person and would be expected to infect individuals in direct contact. Finally, viral particles could land on surfaces, where influenza viruses remain infectious [80, 81] and could infect others through indirect contact. There is substantial evidence for all three modes of transmission in experimental studies and epidemiologic observations, but the relative roles of each mode of transmission are uncertain and remain controversial, with obvious implications for infection control practices and for potential interventions to mitigate pandemics.
Small particle aerosols are generated by infected humans, and influenza genome can be detected in these small particles by polymerase chain reaction techniques [82, 83]. It has not been proven that these aerosols contain significant amounts of infectious virus, but experimental studies in humans have shown that vary small amounts (~5 infectious particles) may be sufficient to infect humans by the aerosol route [84, 85]. Aerosol transmission has also been demonstrated in animal models in which infected and exposed ferrets or guinea pigs are separated by several meters, with transmission occurring in the direction of airflow [56, 86].
Airborne transmission has also been implicated in multiple observations of outbreaks where an airborne route of transmission appears to be the most plausible explanation for the characteristics of the outbreak. The most often cited such outbreak occurred in a commercial airliner that was delayed for approximately 4½ h with a poorly functioning ventilation system. The risk of transmission of influenza A from the index case to other passengers was related to the amount of time passengers spent on the aircraft and not on their seating proximity to the index case. Since most of the passengers did not have direct contact with the index case, airborne transmission appears to be likely [87]. In a well-investigated hospital outbreak, nosocomial cases occurred significantly less frequently in a hospital ward where the air was treated with UV light than in an otherwise similar ward without UV light treatment [88]. In an outbreak in a long-term care facility, there appeared to be an association between the risk of nosocomial influenza and the air handling systems in several wards [89].
Additional observations consistent with airborne transmission were made during an early study of zanamivir prophylaxis of influenza in families. In this study [90], subjects who received short-term prophylaxis with inhaled zanamivir were protected compared to placebo recipients, but recipients of zanamivir administered by nasal spray were not. In addition, the combination of nasal and inhaled zanamivir was no better than inhaled zanamivir alone.
In most of these outbreaks, there are alternative explanations for the observations that could at least partially explain the epidemic behavior without requiring aerosol transmission [91], and the real role of aerosol transmission remains controversial. If aerosol transmission plays a dominant role in influenza, then health-care workers would need to wear filtering face masks, and patients would require negative pressure isolation, to prevent nosocomial transmission of influenza. This has prompted several studies that have attempted to evaluate the role of facemasks in infection prevention in hospitals. In one large, randomized trial, nursing staff who were randomly assigned to wear N-95 respirators had the same rate of influenza as staff assigned to wear simple surgical masks while caring for patients with influenza [92]. This study suggests that airborne transmission does not play a major role at least in nosocomial influenza, although it has been pointed out that cases in the N-95 group could have been acquired outside the hospital and that compliance with these masks is frequently poor. In contrast, hand hygiene and simple surgical masks were reported to be modestly effective in the prevention of influenza transmission in households [93] suggesting that in this setting, droplet spread was the predominant modality.
6 Pathogenesis and Immunity
6.1 Pathogenesis
Once virus is deposited on the respiratory tract epithelium, it can attach to and penetrate columnar epithelial cells if not prevented from doing so by specific secretory antibody (IgA), by nonspecific mucoproteins to which virus may attach, or by the mechanical action of the mucociliary apparatus. After adsorption, virus replication begins, leading to cell death by inhibiting cellular protein synthesis and disrupting other cellular functions and by inducing apoptosis. Virus release continues for several hours before cell death ensues. Released virus then may initiate infection in adjacent and nearby cells, so within a few replication cycles, a large number of cells in the respiratory tract are releasing virus and dying as a result of the virus replication. The time between the incubation period and the onset of illness and virus shedding varies from 18 to 72 h depending in part on the inoculum dose [94].
Quantitation of virus in respiratory tract specimens from otherwise healthy young adults reveals a characteristic pattern (Fig. 19.5). Virus is first detected just before the onset of illness (within 24 h), rapidly rises to a peak of 3.0–7.0 log10 TCID50/mL, remains elevated for 24–48 h, and then rapidly decreases to low titers [95]. Usually, virus is no longer detectable after 5–10 days of virus shedding. However, because of the relative lack of immunity in the young, more prolonged shedding of higher titers of virus is expected in children.
Time course of influenza in healthy adults who were experimentally infected with influenza A/Texas/91. Symptoms are temporally correlated with the peak of virus shedding and production of a variety of cytokines (Data adapted from Hayden et al. [95])
fections of healthy adults, the severity of illness correlates well with the quantities of virus shed, suggesting that a major mechanism in the production of illness is cell death resulting from viral replication. Although the clinical manifestations of influenza are dominated by systemic symptoms, viral replication is limited to the respiratory tract. Instead, systemic symptoms are probably due to the release of potent cytokines, such as type I interferons, tumor necrosis factor, and interleukins (ILs), by infected cells and responding lymphocytes [95]. In fact it has been suggested that an overly vigorous cytokine response to infection may contribute to the high fatality rate seen with H5N1 influenza [96, 97].
Bronchoscopy of individuals with typical, uncomplicated acute influenza has revealed diffuse inflammation of the larynx, trachea, and bronchi, with mucosal injection and edema [98, 99]. Biopsy in these cases has revealed a range of histologic findings, from vacuolization of columnar cells with cell loss to extensive desquamation of the ciliated columnar epithelium down to the basal layer of cells [99]. Individual cells show shrinkage, pyknotic nuclei, and a loss of cilia. Viral antigen can be demonstrated in epithelial cells [100]. Generally, the tissue response becomes more prominent as one moves distally in the airway [99]. Epithelial damage is accompanied by cellular infiltrates primarily composed of lymphocytes and histiocytes. Histologic findings on autopsy in more severe cases show extensive necrotizing tracheobronchitis, with ulceration and sloughing of the bronchial mucosa [101], extensive hemorrhage, hyaline membrane formation, and a paucity of PMN infiltration. Patients with secondary bacterial pneumonia have the changes characteristic of bacterial pneumonia in addition to the tracheobronchial findings of influenza. Recovery is associated with rapid regeneration of the epithelial cell layer and with pseudometaplasia.
Abnormalities of pulmonary function are frequently demonstrated in otherwise healthy, nonasthmatic young adults with uncomplicated (nonpneumonic) acute influenza. Demonstrated defects include diminished forced flow rates, increased total pulmonary resistance, and decreased density-dependent forced flow rates consistent with generalized increased resistance in airways less than 2 mm in diameter [102, 103], as well as increased responses to bronchoprovocation [102]. In addition, abnormalities of carbon monoxide diffusing capacity [104] and increases in the alveolar-arterial oxygen gradient [105] have been seen. Of note, pulmonary function defects can persist for weeks after clinical recovery. Influenza in asthmatics [106] or in patients with chronic obstructive disease [107] may result in acute declines in forced expiratory vital capacity (FVC) or forced expiratory volume in 1 s (FEV1). Individuals with acute influenza may be more susceptible to bronchoconstriction from air pollutants such as nitrates [108].
Primary viral pneumonia is an uncommon but frequently severe complication of acute influenza. In this situation, virus infection reaches the lung either by contiguous spread from the upper respiratory tract or by inhalation. The trachea and bronchi contain bloody fluid, and the mucosa is hyperemic [109]. Tracheitis, bronchitis, and bronchiolitis are seen, with loss of normal ciliated epithelial cells. Submucosal hyperemia, focal hemorrhage, edema, and cellular infiltrate are present. The alveolar spaces contain varying numbers of neutrophils and mononuclear cells admixed with fibrin and edema fluid. The alveolar capillaries may be markedly hyperemic with intra-alveolar hemorrhage. Acellular hyaline membranes line many of the alveolar ducts and alveoli [109]. Pathologic findings seen by biopsy of lung in nonfatal cases are similar to those described in fatal cases [110].
Bacterial superinfection is a well-recognized complication of influenza that may account for a substantial proportion of morbidity and mortality, especially in adults. For example, the frequency of pneumococcal hospitalizations has been shown to increase in association with influenza epidemics [111]. Consequently, the spectrum of disease and pathophysiology of bacterial superinfection has been studied intensively, and a number of factors have been identified in viral respiratory disease that could play a role in increasing the risk of bacterial infection. Uncomplicated influenza is associated with significant abnormalities in ciliary clearance mechanisms [112]. In addition, increased adherence of bacteria to virus-infected epithelial cells has been demonstrated [113]. The disruption of the normal epithelial cell barrier to infection and loss of mucociliary clearance undoubtedly enhance bacterial pathogenesis. In addition, influenza infection may upregulate certain cell surface receptors involved in bacterial adherence [114]. Alterations in PMNs and mononuclear cells may also contribute to enhanced bacterial infection.
6.2 Immunity
Epidemiologic and experimental observations in humans have shown that infection with influenza virus results in long-lived resistance to reinfection with the homologous virus. In addition, variable degrees of cross-protection within a subtype have been observed [115]. Infection induces both systemic and local antibody, as well as cellular immune responses, each of which plays a role in recovery from infection and resistance to reinfection (Fig. 19.6) [116].
Immune response to influenza infection and reinfection (Adapted from Subbarao et al. [116])
6.2.1 Systemic Antibody Responses
Infection with influenza virus results in the development of antibody to the influenza virus envelope glycoproteins HA and NA, as well as to the structural M and NP proteins. Some individuals may develop antibody to the M2 protein as well [117]. The antibody response is more rapid after reinfection. Peak antibody responses after primary infection are seen at approximately 4–7 weeks and decline slowly thereafter. Antibody titers can sometimes be detected years after exposure, for example, persons born before 1968 frequently have detectable titers of antibody against H2 viruses. The mechanisms that allow such persistent antibody are not known.
As implied by the name, the HA protein of influenza is defined by the ability to hemagglutinate red blood cells, a function which is directly related to binding to cellular receptors. Thus, antibody that can block hemagglutination, or so-called hemagglutination-inhibiting antibody (HAI), has been studied intensively and is generally accepted as a surrogate for virus neutralization and protection against infection. Serum antibody to the HA has been demonstrated to have a protective role against influenza infection and disease in both animal models as well as in experimental infection of humans and in epidemiologic observations. An increased risk of laboratory-documented influenza among those with the lowest titers of preexposure HAI or neutralizing antibody is a consistent finding of most but not all studies. However, there is considerable uncertainty about the actual level of HA antibody that is the best predictor of protection, with estimates ranging from HAI titers of 1:8 to 1:160 or higher [118]. Given the substantial variation from laboratory to laboratory in the estimation of the HAI titer on the same set of samples [119], the inability to use an absolute value for protection is not unexpected. In addition, the amounts of antibody needed to mediate protection could vary by population, degree of exposure, age, and specific influenza type or subtype, although this has not been analyzed comprehensively.
B cells secreting HA-specific antibody that binds to the stem region rather than the head of the HA have been detected in the blood of individuals experiencing infections with novel influenza viruses such as pH1N1 or H5N1 [120, 121]. These antibodies exhibit neutralizing activity in some assays but do not inhibit hemagglutination. Stalk-binding antibody can be detected in serum as well [122]. Because the stalk region is well conserved, these antibodies can be cross-neutralizing among HA subtypes and have increased interest in the potential creation of a universal influenza vaccine.
In contrast to anti-HA antibody, anti-NA antibody does not neutralize virus infectivity but instead reduces efficient release of virus from infected cells, resulting in decreased plaque size in in vitro assays [123] and in reductions in the magnitude of virus shedding in infected animals [124, 125]. Observations on the relative protection of those with anti-N2 antibody during the A/Hong Kong/68 (H3N2) pandemic [26, 126], as well as experimental challenge studies in humans [127], have shown that anti-NA antibody can also be protective against disease and results in decreased virus shedding and severity of illness but that it is infection permissive [128].
Antibody to other influenza viral proteins has also been evaluated for potential protection. Antibody to M2 reduces plaque size in vitro, and passive transfer studies in mice have also suggested that antibody to the M2 protein of influenza A viruses may be partially protective if present in large enough amounts [129]. The mechanism of protection in vivo is related to mediation of antibody-dependent cytotoxicity [130]. Antibody to internal viral proteins such as M or NP is also cross-reactive among type A viruses, but they are non-neutralizing. Studies in mice have suggested that such non-neutralizing but cross-reactive antibody may mediate protection under some circumstances [131]. The mechanism by which antibody to viral proteins that are not exposed on the surface can mediate protection is unclear.
6.2.2 Mucosal Antibody Responses
Both natural viral infection and live attenuated vaccines have been found to induce significant mucosal antibody responses. Nasal HA-specific IgG is predominantly IgG1, and its levels correlate well with serum levels of HA-specific IgG1, suggesting that nasal IgG originates by passive diffusion from the systemic compartment [132]. Nasal HA-specific IgA is predominantly polymeric and IgA1, suggesting local synthesis. Studies in mice and ferrets have emphasized the importance of local IgA antibody in resistance to infection, particularly in protection of the upper respiratory tract. Studies in humans have also suggested that the resistance to reinfection induced by virus infection is mediated predominantly by local HA-specific IgA, whereas that induced by parenteral immunization with inactivated virus depends also on systemic IgG [133]. Importantly, either mucosal or systemic antibody alone can be protective if present in high enough concentrations, and optimal protection occurs when both serum and nasal antibodies are present [134, 135].
6.2.3 Cellular Immune Responses
The induction of cellular immune response to influenza virus infection has been studied intensively in murine models, and such studies suggest that B cell, CD4+ T cell, and CD8+ T cell responses all can play a role in protection against disease and recovery from infection. A large number of HLA class I-restricted (CD8+ T cell) and HLA class II-restricted (CD4+ T cell) epitopes have been described, and in situations where those epitopes are on relatively well-conserved influenza proteins such as the polymerase, NP, and M proteins, the cellular responses are cross-reactive between subtypes, but not between types A and B.
Cellular immune responses to influenza vaccination and infection have not been studied as extensively in humans, but B cell (memory B cell and antibody-secreting cell), CD4+ T cell, and CD8+ T cell responses in peripheral blood have been described after infection or vaccination [136]. It can be difficult to capture the peak of the response and detectable increases in antigen-specific cells may only be seen on a few days after exposure. Generally, the peak cellular response occurs somewhere between 5 and 14 days depending on the status of the subject and the nature of the response. As seen in murine models, a major component of the cellular response is directed at conserved peptides to which the subject has already been exposed during previous infections or vaccinations.
Antibody-secreting cells (ASC) appear in blood and tonsils as early as 2 days after vaccination and are detected in the blood of adults and older children more frequently than in young children after immunization [137]. An increase in cytotoxic T lymphocytes, directed primarily at the conserved internal proteins, has been shown in healthy adults with a peak at 14 and 21 days after vaccination and return to baseline at 6 months. An increase in HA-specific CD8+ T cells on day 7 after vaccination has also been detected by tetramer staining in adults receiving inactivated influenza vaccine [138].
An important role of the cellular immune response in recovery from influenza infection in humans is strongly supported by the observation of prolonged illness and viral shedding in individuals who are lymphopenic as a result of disease or chemotherapy. However, it has been difficult to develop specific markers of T cell immunity as correlates of protective immunity. Activated T cells, in the form of granzyme B-positive T cells, have been associated with protection in older subjects [139]. In the human challenge model, early studies identified the early induction of virus-specific cytotoxic T cells as measured by cytochrome release assays as correlated with reductions in the duration and level of virus replication in adults [140]. In a subsequent study done many years later by the same group, prechallenge CD4+ T cells, but not CD8+ T cells, correlated with relatively lower levels of viral shedding and symptoms following experimental infection [141]. In a large study of the efficacy of live attenuated vaccine in children, it was shown that higher levels of influenza-specific T cells assayed by gamma-interferon ELISPOT was associated with a decreased risk of PCR-documented influenza [142]. The development of more sophisticated markers that can specifically identify reactive cells in peripheral blood will help to define the role of cellular immunity in protection, but the field remains limited by the lack of convenient access to compartments other than peripheral blood in humans.
7 Patterns of Host Response
Typical uncomplicated influenza often begins with an abrupt onset of symptoms after an incubation period of 1–2 days. Many patients can pinpoint the hour of onset. Initially, systemic symptoms predominate, including feverishness, chilliness or frank shaking chills, headaches, myalgia, malaise, and anorexia. Usually, myalgia or headache is the most troublesome symptom, and the severity is related to the height of the fever. Respiratory symptoms, particularly cough and sore throat, are usually also present at the onset of illness. The predominance of systemic symptoms is a major feature distinguishing influenza from other viral upper respiratory infections. Older adults may simply present with high fever, lassitude, and confusion without the characteristic respiratory complaints, which may not occur at all. Generally there is a wide range of symptomatology in healthy adults, ranging from classic influenza to mild illness or asymptomatic infection.
Two manifestations of pneumonia associated with influenza are well recognized: primary influenza viral pneumonia and secondary bacterial infection. The syndrome of primary influenza viral pneumonia was first well documented in the 1957–1958 pandemic, predominantly among persons with cardiovascular disease, especially rheumatic heart disease with mitral stenosis, and to a lesser extent in others with chronic cardiovascular and pulmonary disorders [109]. The illness begins with a typical onset of influenza, followed by a rapid progression of fever, cough, dyspnea, and cyanosis. Secondary bacterial pneumonia classically presents after a brief period of improvement [143, 144], although most patients do not fit this classic pattern. Bacteria frequently associated with influenza include Streptococcus pneumoniae or Haemophilus influenzae and, notably, an increased frequency of Staphylococcus aureus. An increased frequency of community-acquired, methicillin-resistant S. aureus has been seen in children and adults following influenza outbreaks [145].
In addition to pneumonia, other pulmonary complications of influenza include croup [146] and exacerbations of chronic bronchitis or asthma [147, 148]. Recognized non-pulmonary complications include myositis and myoglobinuria [149], myocarditis and pericarditis [150, 151], toxic shock syndrome [152, 153], Guillain-Barré syndrome and transverse myelitis [154], and Reye’s syndrome particularly in children who have been given aspirin to treat fever.
8 Control and Prevention
8.1 Antiviral Drugs
Two classes of antiviral drugs have been used clinically to treat and prevent influenza. The adamantanes amantadine and rimantadine are related primary symmetrical amines whose mechanism of action involves inhibition of the M2 ion channel activity of susceptible viruses. The function of the M2 ion channel in viral replication is to acidify the interior of the virion, disrupting the interaction between the matrix and nucleoproteins and allowing the ribonucleoproteins to be transported to the nucleus, where replication occurs [155]. Similar ion channels have been described for influenza B and C viruses; however, at clinically achievable levels, these drugs are active against only influenza A.
Amantadine and rimantadine are effective in the therapy of both experimentally induced and naturally occurring influenza A, with more rapid decrease in fever, more rapid improvement in symptoms, and decreased shedding of virus [156, 157]. Rimantadine has also been shown to be effective in the treatment of influenza A in children [158].
Drug resistance has been a factor in limiting the use of these antiviral agents. Resistant viruses emerge frequently in treated individuals, particularly children, in whom subpopulations of resistant virus can be detected following treatment in virtually all cases [159]. Resistance is the result of single point mutations in the membrane-spanning region of the M2 protein, and it confers complete cross-resistance between amantadine and rimantadine [160]. Resistant virus can be transmitted to, and can cause disease in, susceptible contacts. Prolonged shedding of resistant viruses may occur in immunocompromised patients, particularly children, and may continue even after therapy is terminated [161], consistent with the relative fitness of these resistant viruses. While previously rare, a rapid increase in the prevalence of de novo resistance to M2 inhibitors was noted in 2005, and essentially all H3N2 viruses are now resistant to these agents [162, 163]. Although previously circulating seasonal H1N1 viruses remained sensitive to these agents, the emerging pH1N1 viruses are also completely resistant to the M2 inhibitors, which now lack activity against all strains of influenza virus currently circulating.
The neuraminidase inhibitors act by inhibiting the functioning of the influenza virus neuraminidase, which is critical in allowing newly formed viruses to egress from the cell and spread to other cells [164]. The two licensed inhibitors, zanamivir and oseltamivir, have shown very similar results in clinical trials. Both drugs reduce the duration of symptoms and enhance the return to normal activities when used within the first 36 h of symptoms in otherwise uncomplicated influenza in healthy adults [165–168]. Both drugs have also been shown to be effective in reducing the duration and severity of influenza in children [169, 170].
Antiviral resistance to these agents has also been detected both in treated and untreated individuals. Mutations within the catalytic framework of the NA that abolish binding of the drugs have been described [171, 172]. The specific mutations conferring resistance are dependent on the specific NA, that is, common resistance mutations in N1 (e.g., H274Y) are different than the ones seen in the N2 (e.g., R292K or E119V) or influenza B (e.g., D198N). In addition, depending on the location of the mutation, these viruses may be specifically resistant to only one inhibitor [173]. Resistance mutations in the NA may be associated with altered characteristics of the enzyme with significantly reduced activity [174, 175].
Some resistant viruses appear to have reduced fitness, with reduced levels of replication, attenuation in animals, and reduced ability to be transmitted from animal to animal [176–179]. Drug resistant viruses were also isolated very infrequently from oseltamivir-treated individuals in clinical trials, being seen in less than 2 % of treated adults and detected in 5.6 % of children [169]. However, subsequent studies have demonstrated that resistant viruses can be detected in up to 18 % of treated children when using sensitive PCR techniques to pick up minor subpopulations [180].
Beginning in 2006, spontaneously resistant H1N1 viruses carrying the H274Y mutation began to be detected in viruses from individuals who did not have a history of exposure to oseltamivir [181]. By 2008, all H1N1 viruses isolated in the United States were resistant to oseltamivir. The mechanism that led to the selection of seasonal H1N1 clades resistant to oseltamivir as the predominant circulating H1N1 viruses is unclear. However, the emerging pH1N1 virus has remained largely susceptible to oseltamivir.
8.2 Vaccines
Two types of influenza vaccines are currently licensed and are used in various countries, inactivated influenza vaccines (IIV) and live attenuated influenza vaccines (LAIV). Shortly after these vaccines were introduced, it was recognized that their efficacy might depend on the antigenic match between the strain(s) contained within the vaccine and the circulating viruses [182], and since that time, the vaccines have been continuously reformatted to keep pace with the ongoing evolution of influenza viruses. Since 1977, influenza vaccines have contained a representative A/H3N2, A/H1N1, and B virus, so-called trivalent influenza vaccine. As mentioned above, two antigenically distinct lineages of influenza B viruses have cocirculated in humans since 2004, and quadrivalent formulations of vaccine are currently being considered.
8.2.1 Safety
IIV are chemically inactivated and have been administered either as the so-called “whole-virus” preparations or as detergent-disrupted and partially purified “split product” or “subunit” vaccines. Hundreds of millions of doses of IIV are administered each year, and the safety of these vaccines has been repeatedly confirmed. For example, no increase in clinically important medically attended events has been noted among over 251,000 children <18 years of age who were enrolled in one of the five health maintenance organizations within the Vaccine Safety Datalink, the largest published post-licensure population-based study of vaccine safety [183]. The most common adverse events reported following immunization with IIV are tenderness and/or pain at the injection site. Most injection site reactions are mild and rarely interfere with daily activities. Systemic reactions following immunization of adults with inactivated vaccine are uncommon. In placebo-controlled clinical trials in younger and elderly adults, rates of systemic reactions were similar among groups given inactivated vaccine or placebo [184, 185]. However, whole-virus IIV are associated with fever in children [186] and are not recommended in this age group.
Recently, an increased frequency of fever and febrile seizures was observed among young children given one specific IIV during the 2010 influenza season in Australia [187]. The reasons for this unexpected reactogenicity are unclear, but preliminary studies have suggested that this vaccine preparation stimulated unusually high cytokine responses in in vitro assays. In addition, concomitant immunization of young children with IIV and pneumococcal conjugate vaccine (PCV) was shown to be associated with an increased risk of developing febrile seizures.
Immediate hypersensitivity reactions (hives, wheezing, angioedema, or anaphylactic shock) following inactivated vaccine can also occur, and vaccine is considered contraindicated for persons who experienced a previous anaphylactic reaction following vaccine. Clinical protocols have been proposed to administer inactivated vaccine to persons who are at high risk for severe or complicated influenza who also have a history of immediate hypersensitivity to eggs, if the benefit of immunization is judged to outweigh the risk [188].
Several unusual syndromes have been associated with IIV. The Guillain-Barré syndrome (GBS), an acute inflammatory demyelinating polyneuropathy, was associated with the 1976 swine influenza vaccination campaign, with an increased risk of approximately 1 per 100,000 vaccinees. More recent studies have suggested a statistically significant but very slight increased relative risk of GBS within 7 weeks of influenza vaccination [189]. The oculorespiratory syndrome (ORS) is a syndrome of red eyes, facial edema, and/or respiratory symptoms such as coughing, wheezing, sore throat, hoarseness, difficulty breathing, or chest tightness that develop within 2–24 h after vaccination, associated with a specific influenza vaccine used in Canada, but not elsewhere [190]. The specific mechanism underlying this phenomenon is unknown.
Although not studied as extensively, LAIV also appear to be quite well tolerated. Nasal symptoms (runny nose, nasal congestion, or coryza) and sore throat have been the most frequently identified adverse symptoms following LAIV. Children under 8 have had slightly increased rates of low-grade fever, runny nose, and abdominal symptoms in the 7 days following vaccination compared to placebo recipients. However, when considering all the pediatric studies in aggregate, no consistent symptom was significantly more common in LAIV compared to placebo recipients. In older children, 11 to <16 years of age, sore throat was observed slightly more frequently in LAIV recipients than IIV recipients.
In larger studies, wheezing has been associated with LAIV in young children, although occurring at low rates. In the largest trial, medically significant wheezing within 42 days of vaccination was reported in 3.8 % of children <2 years old after receipt of LAIV compared to 2.1 % in those who received IIV [191]. Wheezing generally occurs in the youngest, previously unvaccinated children following the first dose of vaccine. Because of this observation, LAIV is currently approved for use in the United States for children ≥2 years old who do not have a history of asthma.
LAIV can be recovered from nasal secretions of about half of adult recipients, although generally shedding of LAIV by adults is of low titer and short duration [192]. Although young children shed much higher levels of vaccine virus, no transmission of LAIV from vaccine recipients to susceptible contacts was detected in studies of young children involved in day-care-like settings where LAIV and placebo recipients played together for up to 8 h a day for 7–10 days after vaccination. In the largest study, 197 children between 8 and 36 months of age were randomized to receive LAIV or placebo, and vaccine virus shedding was assessed for 21 days after vaccination. Although 80 % of LAIV recipients shed at least one vaccine strain, for a mean of 7.6 days, clear evidence of transmission was detected in only one placebo recipient [193].
8.2.2 Efficacy and Effectiveness
The ability of influenza vaccines to prevent influenza has been assessed in numerous clinical studies which vary greatly in design, populations, and endpoints. These studies have included prospective, randomized controlled studies, in which case they are referred to as efficacy studies, as well as a wide variety of nonrandomized cohort and retrospective study designs which assess vaccine effectiveness. Endpoints evaluated in these studies have included both laboratory-confirmed influenza and non-laboratory-confirmed respiratory illnesses. In this regard, it has been recognized that studies that utilize a serologic definition of influenza infection may overestimate the efficacy of influenza vaccine, since it will be harder to demonstrate postvaccination to post-season antibody increases in the vaccinated group [194].
Randomized studies of IIV efficacy against laboratory-confirmed influenza have mostly been conducted in healthy adults. These studies have shown a wide range of efficacy, from approximately 40–80 %, with lower levels of efficacy typically seen in years with apparent antigenic mismatch. A recent meta-analysis of 8 randomized, controlled trials in healthy adults during 2004–2008 estimated the pooled efficacy of IIV against culture-confirmed influenza to be 59 % (95 % CI 51–67) among those aged 18 through 64 years [195]. The role of antigenic mismatch in the efficacy observed in these trials is unclear, and some studies in young adults have demonstrated high levels of efficacy (76 %) despite a degree of antigenic mismatch. Recent studies using virus culture and/or PCR endpoints have demonstrated similar levels of efficacy for both egg-grown and cell culture-grown IIVs [196].
Relatively few placebo-controlled trials of the efficacy of LAIV have been conducted in adults. In the human challenge model, cold-adapted and inactivated influenza vaccines were of approximately equal efficacy in the prevention of experimentally induced influenza A (H1N1), A (H3N2), and B. The combined efficacy in preventing laboratory-documented influenza illness due to the three wild-type influenza strains was 85 % for LAIV [197]. In a randomized, controlled study in healthy persons aged 1 through 64 years, of whom most of the participants were adults, the efficacy of a pre-licensure, bivalent preparation of LAIV for preventing culture-confirmed influenza A illness in adults was 85 % (95 % CI 70–92 %) for H1N1 and 58 % (95 % CI 29–75 %) for H3N2 [198]. LAIV was also evaluated in a large study against clinical endpoints performed in 4,561 healthy working adults [199]. In this study, the effectiveness of LAIV-T in preventing severe febrile respiratory illness of any cause during the influenza season was 29 %.
Relatively few recent prospective trials have assessed IIV efficacy in children. In one randomized, controlled trial in healthy children aged 6 through 23 months, vaccine efficacy was 66 % (95 % CI 34–82) in the first year, but efficacy could not be assessed in the second year due to a very low influenza attack rate (efficacy −7 %: 95 % CI −247–67) [200]. Immunization of asthmatic children has also been shown to reduce the incidence of influenza. More recently, the efficacy of IIV against PCR-confirmed influenza was assessed in a randomized, placebo-controlled trial in healthy children between the ages of 6 and 72 months [201]. The efficacy of IIV against all influenza strains was 43 % compared with the placebo group.
LAIV was demonstrated to be efficacious in the prevention of influenza in a pivotal 2-year, randomized placebo-controlled trial conducted in 1,314 children aged 15 to <72 months [202]. The efficacy against culture-confirmed influenza illness in the first year of this trial was 95 % (95 % C.I. 88–97 %) for influenza A/H3N2 and 91 % (95 % C.I. 79–96 %) for influenza B. In the second year of the trial, the H3 component of the vaccine (A/Wuhan/93) was not a close match with the predominant H3 virus that season, A/Sydney/95. However, the efficacy of LAIV against this variant was 86 % (95 % C.I. 75–92 %) [203]. Overall, the efficacy of LAIV to prevent any influenza illness during the 2-year period of surveillance in this field study was 92 % (95 % C.I. 88–94 %). The overall efficacy of LAIV against culture-confirmed influenza among children 6 to <36 months who were attending day care was recently shown to be 85 and 89 % in the first and second year of the study, respectively [204]. Significant protection against flu-associated acute otitis media also was demonstrated (>90 % in both years). Studies done in Asia have reached similar conclusions, with an efficacy of LAIV compared to placebo of between 64 and 84 % over multiple seasons, depending on the antigenic match with the vaccine [205].
Although annual vaccination of elders and other high-risk persons has been recommended for many years, there are very few randomized trials demonstrating efficacy in these groups, in part because the existing vaccine recommendations make it difficult to do studies using a placebo group. In the most commonly referenced study, TIV was 52 % (95 % CI 29–67) efficacious in preventing serologically documented influenza illness in a population of adults 60 years of age and older [184]. When the groups were further stratified by age, efficacy estimates against serologically documented influenza illness were 57 % (95 % CI 33–72 %) in those 60 through 69 years old but only 23 % (95 % CI −51–61 %) in those ≥70 years old.
In a recently reported randomized, double-blind, placebo-controlled clinical trial of LAIV among community-dwelling ambulatory adults ≥65 years old, the overall efficacy of LAIV against viruses that were antigenically similar to the vaccine was 42 % [206], similar to the protection seen with inactivated vaccine. However, LAIV is not currently licensed for use in individuals over 49 years of age.
Monitoring influenza vaccine efficacy on an annual basis by conducting randomized placebo-controlled studies would clearly be a very difficult undertaking and is probably not possible in children, elders, and other high-risk groups. Therefore, a number of observational study designs have been used for this purpose. Many recent studies have utilized a test-negative, case–control design, in which individuals meeting a particular case definition are tested for influenza using a highly sensitive and specific diagnostic test, and the vaccination exposure of test-positive cases and test-negative controls is determined [207]. Large surveillance networks for this purpose have been established in Canada, the United States, Europe, and Australia for purposes of making interim and end-of-season estimates of vaccine effectiveness.
Studies using this design have shown variable results with estimates generally ranging from as low as 20 %, or in some cases, no effectiveness, to as high as 70 % [195]. While the various networks vary in their study design and the specific selection criteria for subject inclusion, a few overall generalizations can be stated. Failure to detect VE has typically occurred in studies with very low prevalence of influenza in the study population, or in years with substantial antigenic mismatch between the vaccine and circulating strains, most often involving influenza B lineage mismatch. The relationship of antigenic mismatch with vaccine effectiveness for influenza A/H1N1 and A/H3N2 viruses is not as consistent, but even in situations of antigenically matched viruses, VE remains in the 50–60 % range [208]. In some cases, viruses recovered from subjects in studies with low vaccine effectiveness have been shown to have substantial changes on a HA sequence level despite appearing well matched by traditional HAI tests [209].
Most studies have not enrolled enough subjects in a single season to make age-specific estimates of VE. However, there is a trend towards decreased VE in elderly, not surprising given their diminished immune response to vaccination. After accumulating cases over several seasons, it was recently possible to use the same test-negative case–control design to demonstrate VE of approximately 60 % against influenza-related hospitalization in a population of community-dwelling older adults [210].
While the use of a study design in which testing is performed without knowledge of vaccination status may eliminate some biases related to health-care access and health-seeking behavior, the results are influenced by the accuracy of the diagnostic testing, since errors in assignment to the case or control group will bias VE towards nil. Recently, in a study done in children, it was demonstrated that using the test-negative case–control approach, estimates of VE were substantially higher when children with documented infections with viruses other than influenza were used as a control group, rather than using all children who were test negative for influenza [211].
A larger body of data exists from nonrandomized or observational studies of vaccine effectiveness. These studies have suggested that influenza vaccination can reduce pneumonia and influenza (P&I) hospitalizations and death among the elderly regardless of whether they have other conditions that place them at high risk for complications following influenza [212]. While post-licensure observational studies are important tools for monitoring vaccine effectiveness, such studies relating to the elderly are particularly challenging to perform and interpret. Frailty selection bias (a higher baseline risk of hospitalization and death among unvaccinated vs. vaccinated subjects) and nonspecific endpoints may overestimate vaccine effectiveness in cohort studies [213].
Infants less than 6 months of age are at substantial risk for influenza-related morbidity but are too young to receive influenza vaccine. One strategy to protect vulnerable infants is maternal immunization, with protection mediated by both transfer of maternal antibody and reduced potential for contact with an influenza-infected mother. In a randomized study of maternal immunization, infants born to mothers immunized with influenza vaccine had substantially lower rates of laboratory-documented influenza in the first 6 months of life than did infants born to mothers immunized with pneumococcal vaccine [214]. Similarly, in a retrospective case–control study, the frequency of influenza immunization was substantially lower in the mothers of infants hospitalized with PCR-confirmed influenza than in mothers infants hospitalized who were PCR negative, with an estimated protective effect of 92 % [215].
There has been considerable interest in potential strategies to protect such individuals indirectly by preventing illness and viral transmission within highly susceptible populations that probably play a role on community transmission, such as schoolchildren. Several studies have suggested that this may be possible. During the 1968 pandemic, it was observed that the incidence of respiratory illness during the period of influenza A circulation was among unvaccinated adults was substantially lower in a community in which schoolchildren were immunized than in a comparison community with no school immunization. Influenza B was not contained in the vaccine, and during a subsequent influenza B epidemic, there was no difference in adjusted respiratory illness rates in adults in the two communities [216]. In a recent study, closed agricultural communities of Hutterites were randomized to vaccination of schoolchildren with influenza vaccine or with hepatitis A vaccine as a control. In the subsequent influenza A epidemic, the rate of laboratory-documented influenza A in unvaccinated adults residing in school-vaccinated communities was reduced by 61 % (95 % CI 8–83 %) compared to adults in unvaccinated communities [217]. Observations in Japan, where it appeared that substantial fluctuations in overall influenza-related mortality (occurring mostly in the elderly) were directly related to the rate of school-aged influenza vaccination, also support a potential role for school vaccination in protection of elders [218].
8.2.3 Adjuvants
For many years, licensed inactivated influenza vaccines have been administered without adjuvants. However, it is clear that the immune response to vaccination and the protective effects of vaccine are suboptimal, particularly in some groups at high risk for influenza-related complications including young children and elders. Thus, there has been significant interest in the potential use of adjuvants to enhance the protective effects of vaccination. Although influenza vaccines adsorb well to aluminum salts, these compounds have not been effective adjuvants for influenza, for unknown reasons. Early studies suggested that water in oil emulsions using mineral oil could very substantially improve antibody responses in military recruits allowing the use of lower doses [219]. However, the use of these adjuvants was restricted by substantial local side effects including the development of sterile abscesses at the site of administration; [220] the use of peanut oil appeared to reduce the observed toxicity [221].
Subsequently, oil-in-water emulsions based on the metabolizable oil squalene have been shown to substantially improve immune responses with an excellent safety profile. While there is relatively little effect on the immune response to seasonal vaccines in healthy adults [222], the oil-in-water emulsion MF59 results in an approximately 50 % increase in antibody titers in older adults [223], and MF59 adjuvanted seasonal IIV has been licensed for use in elders in Italy for several years. Recently, a large randomized trial in young children compared MF59-adjuvanted IIV with unadjuvanted IIV over two seasons. Absolute efficacy against all influenza strains was 86 % in the group given IIV-MF59 and 43 % in the group given unadjuvanted IIVs when compared with the placebo group [201].
Oil-in-water emulsions have demonstrated especially striking improvements in the antibody responses to candidate H5 pandemic vaccines. These studies have shown higher titers of antibody against the vaccine virus, as well as against antigenic variants, the development of B cells that recognize a larger variety of HA epitopes, and broadened and more vigorous CD4 T cell responses [224, 225].
8.2.4 Comparisons of Inactivated and Live Influenza Vaccines
While relatively few randomized direct comparisons of the efficacy of live and inactivated vaccines have been performed, the available studies are consistent with the observed effects of age and prior influenza experience on immunogenicity. When these vaccines have been compared in young children 12 months through 59 months of age, LAIV has shown consistently superior protection, with an approximately 50 % greater protective efficacy than inactivated vaccine [191, 226]. Studies conducted in healthy young children have generally concluded that LAIV may be more efficacious than IIV, including both against viruses which are well matched antigenically to the vaccine virus and those which are antigenically drifted [191].
In contrast to young children, studies that have directly compared the vaccines in adults have suggested that the vaccines have similar efficacy or that IIV vaccine is slightly more efficacious than live vaccine. In one 3-armed study, the efficacy of LAIV compared to placebo for prevention of laboratory-confirmed influenza in healthy adults was 57 %, while the efficacy of the IIV was 77 %, but the difference between the two vaccines was not statistically significant [227]. In a subsequent season in the same population, the absolute efficacies of IIV and LAIV were 68 and 36 %, respectively [228]. Similar results have been reported from a retrospective study evaluating the effectiveness of IIV and LAIV against medical visits for pneumonia- and influenza-related diagnoses in the US military [229]. Generally, rates of such visits were lower in recipients of IIV, except for personnel who had not been vaccinated in previous years, in which there was not apparent difference in the effectiveness of the two types of vaccines. In a recent effectiveness study that included both LAIV and IIV recipients, there was no clear-cut difference in effectiveness between the two vaccines [208].
9 Unresolved Problems
Despite many years of research and vast energies devoted to uncovering its mysteries, influenza remains an enigma and effective strategies for its control are elusive. Influenza vaccines, including unadjuvanted inactivated vaccines of various formulations and live attenuated vaccines based on cold-adapted master donor viruses, have clearly been demonstrated to reduce the risk of influenza in selected populations. However, these vaccines have never been shown to have any substantial impact on the overall disease burden of influenza. In part, this is a problem of inconsistent supply and limited uptake, compounded by the need for annual reformulation and administration, but in part this also reflects our growing recognition of the limited effectiveness of these vaccines and an incomplete understanding of the nature of protective immunity.
Development of more effective vaccines, perhaps to include broadly cross-protective vaccines, is an obvious goal. The specific types of immune responses that could contribute to more effective vaccination are unclear, and the best strategies for generating these responses remain to be determined. Many exciting new discoveries have recently been revealed, and multiple innovative approaches for new vaccines and adjuvant systems are underway. Each of these pathways will need to carefully balance safety and efficacy in the context of the existing very safe but modestly effective options.
Antiviral agents are another strategy to mitigate the impact of seasonal and pandemic influenza, but their real utility in this disease has also been quite limited. The rapid emergence of resistance is a major obstacle facing effective use of antiviral strategies, but the basic pathogenesis of influenza creates an extremely narrow time window for antiviral intervention in most cases, further complicating antiviral therapy. Novel approaches that focus on modifying the host or the host innate immune response may offer one alternative that might overcome some of these problems.
Although it should be recognized that the cumulative disease burden of seasonal influenza is generally larger than that of pandemic influenza, the threat of a severe influenza pandemic represents another unresolved issue in the field. Extensive efforts have been devoted to comprehensive surveillance for novel influenza viruses and potential pandemic threats in a wide variety of domestic and wild animal populations, and our understanding of the ecology of influenza is expanding rapidly. However, our ability to actually predict the emergence of any specific pandemic remains extremely limited.
References
Hirsch A. Handbook of geographical and historical pathology. 2nd ed. London: New Syndenham Society; 1883.
Crosby AW. Epidemic and peace, 1918. Part IV. Westport: Greenwood Press; 1976.
Farr W. Tenth annual report of the registrar general. London: HMSO; 1847.
Frost WH. The epidemiology of influenza. Public Health Rep. 1919;34:1823–6.
Collins SD, Lehmann J. Trends and epidemics of influenza and pneumonia, 1918–1951. Public Health Rep. 1951;66:1407–505.
Collins SD. Influenza in the United States. Government Printing Office; 1957.
Serfling RE. Methods for current statistical analysis of excess pneumonia-influenza deaths. Public Health Rep. 1963;78:494–506.
Shope RE. Swine influenza I. Experimental transmission and pathology. J Exp Med. 1931;54:349–60.
Shope RE. Swine influenza III. Filtration experiments and etiology. J Exp Med. 1931;54:373–85.
Smith W, Andrewes CH, Laidlaw PP. A virus obtained from influenza patients. Lancet. 1933;2:66–8.
Francis Jr T. A new type of virus from epidemic influenza. Science. 1940;92:405–8.
Taylor RM. A further note on 1233 (“influenza C”) virus. Arch Gesamte Virusforsch. 1951;4:485–95.
Burnet FM. Influenza virus on the developing egg I. changes associated with the development of an egg-passage strain of virus. Br J Exp Pathol. 1936;17:282–95.
Mogabgab WJ, Green IJ, Dierkhising OC. Primary isolation and propagation of influenza virus in cultures of human embryonic renal tissue. Science. 1954;120:320–1.
Hirst GK. The agglutination of red cells by allantoic fluid of chick embryos infected with influenza virus. Science. 1941;94:22–3.
Francis Jr T, Salk JE, Pearson HE, Brown PN. Protective effect of vaccination against influenza A. Proc Soc Exp Biol Med. 1944;55:104–5.
Smorodintseff AA, Tushinsky KMD, Drobyshevskaya AI, Korovin AA, Osetroff AI. Investigation of volunteers infected with the influenza virus. Am J Med Sci. 1937;194:159–70.
Tong S, Li Y, Rivailler P, et al. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci. 2012;109:4269–74.
Zhu X, Yang H, Guo Z, et al. Crystal structures of two subtype N10 neuraminidase-like proteins from bat influenza A viruses reveal a diverged putative active site. Proc Natl Acad Sci. 2012;109:18903–8.
Xu X, Lindstrom SE, Shaw MW, et al. Reassortment and evolution of current human influenza A and B viruses. Virus Res. 2004;103:55–60.
Osterhaus AD, Rimmelzwaan GF, Martina BE, Bestebroer TM, Fouchier RA. Influenza B virus in seals. Science. 2000;288:1051–3.
Scholtissek C, Stech J, Krauss S, Webster RG. Cooperation between the hemagglutinin of avian viruses and the matrix protein of human influenza A viruses. J Virol. 2002;76:1781–6.
Chen W, Calvo PA, Malide D, et al. A novel influenza A virus mitochondrial protein that induces cell death. Nat Med. 2001;7:1306–12.
Jagger BW, Wise HM, Kash JC, et al. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science. 2012;337:199–204.
Kilbourne ED, Loge JP. Influenza A prime: a clinical study of an epidemic caused by a new strain of virus. Ann Intern Med. 1950;33:371–82.
Murphy BR, Kasel JA, Chanock RM. Association of serum antineuraminidase antibody with resistance to influenza in man. N Engl J Med. 1972;286:1329–32.
Dowdle WR. Influenza A, virus recycling revisited. Bull World Health Organ. 1999;77:820–8.
Kawaoka Y, Krauss S, Webster RG. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol. 1989;63:4603–8.
Taylor HR, Turner AJ. A case report of fowl plague keratoconjunctivitis. Br J Ophthalmol. 1977;61:86–8.
Kurtz J, Manvell RJ, Banks J. Avian influenza virus isolated from a woman with conjunctivitis. Lancet. 1996;384:901–2.
Webster RG, Geraci J, Petursson G, Skirnisson K. Conjunctivitis in human beings caused by influenza A virus of seals. N Engl J Med. 1981;304:911.
Koopmans M, Wilbrink B, Conyn M, et al. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet. 2004;363:587–93.
Fouchier RAM, Schneeberger PM, Rozendaal FW, et al. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci. 2004;101:1356–61.
Gao R, Cao B, Hu Y, et al. Human infection with a novel avian-origin influenza a (H7N9). Virus N Engl J Med 2013;368:1888–97.
Gao H-N, Lu H-Z, Cao B, et al. Clinical findings in 111 cases of influenza a (H7N9). Virus Infection N Engl J Med 2013;368:2277–85.
Ai J, Huang Y, Xu K, et al. Case-control study of risk factors for human infection with influenza a (H7N9). Virus in Jiangsu province, China, 2013 Euro Surveillance 2013;18:205–10.
Peiris M, Yuen KY, Leung CW, et al. Human infection with influenza H9N2. Lancet. 1999;354:916–7.
Uyeki TM, Chong Y-H, Katz JM, et al. Lack of evidence for human-to-human transmission of avian influenza A(H9N2) viruses in Hong Kong, China, 1999. Emerg Infect Dis. 2002;8: 154–9.
Guo YJ, Krauss S, Senne DA, et al. Characterization of the pathogenicity of members of the newly established H9N2 influenza virus lineages in Asia. Virology. 2000;267:279–88.
Claas ECJ, Osterhous ADME, van Beek R, et al. Human influenza A H5N1 related to a highly pathogenic avian influenza virus. Lancet. 1998;351:472–7.
WHO. Epidemiology of WHO-confirmed human cases of avian influenza A(H5N1) infection. Wkly Epidemiol Rec. 2005;81:249–57.
WHO. Avian influenza A (H5N1) infection in humans. N Engl J Med. 2005;353:1374–85.
VanKerkhove MD, Mumford E, Mounts AW, et al. Highly pathogenic avian influenza (H5N1): pathways of exposure at the animal-human interface, a systematic review. PLoS One. 2011;6:e14582.
Olsen SJ, Ungchusak K, Sovann T, et al. Family clustering of avian influenza A (H5N1). Emerg Infect Dis. 2005;11:1799–801.
Butler D. Pandemic “dry run” is cause for concern. Nature. 2006;441:554–5.
Ungchusak K, Auewarakul P, Dowell SF, et al. Probable person-to-person transmission of avian influenza A (H5N1). N Engl J Med. 2005;352:333–40.
Garten RJ, Davis CT, Russell CA, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325:197–201.
Ito T, Couceiro JN, Kelm S, et al. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol. 1998;72:7367–73.
Dawood FS, Jain S, Finelli L, et al. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360:2605–15.
Chou Y-Y, Albrecht RA, Pica N, et al. The M segment of the 2009 new pandemic H1N1 influenza virus is critical for its high transmission efficiency in the guinea pig model. J Virol. 2011;85:11235–41.
Hancock K, Veguilla V, Lu X, et al. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med. 2009;361:1945–52.
CDC. Swine-origin influenza A (H3N2) virus infection in two children - - - Indiana and Pennsylvania, July-August 2011. MMWR Morb Mortal Wkly Rep. 2011;60:1213–5.
CDC. Antibodies cross-reactive to influenza A (H3N2) variant virus and impact of 2010–11 seasonal influenza vaccine on cross-reactive antibodies – United States. MMWR Morb Mortal Wkly Rep. 2012;61:237–41.
Azziz Baumgartner E, Dao CN, Nasreen S, et al. Seasonality, timing, and climate drivers of influenza activity worldwide. J Infect Dis. 2012;206:838–46.
Schaffer FL, Soergel ME, Straube DC. Survival of airborne influenza virus: effects of propagating host, relative humidity, and composition of spray fluids. Arch Virol. 1976;54:263–73.
Lowen AC, Mubareka S, Steel J, Palese P. Influenza virus transmission is dependent on relative humidity and temperature. PLoS Pathog. 2007;3:e151.
Simonsen L, Taylor R, Viboud C, Dushoff J, Miller BA. US flu mortality estimates are based on solid science. Br Med J. 2006;332: 177–8.
Thompson WW, Shay DK, Weintraub E, et al. Influenza-associated hospitalizations in the United States. JAMA. 2004;292:1333–40.
Thompson WW, Shay DK, Weintraub E, et al. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289:179–86.
Simonsen L, Clarke MJ, Williamson DW, Stroup DF, Arden NH, Schonberger LB. The impact of influenza epidemics on mortality: introducing a severity index. Am J Public Health. 1997;87: 1944–50.
Bhat N, Wright JG, Broder KR, et al. Influenza-associated deaths among children in the United States, 2003–2004. N Engl J Med. 2005;353:2559–67.
Neuzil KM, Reed GW, Mitchel Jr EF, Griffin MR. Influenza-associated morbidity and mortality in young and middle-aged women. JAMA. 1999;281:901–7.
Lin JC, Nichol KL. Excess mortality due to pneumonia or influenza during influenza seasons among persons with acquired immunodeficiency syndrome. Arch Intern Med. 2001;161:441–6.
Siston AM, Rasmussen SA, Honein MA, et al. Pandemic 2009 influenza A(H1N1) virus illness among pregnant women in the United States. JAMA. 2010;303:1517–25.
Neuzil KM, Reed GW, Mitchel EF, Simonsen L, Griffin MR. The impact of influenza on acute cardiopulmonary hospitalizations in pregnant women. Am J Epidemiol. 1998;148:1094–102.
Couch RB, Kasel JA, Glezen WP, et al. Influenza: its control in persons and populations. J Infect Dis. 1986;153:431–40.
Glezen WP, Couch RB. Interpandemic influenza in the Houston area, 1974–1976. N Engl J Med. 1978;298:587–93.
Neuzil KM, Mellen BG, Wright PF, Mitchel EF, Griffin MR. The effect of influenza on hospitalizations, outpatient visits, and courses of antibiotics in children. N Engl J Med. 2000;342:225–31.
Izurieta HS, Thompson WW, Kramarz P, et al. Influenza and the rates of hospitalization for respiratory disease among infants and young children. N Engl J Med. 2000;342:232–9 [see comments].
Ampofo K, Gesteland PH, Bender J, et al. Epidemiology, complications, and cost of hospitalization in children with laboratory-confirmed influenza infection. Pediatrics. 2006;118:2409–17. doi:10.1542/peds.2006-1475.
Silberry GK. Complications of influenza infection in children. Pediatr Ann. 2000;29:683–90.
Poehling KA, Edwards KM, Weinberg GA, et al. The underrecognized burden of influenza in young children. N Engl J Med. 2006;355:31–40.
Kavet J. A perspective on the significance of pandemic influenza. Am J Public Health. 1977;67:1063–70.
Nichol KL, Lind A, Margolis KL, et al. The effectiveness of vaccination against influenza in healthy, working adults. N Engl J Med. 1995;333:889–93.
Neuzil KM, Hohlbein C, Zhu Y. Illness among schoolchildren during influenza season: effect on school absenteeism, parental absenteeism from work, and secondary illness in families. Arch Pediatr Adolesc Med. 2002;156:986–91.
Keech M, Scott AJ, Ryan PJJ. The impact of influenza and influenza-like illness on productivity and healthcare resource utilization in a working population. Occup Med. 1998;48:85–90.
Barker WH, Borisute H, Cox C. A study of the impact of influenza on the functional status of frail older people. Arch Intern Med. 1998;158:645–50.
Sullivan KM, Monto AS, Longini IM. Estimates of the US health impact of influenza. Am J Public Health. 1993;83:1712–6.
Buxton-Bridges C, Kuehnert MJ, Hall CB. Transmission of influenza: implication for control in health care settings. J Infect Dis. 2003;37:1094–101.
Bean B, Moore BM, Sterner B, Peterson LR, Gerding DN, Balfour HHJ. Survival of influenza viruses on environmental surfaces. J Infect Dis. 1982;146:47–51.
Boone SA, Gerba CP. The occurrence of influenza A virus on household and day care center fomites. J Infect. 2005;51:103–9.
Blachere FM, Lindsley WG, Pearce TA, et al. Measurement of airborne influenza virus in a hospital emergency department. Clin Infect Dis. 2009;48:438–40.
Yang W, Elankumaran S, Marr L. Concentrations and size distributions of airborne influenza A viruses measured indoors at a health center, a day-care center, and on airplanes. J R Soc Interface. 2011;8:1176–84.
Alford RH, Kasel JA, Gerone PJ, Knight V. Human influenza resulting from aerosol inhalation. Proc Soc Exp Biol Med. 1966;122:800–4.
Little JW, Douglas Jr RG, Hall WJ, Roth FK. Attenuated influenza produced by experimental intranasal inoculation. J Med Virol. 1979;3:177–88.
Andrewes C, Glover R. Spread of infection from the respiratory tract of the ferret: I. Transmission of influenza A virus. Br J Exp Pathol. 1941;22:91–7.
Moser MR, Bender TR, Margolis HS, Noble GR, Kendal AP, Ritter DG. An outbreak of influenza aboard a commercial airliner. J Epidemiol. 1979;110:1–6.
McLean R. The effect of ultraviolet radiation upon the transmission of epidemic influenza in long-term hospital patients. Am Rev Respir Dis. 1961;83:36–8.
Drinka PJ, Krause P, Schilling M, Miller BA, Shult PA, Gravenstein S. Report of an outbreak: nursing home architecture and influenza A attack rates. J Am Geriatr Soc. 1996;44:910–3.
Kaiser L, Henry D, Flack NP, Keene O, Hayden FG. Short-term treatment with zanamivir to prevent influenza: results of a placebo-controlled study. Clin Infect Dis. 2000;30:587–9.
Brankston G, Gitterman L, Hirji Z, Lemieux C, Gardam M. Transmission of influenza A in human beings. Lancet Infect Dis. 2007;7:257–65.
Loeb M, Dafoe N, Mahony J, et al. Surgical mask vs N95 respirators for preventing influenza among health care workers: a randomized trial. JAMA. 2009;302:1865–71.
Cowling BJ, K-H C, Fang V, et al. Facemasks and hand hygiene to prevent influenza transmission in households. Ann Intern Med. 2009;151:437–46.
Jordan WS, Badger GF, Dingle JH. A study of illness in a group of Cleveland families. XVI. The epidemiology of influenza 1948–1953. Am J Hyg. 1958;68:169–89.
Hayden FG, Fritz R, Lobo MC, Alvord W, Strober W, Straus SE. Local and systemic cytokine responses during experimental human influenza A virus infection. Relation to symptom formation and host defense. J Clin Invest. 1998;101:643–9.
Peiris JSM, Yu WC, Leung CW, et al. Re-emergence of fatal human influenza A subtype H5N1 disease. Lancet. 2004;363:617–9.
de Jong MD, Simmons CP, Thanh TT, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12:1203–7.
Martin CM, Kunin CM, Gottlieb LS, Barnes MW, Liu C, Finland M. Asian influenza A in Boston, 1957–1958. Arch Intern Med. 1959;103:516–31.
Walsh JJ, Dietlein LF, Low FN, Burch GE, Mogabgab WJ. Bronchotracheal response in human influenza. Arch Intern Med. 1961;108:376–88.
Guarner J, Shieh WJ, Dawson J, et al. Immunohistochemical and in situ hybridization studies of influenza A virus infection in human lungs. Am J Clin Pathol. 2000;114:227–33.
Oseasohn R, Adelson L, Kaji M. Clinicopathologic study of thirty-three fatal cases of Asian influenza. N Engl J Med. 1959;11:509–18.
Little JW, Hall WJ, Douglas Jr RG, Mudholkar GS, Speers DM, Patel K. Airway hyperreactivity and peripheral airway dysfunction in influenza A infection. Am Rev Respir Dis. 1978;118:295–303.
Hall WJ, Douglas Jr RG, Hyde RW, Roth FK, Cross AS, Speers DM. Pulmonary mechanics after uncomplicated influenza A infection. Am Rev Respir Dis. 1976;113:141–7.
Horner GJ, Gray Jr FD. Effect of uncomplicated, presumptive influenza on the diffusing capacity of the lung. Am Rev Respir Dis. 1973;108:866–9.
Johanson WGJ, Pierce AK, Sanford JP. Pulmonary function in uncomplicated influenza. Am Rev Respir Dis. 1969;100:141–6.
Kondo S, Abe K. The effects of influenza virus infection on FEV1 in asthmatic children. Chest. 1991;100:1235–8.
Smith CB, Kanner RE, Goldern CA, Klauber MR, Renzetti Jr AD. Effect of viral infections on pulmonary function in patients with chronic obstructive pulmonary diseases. J Infect Dis. 1980;141:271–9.
Utell MJ, Aquilina AT, Hall WJ, et al. Development of airway reactivity to nitrates in subjects with influenza. Am Rev Respir Dis. 1980;121:233–41.
Louria DB, Blumenfeld HL, Ellis JT, Kilbourne ED, Rogers DE. Studies on influenza in the pandemic of 1957–1958. II. Pulmonary complications of influenza. J Clin Invest. 1959;38:213–65.
Yelandi AV, Colby TV. Pathologic features of lung biopsy specimens from influenza pneumonia cases. Hum Pathol. 1994;25:47–53.
Weinberger DM, Simonsen L, Jordan R, Steiner C, Miller MA, Viboud C. Impact of the 2009 influenza pandemic on pneumococcal pneumonia hospitalizations in the United States. J Infect Dis. 2012;205:458–65.
Levandowski RA, Gerrity TR, Garrard CS. Modifications of lung clearance mechanisms by acute influenza A infection. J Lab Clin Med. 1985;106:428–32.
George RC, Broadbent DA, Drasar BS. The effect of influenza virus on the adherence of Haemophilus influenzae to human cells in tissue culture. Br J Exp Pathol. 1983;64:655–9.
McCullers JA, Rehg JE. Lethal synergism between influenza virus and Streptococcus pneumoniae: characterization of a mouse model and the role of platelet-activating factor receptor. J Infect Dis. 2002;186:341–50.
Epstein SL. Prior H1N1 influenza infection and susceptibility of Cleveland Family Study participants during the H2N2 pandemic of 1957: and experiment of nature. J Infect Dis. 2006;193:49–53.
Subbarao K, Murphy BR, Fauci AS. Development of effective vaccines against pandemic influenza. Immunity. 2006;24:5–9.
Black RA, Rota PA, Gorodkova N, Klenk H-D, Kendal AP. Antibody response to the M2 protein of influenza A virus expressed in insect cells. J Gen Virol. 1993;74:143–6.
Black S, Nicolay U, Mesikari T, et al. Hemagglutination inhibition antibody titers as a correlate of protection for inactivated influenza vaccines in children. Pediatr Infect Dis J. 2011;30:1081–5.
Stephenson I, Das RG, Wood JM, Katz JM. Comparison of neutralising antibody assays for detection of antibody to influenza A/H3N2 viruses: an international collaborative study. Vaccine. 2007;25:4056–63.
Wrammert J, Koutsnanos D, Li G-M, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med. 2011;208:181–93.
Simmons CP, Bernasconi NL, Suguitan AL, et al. Prophylactic and therapeutic efficacy of human monoclonal antibodies against H5N1 influenza. PLoS Med. 2007;4:e178.
Sui J, Sheehan J, Hwang WC, et al. Wide prevalence of heterosubtypic broadly neutralizing human anti-influenza A antibodies. Clin Infect Dis. 2011;52:1003–9.
Webster RG, Reay PA, Laver WG. Protection against lethal influenza with neuraminidase. Virology. 1988;164:230–7.
Schulman JL, Khakpour M, Kilbourne ED. Protective effects of specific immunity to viral neuraminidase on influenza virus infection of mice. J Virol. 1968;2:778–86.
Schulman JL, Khakpour M, Kilbourne ED. Protective effects of hemagglutinin and neuraminidase antigens on influenza virus: distinctiveness of hemagglutinin antigens of Hong Kong – 68 virus. J Virol. 1968;2:778.
Monto AS, Kendal AP. Effect of neuraminidase antibody on Hong Kong influenza. Lancet. 1973;7804:623–5.
Clements ML, Betts RF, Tierney EL, Murphy BR. Resistance of adults to challenge with influenza A wild-type virus after receiving live or inactivated virus vaccine. J Clin Microbiol. 1986;23:73–6.
Johansson BE, Grajower B, Kilbourne ED. Infection-permissive immunization with influenza virus neuraminidase prevents weight loss in infected mice. Vaccine. 1993;11:1037–9.
Treanor JJ, Tierney EL, Zebedee SL, Lamb RA, Murphy BR. Passively transferred monoclonal antibody to the M2 protein inhibits influenza A virus replication in mice. J Virol. 1990;64:1375–7.
Jegerlehner A, Schmitz N, Storni T, Bachmann MF. Influenza A vaccine based on the extracellular domain of M2: weak protection mediated via antibody-dependent NK cell activity. J Immunol. 2004;172:5598–605.
Carragher DM, Kaminski DA, Moquin A, Hartson L, Randall TD. A novel role for non-neutralizing antibodies against nucleoprotein in facilitating resistance to influenza virus. J Immunol. 2008;181:4168–76.
Wagner DK, Clements ML, Reimer CB, Snydr M, Nelson DL, Murphy BR. Analysis of immunoglobulin G antibody responses after administration of live and inactivated influenza A vaccine indicates that nasal wash immunoglobulin G is a transudate from serum. J Clin Microbiol. 1987;25:559–62.
Clements ML, Betts RF, Tierney EL, Murphy BR. Serum and nasal wash antibodies associated with resistance to experimental challenge with influenza A wild-type virus. J Clin Microbiol. 1986;24:157–60.
Belshe RB, Gruber WC, Mendelman PM, et al. Correlates of immune protection induced by live attenuated, cold-adapted, trivalent, intranasal influenza virus vaccine. J Infect Dis. 2000;181:1133–7.
Treanor J, Wright PF. Immune correlates of protection against influenza in the human challenge model. Dev Biol. 2003;115:97–105.
Hillaire ML, van Trierum SE, Bodewes R, et al. Characterization of the human CD8+ T-cell response following infection with 2009 pandemic influenza H1N1 virus. J Virol. 2011;85:12057–61.
Sasaki S, Jaimes MC, Holmes TH, et al. Comparison of the influenza-specific effector and memory B cell responses to immunization of children and adults with live attenuated or inactivated influenza vaccines. J Virol. 2007;81:215–28.
Kosor Krnic E, Gagro A, Drazenovic V, et al. Enumeration of haemagglutinin-specific CD8+ T cells after influenza vaccination using MHC class I peptide tetramers. Scand J Immunol. 2008;67:86–94.
McElhaney JE, Ewen C, Zhou X, et al. Granzyme B: correlates with protection and enhanced CTL response to influenza vaccination in older adults. Vaccine. 2009;27:2418–25.
McMichael AJ, Gotch FM, Noble GR, Beare PAS. Cytotoxic T-cell immunity to influenza. N Engl J Med. 1983;309:13–7.
Wilkinson TM, Li CKF, Chui CSC, et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med. 2012;18:274–80.
Forrest BD, Pride MW, Dunning AJ, et al. Correlation of cellular immune responses with protection against culture-confirmed influenza virus in young children. Clin Vaccine Immunol. 2008;15:1042–53.
Schwarzmann SW, Adler JL, Sullivan RFJ, Marine WM. Bacterial pneumonia during the Hong Kong influenza epidemic of 1968–1969. Arch Intern Med. 1971;127:1037–41.
Bisno AL, Griffin JP, VanEpps KA. Pneumonia and Hong Kong influenza: a prospective study of the 1968–1969 epidemic. Am J Med Sci. 1971;261:251–74.
CDC. Severe methicillin-resistant Staphylococcus aureus community-acquired pneumonia associated with influenza – Louisiana and Georgia, December 2006. MMWR Morb Mortal Wkly Rep. 2007;56:325–39.
Howard JB. Influenza A2 virus as a cause of croup requiring tracheostomy. J Pediatr. 1972;81:1148–50.
Monto AS, Ross HW. The Tecumseh study of respiratory illness. X. Relation of acute infections to smoking, lung function, and chronic symptoms. Am J Epidemiol. 1978;107:57–64.
Ferson MJ, Morton JR, Robertson PW. Impact of influenza on morbidity in children with cystic fibrosis. J Paediatr Child Health. 1991;27:308–11.
Crum-Cianflone NF. Bacterial, fungal, parasitic, and viral myositis. Clin Microbiol Rev. 2008;21:473–94.
Karjalainen J, Nieminen MS, Heikkila J. Influenza A1 myocarditis in conscripts. Acta Med Scand. 1980;207:27–30.
Craver RD, Sorrells K, Gohd R. Myocarditis with influenza B infection. Pediatr Infect Dis J. 1997;16:629–30.
MacDonald KL, Osterholm MT, Hedberg CW, et al. Toxic shock syndrome: a newly recognized complication of influenza and influenza like illness. JAMA. 1987;257:1053–8.
Sperber SJ, Francis JB. Toxic shock during an influenza outbreak. JAMA. 1987;257:1086–9.
Edelen JS, Bender TR, Chin TDY. Encephalopathy and pericarditis during an outbreak of influenza. Am J Epidemiol. 1974;100:79–83.
Bui M, Whittaker G, Helenius A. Effect of M1 protein and low pH on nuclear transport of influenza virus ribonucleoproteins. J Virol. 1996;70:8391–401.
Van Voris LP, Betts RF, Hayden FG, Christmas WA, Douglas Jr RG. Successful treatment of naturally occurring influenza A/USSR/77 H1N1. JAMA. 1981;245:1128–31.
Younkin SW, Betts RF, Roth FK, Douglas Jr RG. Reduction in fever and symptoms in young adults with influenza A/Brazil/78 H1N1 infection after treatment with aspirin or amantadine. Antimicrob Agents Chemother. 1983;23:577–82.
Hall CB, Dolin R, Gala CL, et al. Children with influenza A infection: treatment with rimantadine. Pediatrics. 1987;80:275–82.
Shiraishi K, Mitamura K, Sakai Y, Goto H, Sugaya N, Kawaoka Y. High frequency of resistant viruses harboring different mutations in amantadine-treated children with influenza. J Infect Dis. 2003;188:57–61.
Hay AJ, Wolstenholme AJ, Skehel JJ, Smith MH. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 1985;4:3021–4.
Boivin G, Goyette N, Bernatchez H. Prolonged excretion of amantadine-resistant influenza a virus quasi species after cessation of antiviral therapy in an immunocompromised patient. Clin Infect Dis. 2002;34:E23–5.
Bright RA, Medina M-J, Xu X. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet. 2005;366:1175–81.
Bright RA, Shay DK, Shu B, Cox NJ, Klimov AI. Adamantane resistance among influenza A viruses isolated early during the 2005–2006 influenza season in the United States. JAMA. 2006;295:891–4.
Mitnaul LJ, Castrucci MR, Murti KG, Kawaoka Y. The cytoplasmic tail of influenza A virus neuraminidase (NA) affects NA incorporation into virions, virion morphology, and virulence in mice but is not essential for virus replication. J Virol. 1996;70:873–9.
Treanor JJ, Hayden FG, Vrooman PS, et al. Efficacy and safety of the oral neuraminidase inhibitor oseltamivir in treating acute influenza: a randomized, controlled trial. JAMA. 2000;283:1016–24.
Nicholson KG, Aoki FY, Osterhaus ADME, et al. Efficacy and safety of oseltamivir in treatment of acute influenza: a randomized controlled trial. Lancet. 2000;355:1845–50.
Hayden FG, Osterhaus ADME, Treanor JJ, et al. Efficacy and safety of the neuraminidase inhibitor zanamivir in the treatment of influenza virus infections. N Engl J Med. 1997;337:874–80.
MIST. Randomised trial of efficacy and safety of inhaled zanamivir in treatment of influenza A and B virus infections. Lancet. 1998;352:1877–81.
Whitley RJ, Hayden FG, Reisinger KS, et al. Oral oseltamivir treatment of influenza in children. Pediatr Infect Dis J. 2001;20:127–33.
Hedrick JA, Barzilai A, Behre U, et al. Zanamivir for treatment of symptomatic influenza A and B infection in children five to twelve years of age: a randomized controlled trial. Pediatr Infect Dis J. 2000;19:410–7.
Gubareva LV, Bethell R, Hart GJ, Murti KG, Penn CR, Webster RG. Characterization of mutants of influenza A selected with the neuraminidase inhibitor 4-guanidino-Neu5Ac2en. J Virol. 1996;70:1818–27.
Gubareva LV, Robinson MJ, Bethell RC, Webster RG. Catalytic and framework mutations in the neuraminidase active site of influenza viruses that are resistant to 4-guanidino-neu5ac2en. J Virol. 1997;71:3385–90.
Moscona A. Oseltamivir resistance – disabling our influenza defenses. N Engl J Med. 2005;353:2633–6.
McKimm-Breschkin JL, Sahasrabudhe A, Blick TJ, et al. Mutations in a conserved residue in the influenza virus neuraminidase active site decreases sensitivity to neu5acen-derived inhibitors. J Virol. 1998;72:2456–62.
Goto H, Bethell RC, Kawaoka Y. Mutations affecting the sensitivity of the influenza virus neuraminidase to 4-guanidino-2,4-dideoxy-2,3-dehydro-N-acetylneuraminic acid. Virology. 1997;238:265–72.
Ives JA, Carr JA, Mendel DB, et al. The H274Y mutation in the influenza A/H1N1 neuraminidase active site following oseltamivir phosphate treatment leave virus severely compromised both in vitro and in vivo. Antiviral Res. 2002;55:307–17.
Carr J, Ives J, Kelly L, et al. Influenza virus carrying neuraminidase with reduced sensitivity to oseltamivir carboxylate has altered properties in vitro and is compromised for infectivity and replicative ability in vivo. Antiviral Res. 2002;54:79–88.
Herlocher ML, Carr J, Ives J, et al. Influenza virus carrying an R292K mutation in the neuraminidase gene is not transmitted in ferrets. Antiviral Res. 2002;54:99–111.
Herlocher ML, Truscon R, Elias S, et al. Influenza viruses resistant to the antiviral drug oseltamivir: transmission studies in ferrets. J Infect Dis. 2004;190:1627–30.
Kiso M, Mitamura K, Sakai-Tagawa Y, et al. Resistant influenza A viruses in children treated with oseltamivir: descriptive study. Lancet. 2004;364:759–65.
Dharan JN, Gubareva LV, Meyer JJ, et al. Infections with oseltamivir-resistant influenza A(H1N1) virus in the United States. JAMA. 2009;301:1034–41.
Francis T, Salk JE, Quilligan JJ. Experience with vaccination against influenza in the spring of 1947. Am J Public Health. 1947;37:1013–6.
France EK, Glanz JM, Xu S, et al. Safety of the trivalent inactivated influenza vaccine among children – a population-based study. Arch Pediatr Adolesc Med. 2004;158:1031–6.
Govaert TM, Thijs CT, Masurel N, Sprenger MJ, Dinant GJ, Knottnerus JA. The efficacy of influenza vaccination in elderly individuals. A randomized double-blind placebo-controlled trial. JAMA. 1994;272:1956–61.
Nichol KL, Margolis KL, Lind A, et al. Side effects associated with influenza vaccination in healthy working adults. A randomized, placebo-controlled trial. Arch Intern Med. 1996;156:1546–50.
Wright PF, Jd C, Foy HM, et al. Antigenicity and reactogenicity of influenza A/USSR/77 virus vaccine in children – a multicentered evaluation of dosage and toxicity. Rev Infect Dis. 1983;5:758–64.
CDC. Update: Recommendations of the Advisory Committee on Immunization Practices regarding use of CSL seasonal influenza vaccine (Afluria) in the United States during 2010–11. MMWR Morb Mortal Wkly Rep. 2010;59:989–92.
CDC. Prevention and control of influenza with vaccines: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2010. MMWR Morb Mortal Wkly Rep. 2010;59:1–62.
Lasky T, Tarracciano GJ, Magder L, et al. The Guillain-Barre syndrome and the 1992–1993 and 1993–1994 influenza vaccines. N Engl J Med. 1998;339:1797–802.
Scheifele DW, Dubal B, Russell ML, et al. Ocular and respiratory symptoms attributable to inactivated split influenza vaccine: evidence from a controlled trial involving adults. Clin Infect Dis. 2003;36:850–7.
Belshe RB, Edwards KM, Vesikari T, et al. Live attenuated versus inactivated influenza vaccine in infants and young children. N Engl J Med. 2007;356:685–96.
Talbot TR, Crocker DD, Peters J, et al. Duration of virus shedding after trivalent intranasal live attenuated influenza vaccination in adults. Infect Control Hosp Epidemiol. 2005;26:494–500.
Vesikari T, Karvonen A, Korhonen T, et al. A randomized, double-blind study of the safety, transmissibility, and phenotypic and genotypic stability of cold-adapted influenza virus vaccine. Pediatr Infect Dis J. 2006;25:590–7.
Petrie JG, Ohmit SE, Johnson E, Cross RT, Monto AS. Efficacy studies of influenza vaccines: effect of end points used and characteristics of vaccine failures. J Infect Dis. 2011;203:1309–15.
Osterholm MT, Kelley NS, Sommer A, Belongia E. Influenza vaccine efficacy and effectiveness: a new look at the evidence. Lancet Infect Dis. 2012;12:36–44.
Frey S, Vesikari T, Szymczakiewicz-Multanowska A, et al. Clinical efficacy of cell culture-derived and egg-derived inactivated subunit influenza vaccines in healthy adults. Clin Infect Dis. 2010;51:997–1004.
Treanor JJ, Kotloff K, Betts RF, et al. Evaluation of trivalent, live, cold-adapted (CAIV-T) and inactivated (TIV) influenza vaccines in prevention of virus infection and illness following challenge of adults with wild-type influenza A (H1N1), A (H3N2), and B viruses. Vaccine. 1999;18:899–906.
Edwards KM, Dupont WD, Westrich MK, Plummer WDJ, Palmer PS, Wright PF. A randomized controlled trial of cold-adapted and inactivated vaccines for the prevention of influenza A disease. J Infect Dis. 1994;169:68–76.
Nichol KL, Mendelman PM, Mallon KP, et al. Effectiveness of live, attenuated intranasal influenza virus vaccine in healthy, working adults: a randomized controlled trial. JAMA. 1999;282:137–44.
Hoberman A, Greenberg DP, Paradise JL, et al. Effectiveness of inactivated influenza vaccine in preventing acute otitis media in young children: a randomized controlled trial. JAMA. 2003;290:1608–16.
Vesikari T, Knuf M, Wutzler P, et al. Oil-in-water emulsion adjuvant with influenza vaccine in young children. N Engl J Med. 2011;365:1406–16.
Belshe RB, Mendelman PM, Treanor J, et al. The efficacy of live attenuated cold-adapted trivalent, intranasal influenza virus vaccine in children. N Engl J Med. 1998;358:1405–12.
Belshe RB, Gruber WC, Mendelman PM, et al. Efficacy of vaccination with live attenuated, cold-adapted, trivalent, intranasal influenza virus vaccine against a variant (A/Sydney) not contained in the vaccine. J Pediatr. 2000;136:168–75.
Vesikari T, Fleming DM, Aristegui JF, et al. Safety, efficacy, and effectiveness of cold-adapted influenza vaccine-trivalent against community-acquired, culture-confirmed influenza in young children attending day care. Pediatrics. 2006;118:2298–312. doi:10.1542/peds.2006-0725.
Tam JS, Capeding MR, Lum LC, et al. Efficacy and safety of a live attenuated, cold-adapted influenza vaccine, trivalent against culture-confirmed influenza in young children in Asia. Pediatr Infect Dis J. 2007;26:619–28.
DeVilliers PJT, Steele AD, Hiemstra LA, et al. Efficacy and safety of a live attenuated influenza vaccine in adults 60 years of age and older. Vaccine. 2010;28:228–34.
Orenstein EW, de Serres G, Haber MJ, et al. Methodologic issues regarding the use of three observational study designs to assess influenza vaccine effectiveness. Int J Epidemiol. 2007;36: 623–31.
Treanor JJ, Talbot HK, Ohmit SE, et al. Effectiveness of seasonal influenza vaccine in the United States during a season with circulation of all three vaccine strains. Clin Infect Dis. 2012;55: 951–9.
Skowronski DM, Janjua NZ, de Serres G, et al. A sentinel platform to evaluate influenza vaccine effectiveness and new variant circulation, Canada 2010–2011 season. Clin Infect Dis. 2012;55: 332–42.
Talbot HK, Griffin MR, Chen Q, Zhu Y, Williams JV, Edwards KM. Effectiveness of seasonal vaccine in preventing confirmed influenza-associated hospitalizations in community dwelling older adults. J Infect Dis. 2011;203:500–8.
Kelly H, Jacoby P, Dixon G, et al. Vaccine effectiveness against laboratory-confirmed influenza in healthy young children. Pediatr Infect Dis J. 2011;30:107–11.
Nichol KL, Nordin J, Nelson DB, Mullooly J, Hak E. Effectiveness of influenza vaccine in the community-dwelling elderly. N Engl J Med. 2007;357:1373–81.
Jackson LA, Jackson ML, Nelson JC, Neuzil KM, Weiss NS. Evidence of bias in estimates of influenza vaccine effectiveness in seniors. Int J Epidemiol. 2006;35:337–44.
Zaman K, Roy E, Arifeen SE, et al. Effectiveness of maternal influenza immunization in mothers and infants. N Engl J Med. 2008;359:1555–64.
Benowitz I, Esposito DB, Gracey KD, Shapiro ED, Vazquez M. Influenza vaccine given to pregnant women reduces hospitalization due to influenza in their infants. Clin Infect Dis. 2010;51:1355–61.
Monto AS, Davenport FM, Napier JA, Francis Jr T. Modification of an outbreak of influenza in Tecumseh, Michigan by vaccination of schoolchildren. J Infect Dis. 1970;122:16–25.
Loeb M, Russell ML, Moss L, et al. Effect of influenza vaccination of children on infection rates in Hutterite communities: a randomized trial. JAMA. 2010;303:943–50.
Reichert TA, Sugaya N, Fedson DS, Glezen WP, Simonsen L, Tashiro M. The Japanese experience with vaccinating schoolchildren against influenza. N Engl J Med. 2001;344:889–96.
Francis TJ. Vaccination against influenza. Bull World Health Organ. 1953;8:725–41.
Perkins FT. Experience in the United Kingdom with oil adjuvant influenza vaccines. Ann Allergy. 1972;30:288–91.
Weibel RE, McLean A, Woodhour AF, Fredman A, Heilleman MR. Ten-year follow-up study for safety of adjuvant 65 influenza vaccine in man. Proc Soc Exp Biol Med. 1973;143:1053–6.
Frey SE, Poland GA, Percell S, Podda A. Comparison of the safety, tolerability, and immunogenicity of a MF59-adjuvanted influenza vaccine and a non-adjuvanted vaccine in non-elderly adults. Vaccine. 2003;21:4234–7.
Podda A. The adjuvanted influenza vaccines with novel adjuvants: experience with the MF59-adjuvanted vaccine. Vaccine. 2001;19:2673–80.
Galli G, Hancock K, Hoschler K, et al. Fast rise of broadly cross-reactive antibodies after boosting long-lived human memory B cells primed by an MF59 adjuvanted prepandemic vaccine. Proc Natl Acad Sci U S A. 2009;106:7962–7.
Khurana S, Verma N, Yewdell JW, et al. MF59 adjuvant enhances diversity and affinity of antibody-mediated immune response to pandemic influenza vaccines. Sci Transl Med. 2011;3:85ra48.
Ashkenazi S, Vertruyen A, Aristegui J, et al. Superior relative efficacy of live attenuated influenza vaccine compared with inactivated influenza vaccine in young children with recurrent respiratory tract infections. Pediatr Infect Dis J. 2006;25: 870–9.
Ohmit SE, Victor JC, Rotthoff JR, et al. Prevention of antigenically drifted influenza by inactivated and live attenuated vaccines. N Engl J Med. 2006;355:2513–22.
Monto AS, Ohmit SE, Petrie JG, et al. Comparative efficacy of inactivated and live attenuated influenza vaccines. N Engl J Med. 2009;361:1260–7.
Wang Z, Tobler S, Roayaei J, Eick A. Live attenuated or inactivated influenza vaccines and medical encounters for respiratory illnesses among US military personnel. JAMA. 2009;301: 945–53.
Suggested Reading
Hayden FG, Fritz R, Lobo MC, Alvord W, Strober W, Straus SE. Local and systemic cytokine responses during experimental human influenza A virus infection. Relation to symptom formation and host defense. J Clin Invest. 1998;101:643–9. Comprehensive description of the clinical and innate immune response to influenza infection in the challenge model.
Osterholm MT, Kelley NS, Sommer A, Belongia E. Influenza vaccine efficacy and effectiveness: a new look at the evidence. Lancet Infect Dis. 2012;12:36–44. This thought-provoking review and meta-analysis of published studies of influenza vaccine efficacy and effectiveness provides a thorough review of the challenges of influenza vaccine.
Poehling KA, Edwards KM, Weinberg GA, Szilagyi PG, Staat MA, Iwane MK, Bridges CB, Grijalva CG, Zhu Y, Bernstein DI, Herrara G, Erdman D, Hall CB, Seither R, Griffin MR. The underrecognized burden of influenza in young children. N Engl J Med. 2006;355:31–40. This clearly established the important role that influenza played in young children and was one of the studies that encouraged current recommendations for the vaccination of children.
Thompson WW, Shay DK, Weintraub E, Brammer L, Bridges CB, Cox NJ, Fukuda K. Influenza-associated hospitalizations in the United States. JAMA. 2004;292:1333–40.
Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, Fukuda K. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289:179–86. These two papers comprehensively describe the methodology and the most complete estimates of the overall disease burden of influenza in the United States.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Science+Business Media New York
About this chapter
Cite this chapter
Treanor, J.J. (2014). Influenza Viruses. In: Kaslow, R., Stanberry, L., Le Duc, J. (eds) Viral Infections of Humans. Springer, Boston, MA. https://doi.org/10.1007/978-1-4899-7448-8_19
Download citation
DOI: https://doi.org/10.1007/978-1-4899-7448-8_19
Published:
Publisher Name: Springer, Boston, MA
Print ISBN: 978-1-4899-7447-1
Online ISBN: 978-1-4899-7448-8
eBook Packages: MedicineMedicine (R0)