Abstract
We present an extension of pls—called partial least squares correspondence analysis (plsca)—tailored for the analysis of nominal data. As the name indicates, plsca combines features of pls (analyzing the information common to two tables) and correspondence analysis (ca, analyzing nominal data). We also present inferential techniques for plsca such as bootstrap, permutation, and \({\chi }^{2}\) omnibus tests. We illustrate plsca with two nominal data tables that store (respectively) behavioral and genetics information.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Key words
- Partial least squares
- Correspondence analysis
- Multiple correspondence analysis
- Chi-square distance
- Genomics
1 Introduction
With the advent of relatively inexpensive genome-wide sequencing it is now possible to obtain large amounts of detailed genetic information on large samples of participants, and, so, several large sample studies are currently under way whose main goal is to relate genetics to behavior or clinical status. In these studies, the genetic information of each participant is a long list of pairs (one per chromosome) of DNA nucleotides (A, T, C, and G)—which could occur in 24 = 16 different configurations—grouped in 23 chromosomes. However, only genomic locations that show enough variability in a population are used. These locations of variability are called single nucleotide polymorphisms (snps). Each snp has a major allele (e.g., A), which is the most frequent nucleotide (in a population), and a minor allele (e.g., T; rare in a population but required to be found in at least 5% of the population to be considered “relevant”). Thus, in practice only three variants for each location are used: the major homozygote (e.g., AA), the minor homozygote (e.g., TT), and the heterozygote (e.g., AT).
Multivariate data sets of snps are most often re-coded through a process of counting alleles: 0, 1, or 2. While 1 is always the heterozygote, 0 and 2 could be ambiguous. For example, minor homozygotes can be coded according to two different schemes: (1) having 2 minor alleles [1] or (2) having 0 major alleles [2]. In most analyses, the snps are treated as quantitative data because most statistical methods used rely upon quantitative measures [3–5]. Some multivariate approaches for snps include independent components analysis (ica) [6], sparse reduced-rank regression (sRRR) [7], multivariate distance matrix regression (mdmr) [8, 9], and pls regression (plsr) [10, 11]. It should be noted that both sRRR and mdmr are plsr-like techniques. However, these methods depend on the allele counting approach that assumes a uniform linear increase for all snp s from 0 to 1 and from 1 to 2, but snps do not identify how much of an allele is present, only which allele (i.e., nucleotide variation) is present. Because the assumptions of a quantitative coding scheme seem unrealistic, we have decided to use a qualitative coding scheme and to consider that the values 0, 1, and 2 represent three different levels of a nominal variable (e.g., 0 = AA, 1 = AT, and 2 = TT). In studies relating genetics and behavior, behavior is evaluated by surveys or questionnaires that also provide qualitative answers. So the problem of relating genetics and behavior reduces to finding the information common to two tables of qualitative data. Partial least square correlation (plsc, see [12, 14]) would be an obvious solution to this “two-table problem” but it works only for quantitative data. An obvious candidate to analyze one table of qualitative data is correspondence analysis (ca), which generalizes principal component analysis (pca) to qualitative data. In this paper, we present partial least squares-correspondence analysis (plsca): A generalization of plsc—tailored for qualitative data—that integrates features of plsc and ca. We illustrate plsca with an example on genetics and substance abuse.
2 PLSC and PLSCA
2.1 Notations
Matrices are denoted by bold face upper-case letters (e.g.,\(\mathbf{X}\)), vectors by bold face lower case letters (e.g., m). The identity matrix is denoted \(\mathbf{I}\). The transpose operation is denoted \({}^{\mathsf{T}}\) and the inverse of a square matrix is denoted −1. The \(\text{diag}\left \{\right \}\) operator transforms a vector into a diagonal matrix when applied to a vector and extracts the diagonal element of a matrix when applied to a matrix.
2.2 PLSC: A Refresher
Partial least square correlation [12, 13] is a technique whose goal is to find and analyze the information common to two data tables collecting information on the same observations. This technique seems to have been independently (re)discovered by multiple authors and therefore, it exists under different names such as “inter-battery analysis” (in 1958 and probably the earliest instance of the technique, [15]), “PLS-SVD” [12, 17, 18], “intercorrelation analysis,” “canonical covariance analysis,” [19], “robust canonical analysis” [20], or “co-inertia analysis” [21]. In plsc, X and Y denote two \(I\) by \(J\) and \(I\) by \(K\) matrices that describe the \(I\) observations (respectively) by \(J\) and \(K\) quantitative variables. The data matrices are, in general, pre-processed such that each variable has zero mean and unitary norm; the pre-processed data matrices are denoted \(\mathbf{Z_{\mathbf{X}}}\) and \(\mathbf{Z_{\mathbf{Y}}}\). The first step of plsc is to compute the correlation matrix \(\mathbf{R} ={ \mathbf{Z_{\mathbf{X}}}}^{\mathsf{T}}\mathbf{Z_{\mathbf{Y}}},\) whose singular value decomposition (svd, [22–24]) is \(\mathbf{R} = \mathbf{U_{\mathbf{X}}}\varDelta {\mathbf{U_{\mathbf{Y}}}}^{\mathsf{T}}.\) The matrices \(\mathbf{U_{\mathbf{X}}}\) and \(\mathbf{U_{\mathbf{Y}}}\) contain (respectively) the left and right singular vectors of \(\mathbf{R}\). In plsc parlance, the singular vectors are called saliences [25]. The diagonal matrix \(\varDelta\) stores the singular values of \(\mathbf{R}\): each singular value expresses how much a pair of singular vectors “explains \(\mathbf{R}\).” To express the saliences relative to the observations described in \(\mathbf{Z_{\mathbf{X}}}\) and \(\mathbf{Z_{\mathbf{Y}}}\), these matrices are projected onto their respective saliences. This creates two sets of latent variables—which are linear combinations of the original variables— which are denoted \(\mathbf{\mathbf{L}_{\mathbf{X}}}\) and \(\mathbf{\mathbf{L}_{\mathbf{Y}}}\), and are computed as:
A pair of latent variables (i.e., one column from \(\mathbf{\mathbf{L}_{\mathbf{X}}}\) and one column \(\mathbf{\mathbf{L}_{\mathbf{Y}}}\)) is denoted \(\ell_{\mathbf{X},_{\ell}}\) and \(\ell_{\mathbf{Y},_{\ell}}\) and together these two latent variables reflect the relationship between \(\mathbf{X}\) and \(\mathbf{Y}\) where the singular value associated to a pair of latent variables is equal to their covariance (see, e.g., [12]).
2.2.1 What Does PLSC Optimize?
The goal of plsc is to find pairs of latent vectors \(\ell_{\mathbf{X},_{\ell}}\) and \(\ell_{\mathbf{Y},_{\ell}}\) with maximal covariance under the constraints that pairs of latent vectors of different indices are uncorrelated and coefficients of latent variables are normalized [15, 16]. Formally, we want to find:
under the constraints that
(note that \(\ell{_{\mathbf{X},\ell}}^{\mathsf{T}}\ell_{\mathbf{X},\ell^{\prime}}\) and \(\ell{_{\mathbf{Y},\ell}}^{\mathsf{T}}\ell_{\mathbf{Y},\ell^{\prime}}\) are not required to be null) and
2.3 PLSCA
In plsc, X and Y are \(I\) by \(J\) and \(I\) by \(K\) matrices that describe the same \(I\) observations with (respectively) \(N_{X}\) and \(N_{Y }\) nominal variables. These variables are expressed with a 0/1 group coding (i.e., a nominal variable is coded with as many columns as it has levels and a value of 1 indicates that the observation has this level, 0 if it does not). The centroid of \(\mathbf{X}\) (resp., \(\mathbf{Y}\)) is denoted \(\boldsymbol{\bar{\mathbf{x}}}\) (resp., \(\boldsymbol{\bar{\mathbf{y}}}\)), the relative frequency for each column of \(\mathbf{X}\), (resp., \(\mathbf{Y}\)) is denoted \(\mathbf{m_{\mathbf{X}}}\) (resp. \(\mathbf{m_{\mathbf{Y}}}\)). These centroids are computed as:
In plsca, each variable is weighted according to the information it provides. Because a rare variable provides more information than a frequent variable, the weight of a variable is defined as the inverse of its relative frequency. Specifically, the weights of \(\mathbf{X}\) (resp \(\mathbf{Y}\)) are stored as the diagonal elements of the diagonal matrix \(\mathbf{W_{\mathbf{X}}}\) (resp. \(\mathbf{W_{\mathbf{Y}}}\)) computed as: \(\mathbf{W_{\mathbf{X}}} = \text{diag}{\left \{\mathbf{m_{\mathbf{X}}}\right \}}^{-1}\) and \(\mathbf{W_{\mathbf{Y}}} = \text{diag}{\left \{\mathbf{m_{\mathbf{Y}}}\right \}}^{-1}\). The first step in plsca is to normalize the data matrices such that their sum of squares is equal to respectively \(\frac{1} {N_{X}}\) and \(\frac{1} {N_{Y }}\). Then the normalized matrices are centered in order to eliminate their means. The centered and normalized matrices are denoted \(\mathbf{Z_{\mathbf{X}}}\) and \(\mathbf{Z_{\mathbf{Y}}}\) and are computed as: \(\mathbf{Z_{\mathbf{X}}} = \left (\mathbf{X} -\boldsymbol{ 1}\boldsymbol{\bar{{\mathbf{x}}}}^{\mathsf{T}}\right ) \times {I}^{-\frac{1} {2} }N_{X}^{-1}\) and \(\mathbf{Z_{\mathbf{Y}}} = \left (\mathbf{Y} -\boldsymbol{ 1}\boldsymbol{\bar{{\mathbf{y}}}}^{\mathsf{T}}\right ) \times {I}^{-\frac{1} {2} }N_{Y }^{-1}.\) Just like in plsc, the next step is to compute the matrix \(J\) by \(K\) matrix \(\mathbf{R}\) as \(\mathbf{R} ={ \mathbf{Z_{\mathbf{X}}}}^{\mathsf{T}}\mathbf{Z_{\mathbf{Y}}}.\) The matrix \(\mathbf{R}\) is then decomposed with the generalized svd as:
In plsca the saliences, denoted \(\mathbf{S_{\mathbf{X}}}\) and \(\mathbf{S_{\mathbf{Y}}}\), are slightly different from the singular vectors and are computed as \(\mathbf{S_{\mathbf{X}}} = \mathbf{W_{\mathbf{X}}}\mathbf{U_{\mathbf{X}}}\text{ and }\mathbf{S_{\mathbf{Y}}} = \mathbf{W_{\mathbf{Y}}}\mathbf{U_{\mathbf{Y}}}.\) Note that
To express the saliences relative to the observations described in \(\mathbf{Z_{\mathbf{X}}}\) and \(\mathbf{Z_{\mathbf{Y}}}\), these matrices are projected onto their respective saliences. This creates two sets of latent variables—which are linear combinations of the original variables—that are denoted \(\mathbf{\mathbf{L}_{\mathbf{X}}}\) and \(\mathbf{\mathbf{L}_{\mathbf{Y}}}\) and are computed as:
2.4 What Does plsca Optimize?
In plsca, the goal is to find linear combinations of \(\mathbf{Z_{\mathbf{X}}}\) and \(\mathbf{Z_{\mathbf{Y}}}\) called latent variables \(\ell_{\mathbf{X},_{\ell}}\) and \(\ell_{\mathbf{Y},_{\ell}}\) which have maximal covariance under the constraints that pairs of latent vectors with different indices are uncorrelated and that the coefficients of each latent variables are normalized to unit length. Formally, we want to find
under the constraints that
and
It follows from the properties of the generalized svd [22] that \(\mathbf{u}_{\mathbf{X},\ell}\) and \(\mathbf{u}_{\mathbf{Y},\ell}\) are singular vectors of \(\mathbf{R}\). Specifically, the product of the matrix of latent variables can be rewritten as (from Eq. 8):
As a consequence, the covariance of a pair of latent variables \(\ell_{\mathbf{X},\ell}\) and \(\ell_{\mathbf{Y},\ell}\) is equal to their singular value:
So, when ℓ = 1, we have the largest possible covariance between the pair of latent variables. Also, the orthogonality constraint for the optimization is automatically satisfied because the singular vectors constitute an orthonormal basis for their respective matrices. So, when ℓ = 2 we have the largest possible covariance for the latent variables under the constraints that the latent variables are uncorrelated with the first pair of latent variables and so on for larger values of ℓ. So plsca and ca differ mostly by how they scale salience vs. factors scores and latent variables vs. supplementary factor scores. Correspondence analysis lends itself to biplots because the scaling scheme of factors/saliences and factor scores/latent variables allows all of them to be plotted on the same graph as they both have the same scale.
2.4.1 Links to Correspondence Analysis
In this section we show that plsca can be implemented as a specific case of correspondence analysis (ca) which, itself, can be seen as a generalization of pca to nominal variables ([26, 27], for closely related approaches see [21, 28, 29]). Specifically, ca was designed to analyze contingency tables. For these tables, a standard descriptive statistic is Pearson’s \({\varphi }^{2}\) coefficient of correlation whose significance is traditionally tested by the \({\chi }^{2}\) test (recall that the coefficient \({\varphi }^{2}\) is equal to the table’s independence \({\chi }^{2}\) divided by the number of elements of the contingency table). In ca, \({\varphi }^{2}\)—which, in this context, is often called the total inertia of the table—is decomposed into a series of orthogonal components called factors. In the present context, ca will first create, from \(\mathbf{X}\) and \(\mathbf{Y}\), a \(J\) by \(K\) contingency table denoted \(\mathbf{{S}^{{\ast}}}\) and computed as: \(\mathbf{{S}^{{\ast}}} ={ \mathbf{X}}^{\mathsf{T}}\mathbf{Y}.\) This contingency table is then transformed into a correspondence matrix (i.e., a matrix with nonnegative elements whose sum is equal to 1) denoted \(\mathbf{S}\) and computed as \(\mathbf{S} = \mathbf{{S}^{{\ast}}}s_{++}^{-1}\) (with \(s_{++}\) being the sum of all the elements of \(\mathbf{{S}^{{\ast}}}\)). The factors of ca are obtained by performing a generalized svd on the double centered \(\mathbf{S}\) matrix obtained as: \(\left (\mathbf{S} -\mathbf{m_{\mathbf{X}}}{\mathbf{m_{\mathbf{Y}}}}^{\mathsf{T}}\right ).\) Simple algebraic manipulation shows that this matrix is, in fact, equal to matrix \(\mathbf{R}\) of plsca. Correspondence analysis then performs the svd described in Eq. 6. The factor scores for the \(\mathbf{X}\) and \(\mathbf{Y}\) set are computed as
For each set, the factor scores are pairwise orthogonal (under the constraints imposed by \({\mathbf{W_{\mathbf{X}}}}^{-1}\) and \({\mathbf{W_{\mathbf{Y}}}}^{-1}\)) and the variance of the columns (i.e., a specific factor) of each set is equal to the square of its singular value. Specifically:
The original \(\mathbf{X}\) and \(\mathbf{Y}\) matrices can be projected as supplementary elements on their respective factor scores. These supplementary factors scores denoted respectively \(\mathbf{G_{\mathbf{X}}}\) and \(\mathbf{G_{\mathbf{Y}}}\) are computed as
Note that the pre-multiplication by \(N_{X}\) and \(N_{Y }\) transforms the data matrices such that each row represents frequencies (this is called a row profile in correspondence analysis) and so each row now sums to one. This last equation shows that an observation is positioned as the barycenter of the coordinates of its variables. These projections are very closely related to the latent variables (see Eqs. 8 and 16) and are computed as
Both pls and ca contribute to the interpretation of plsca. Pls shows that the latent variables have maximum covariance, ca shows that factors scores have maximal variance and that this variance “explains” a proportion of the \({\varphi }^{2}\) associated to the contingency table. Traditionally ca is interpreted with graphs plotting one dimension against the other. For these graphs, using the factor scores is preferable to the saliences because these plots preserve the similarity between elements. In ca, it is also possible to plot the factor scores of \(\mathbf{X}\) and \(\mathbf{Y}\) in the same graph (because they have the same variance) which is called a symmetric plot. If one set is privileged, it is possible to use an asymmetric plot in which the factor scores of the privileged set have a variance of one and the factor scores of the other set have a variance of δ2.
2.5 Inference
Later in this paper, we present with an example three inferential methods of plsca: (1) a permutation test of the data for an omnibus \({\chi }^{2}\) test to determine if, overall, the structure of the data is not due to chance, (2) a permutation test of the data to determine what, if any factors are not due to chance, and (3) a bootstrap test to determine which measures contribute a significant amount of variance.
3 Illustration
To illustrate how plsca works and how to interpret the results, we have created a small example from a subset of data to be analyzed. The data come from a study on the individual and additive role of specific genes and substance abuse in marijuana users [30]. Here, our (toy) hypothesis is that marijuana abusing participants \((I = 50)\) with specific genotypes are more likely to frequent additional substances (i.e., certain genotypes predispose people to be polysubstance users).
3.1 Data
Each participant is given a survey that asks if they do or do not use certain (other) drugs—specifically, ecstasy (e), crack/cocaine (cc) or crystal meth (cm). Additionally, each participant is genotyped for COMT (which inactivates certain neurotransmitters) and FAAH (modulates fatty acid signals). The data are arranged in matrices \(\mathbf{X}\) (behavior) and \(\mathbf{Y}\) (snps; see Table 1).
Sometimes genotype data cannot be obtained (e.g., COMT for Subject 2). This could happen if, for example, the saliva sample were too degraded to detect which nucleotides are present. Instances of missing data receive the average values from the whole sample. From \(\mathbf{X}\) and \(\mathbf{Y}\) we compute \(\mathbf{R}\) (Table 2), which is a contingency table with the measures (columns) of \(\mathbf{X}\) on the rows and the measures (columns) of \(\mathbf{Y}\) on the columns. The \(\mathbf{R}\) matrix is then decomposed with ca.
3.2 PLSCA Results
With factor scores and factor maps, we can now interpret the results. The factor map is made up of two factors (1 and 2), which are displayed as axes. As in all svd-based techniques, each factor explains a certain amount of variance within the dataset. Factor 1 (horizontal) explains 69% of the variance; Factor 2 explains 21%. Plotted on the factor map we see the rows (survey items, purple) and the columns (snps, green) from the \(\mathbf{R}\) matrix (after decomposition). In ca, the distances between row items are directly interpretable. Likewise, the distances between column items are directly interpretable. However, the distances between row items and column items are not directly interpretable; the distances are relative. That is, “e.yes” is more likely to occur with COMT.GG than other responses.
Factors 1 (horizontal: 69% of variance) and 2 (vertical: 21% of variance). From the relative distances between snps and other drug use, we can infer that faah.aa is more likely to occur with other drug use (besides marijuana) than no drug use, compared to other snps; or, the AA allele of faah may predispose individuals to polysubstance abuse
In Fig. 1 on Factor 1, we see an interesting dichotomy. Marijuana users who have used crystal meth (cm.yes) are unlikely to use other drugs (e.no, cc.no); whereas marijuana users who have not used crystal meth (cm.no) may have used other drugs (e.yes, cc.yes). One explanation for this dichotomy is that ecstasy and cocaine could be considered more “social” drugs, whereas crystal meth is, socially, considerably frowned upon. But on Factor 2 we see that all “yes” responses occur above 0, where all “no” responses occur below 0. In this case, we can call Factor 1 “social drug use”, and Factor 2 “any drug use”. It is important to note that items (both rows and columns) near the origin occur in high frequency and therefore are considered “average.” Items that are not average help with interpretation. Additionally, we see snps with our responses on the factor map. From this map, we know that FAAH.AA, COMT.GG and COMT.AA are rare (small frequency). Furthermore, we can see that FAAH.AA is more likely to occur with other drug use (besides marijuana) than no drug use, compared to other snps.
3.3 Latent Variables
In the pls framework, we compute latent variables from the singular vectors. The latent variables of \(\mathbf{X}\) (\(\mathbf{\mathbf{L}_{\mathbf{X}}}\)) and \(\mathbf{Y}\) (\(\mathbf{\mathbf{L}_{\mathbf{X}}}\)) are computed in order to show the relationships of participants with respect to snps (\(\mathbf{X}\); Fig. 2a) and behaviors (\(\mathbf{Y}\); Fig. 2b). In the latent variable plots, the circle size grows as more individuals are associated to it. That is, for example, in Fig. 2a, the large circle on the bottom left, with the number 13 in it, represents 13 individuals. This dot indicates that 13 individuals have the same patterns of responses to drug use.
3.4 Inferential Results
3.4.1 Permutation Tests
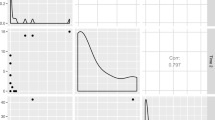
A permutation test of the data can test the omnibus null hypothesis. This test is performed by computing the \({\chi }^{2}\) value (or alternatively, the total inertia) of the entire table for each permutation. The original table has a \({\chi }^{2}\) value of 19.02, which falls outside the 95 %-ile for 1,000 permutations (which is 18.81) and this indicates that the overall structure of the data is significant (see Fig. 3).The same permutation tests are used to determine which components contribute more variance than due to chance. We test the components with the distribution of the eigenvalues. From the toy example, only the third component (not shown above, see Fig. 4) contributes a significant amount of variance (note that this implementation of the permutation test is likely to give correct values only for the first factor, because the inertia extracted by the subsequent factors depend in part upon the inertia extracted by earlier factors; a better approach would be to recompute the permutation test for a given factor after having partialled out the inertia of all previous factors from the data matrices).
Distributions for the permutation tests for each factor (1, 2, and 3, respectively). The red lines show the 95 ‰ (i.e., p < 0.05) for 1,000 permutations and the green lines are the eigenvalues of the factors. Factors 1 and 3 reach significance (p = 0.048 and p = 0.033, respectively) but Factor 2 does not (p = 0.152)
3.4.2 Bootstrap Ratios
Bootstrap resampling [31] of the observations provides distributions of how each of the measures (behavior and snps) changes with resampling. These distributions are used to build bootstrap ratios (also called bootstrap intervals t). When a value falls in the tail of a distribution (e.g., a bootstrap ratio of magnitude > 2), it is considered significant at the appropriate α level (e.g., p < 0.05). Table 3 shows that COMT (AA and GG) and ecstasy use (and non-use) contribute significantly to Factor 1.
The bootstrap tests, in conjunction with the descriptive results, indicate that certain genotypes are related to additional drug use or drug avoidance. More specifically, COMT.AA is more associated to “no ecstasy use” than any other allele and, oppositely, COMT.GG is more associated to “ecstasy use” than any other allele.
4 Conclusion
In this paper, we presented plsca, a new method tailored to the analysis of genetics, behavioral and brain imaging data. Plsca stands apart from current methods, because it directly analyzes snps as qualitative variables. Furthermore, plsca is particularly suited for the concomitant analysis of genetics and high-level behaviors as explored, for example, with surveys. Surveys are essential for the analysis of genetics and behavior as they are often designed and refined to capture the specific behaviors of given populations or psychological constructs. This way, these survey data work as an “anchor” to provide variance for genetics data. Plsca, being the ideal tool to analyze the relationship between survey and genetic data, will help to better understand the genetic underpinnings of brains, behavior, and cognition.
References
J. de Leon, J. C. Correa, G. Ruaño, A. Windemuth, M. J. Arranz, and F. J. Diaz, “Exploring genetic variations that may be associated with the direct effects of some antipsychotics on lipid levels,” Schizophrenia Research 98, pp. 1–3, 2008.
C. Cruchaga, J. Kauwe, K. Mayo, N. Spiegel, S. Bertelsen, P. Nowotny, A. Shah, R. Abraham, P. Hollingworth, D. Harold, et al., “snps associated with cerebrospinal fluid phospho-tau levels influence rate of decline in Alzheimer’s disease,” PLoS Genetics 6, 2010.
D. Y. Lin, Y. Hu, and B. E. Huang, ‘ ‘Simple and efficient analysis of disease association with missing genotype data,” American Journal of Human Genetics 82, pp. 444–452, 2008.
C. Lippert, J. Listgarten, Y. Liu, C. M. Kadie, R. I. Davidson, and D. Heckerman, “FaST linear mixed models for genome-wide association studies,” Nature Methods 8, pp. 833–835, 2011.
C. J. Hoggart, J. C. Whittaker, M. De Iorio, and D. J. Balding, “Simultaneous analysis of all SNPs in Genome-Wide and Re-Sequencing association studies,” PLoS Genetics 4, p. e1000130, 2008.
J. Liu, G. Pearlson, A. Windemuth, G. Ruano, N. I. Perrone-Bizzozero, and V. Calhoun, “Combining fMRI and SNP data to investigate connections between brain function and genetics using parallel ICA,” Human Brain Mapping 30, pp. 241–255, 2009.
M. Vounou, T. E. Nichols, and G. Montana, “Discovering genetic associations with high-dimensional neuroimaging phenotypes: A sparse reduced-rank regression approach,” NeuroImage 53, pp. 1147–1159, 2010.
M. A. Zapala and N. J. Schork, “Multivariate regression analysis of distance matrices for testing associations between gene expression patterns and related variables,” Proceedings of the National Academy of Sciences 103, pp. 19430–19435, 2006.
C. S. Bloss, K. M. Schiabor, and N. J. Schork, “Human behavioral informatics in genetic studies of neuropsychiatric disease: Multivariate profile-based analysis,” Brain Research Bulletin 83, pp. 177–188, 2010.
G. Moser, B. Tier, R. E. Crump, M. S. Khatkar, and H. W. Raadsma, “A comparison of five methods to predict genomic breeding values of dairy bulls from genome-wide SNP markers,” Genetics Selection Evolution 41, p. 56, 2009.
J. Poline, C. Lalanne, A. Tenenhaus, E. Duchesnay, B. Thirion, and V. Frouin, “Imaging genetics: bio-informatics and bio-statistics challenges,” in 19th International Conference on Computational Statistics, Y. Lechevallier and G. Saporta, (eds.), (Paris, France), 2010.
A. Krishnan, L. J. Williams, A. R. McIntosh, and H. Abdi, “Partial least squares (PLS) methods for neuroimaging: A tutorial and review,” NeuroImage 56, pp. 455–475, 2011.
A. McIntosh, F. Bookstein, J. Haxby, and C. Grady, “Spatial pattern analysis of functional brain images using partial least squares,” NeuroImage 3, pp. 143–157, 1996.
A. Krishnan, N. Kriegeskorte, and H. Abdi, “Distance-based partial least squares analysis,” in New perspectives in Partial Least Squares and Related Methods, H. Abdi, W. Chin, V. Esposito Vinzi, G. Russolilo, and L. Trinchera, (eds.), New York, Springeer Verlag, pp. 131–145.
L.R., Tucker, “An inter-battery method of factor analysis.” Psychometrika 23, pp. 111–136, 1958.
H. Abdi and L.J. Williams, “Partial least squares methods: Partial least squares correlation and partial least square regression,” in: Methods in Molecular Biology: Computational Toxicology, B. Reisfeld and A. Mayeno (eds.), pp. 549–579. New York: Springer Verlag. 2013.
F.L. Bookstein, P.L. Sampson, A.P. Streissguth, and H.M. Barr, “Exploiting redundant measurements of dose and developmental outcome: New methods from the behavioral teratology of alcohol,” Developmental Psychology 32, pp. 404–415, 1996.
P.D. Sampson, A.P. Streissguth, H.M. Barr, and F.S. Bookstein, “Neurobehavioral effect of prenatal alcohol: Part II, partial least square analysis,” Neurotoxicology and Teratology 11, pp. 477–491, 1989
A. Tishler, D. Dvir, A. Shenhar, and S. Lipovetsky, “Identifying critical success factors in defense development projects: A multivariate analysis,” Technological Forecasting and Social Change 51, pp. 151–171, 1996.
A. Tishler, and S. Lipovetsky, “Modeling and forecasting with robust canonical analysis: method and application,” Computers and Operations Research 27, pp. 217–232, 2000.
S. Dolédec, and D. Chessel, “Co-inertia analysis: an alernative method for studying species-environment relationships.” Freshwater Biology 31, pp. 277–294, 1994.
H. Abdi, “Singular value decomposition (svd) and generalized singular value decomposition (gsvd),” in Encyclopedia of Measurement and Statistics, N. Salkind, ed., pp. 907–912, Thousand Oaks (CA): Sage, 2007.
M. Greenacre, Theory and Applications of Correspondence Analysis, London, Academic Press, 1984.
H. Yanai, K. Takeuchi, and Y. Takane, Projection Matrices, Generalized Inverse Matrices, and Singular Value Decomposition, New York, Springer, 2011.
F. Bookstein, “Partial least squares: a dose–response model for measurement in the behavioral and brain sciences,” Psycoloquy 5, 1994.
H. Abdi and L. J. Williams, “Correspondence analysis,” in Encyclopedia of Research Design, pp. 267–278, Thousand Oaks, (CA), Sage, 2010.
H. Abdi and D. Valentin, “Multiple correspondence analysis,” in Encyclopedia of Measurement and Statistics, pp. 651–657, Thousand Oaks, (CA),Sage, 2007.
A. Leclerc, “L’analyse des correspondances sur juxtaposition de tableaux de contingence,” Revue de Statistique Appliquée 23, pp. 5–16
L. Lebart, M. Piron, and A. Morineau, Statistiques Exploratoire Multidimensionnelle: Visualisations et Inférences en Fouille de Données, Paris, Dunod, 2006.
F. M. Filbey, J. P. Schacht, U. S. Myers, R. S. Chavez, and K. E. Hutchison, “Individual and additive effects of the cnr1 and faah genes on brain response to marijuana cues,” Neuropsychopharmacology 35, pp. 967–975, 2009.
T. Hesterberg, “Bootstrap,” Wiley Interdisciplinary Reviews: Computational Statistics 3, pp. 497–526, 2011.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer Science+Business Media New York
About this paper
Cite this paper
Beaton, D., Filbey, F., Abdi, H. (2013). Integrating Partial Least Squares Correlation and Correspondence Analysis for Nominal Data. In: Abdi, H., Chin, W., Esposito Vinzi, V., Russolillo, G., Trinchera, L. (eds) New Perspectives in Partial Least Squares and Related Methods. Springer Proceedings in Mathematics & Statistics, vol 56. Springer, New York, NY. https://doi.org/10.1007/978-1-4614-8283-3_4
Download citation
DOI: https://doi.org/10.1007/978-1-4614-8283-3_4
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4614-8282-6
Online ISBN: 978-1-4614-8283-3
eBook Packages: Mathematics and StatisticsMathematics and Statistics (R0)