Abstract
As a major stress-responsive catabolic pathway, autophagy communicates with a variety of signal transduction pathways ranging from proliferative signaling, metabolic pathways, cell death pathways, and multiple cellular stresses. As such, autophagy needs to be able to integrate diverse signaling events and respond to various complex biological conditions in a highly orchestrated manner. Mechanistically, while the molecular basis underlying the function of the core autophagy machinery has been relatively well established, how does autophagy pathway sense diverse upstream signaling pathways? How does autophagy contribute to the eventual biological outcomes of these signaling pathways? Is there any feedback regulation from autophagy to the upstream signaling pathways? And what governs signal transduction within the core autophagy pathway? This chapter discusses the current understanding of these questions. Particularly, an emphasis is placed on the role of the Atg1/ULK1 complex in sensing the upstream nutrient/energy signaling and relaying the upstream signaling to downstream autophagy machinery.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Autophagy
- Signal transduction
- Nutrient signaling
- Regulation
- Autophagic cell death
- ULK1
- Atg1
- mTOR
- AMPK
- Phosphorylation
- Ubiquitin-like reaction
- Acetylation
1 The Basic Autophagy Machinery
Autophagy is a conserved catabolic process that utilizes lysosomal activity to turnover cellular proteins or organelles. Mammalian cells can undergo three types of autophagy: macroautophagy, microautophagy and chaperone-mediated autophagy. These modes of autophagy differ in the type of cargos to be degraded and the methods of delivering them to lysosomes. Chaperone-mediated autophagy makes use of a protein translocation pathway that selectively feeds individual substrate proteins directly into the lysosome. Microautophagy occurs through direct sequestration of cargos, involving the formation of tubular invaginations at the lysosomal membrane. Macroautophagy delivers cargo to the lysosomes through the formation and transport of specific intracellular membrane vesicles termed autophagosomes (Chen and Klionsky 2011; Mizushima and Komatsu 2011; Todde et al. 2009).
Of the three types of autophagy, macroautophagy is thought to occur predominantly and is the most studied (Mizushima and Komatsu 2011). The process of macroautophagy, herein referred to as autophagy, starts by the formation of a cup-shaped membrane structure, termed the phagophore or isolation membrane. The phagophore elongates, sequesters cytosolic components (cargos), and eventually seals to form a double membrane vesicle called the autophagosome. Cargo loading into autophagosomes may be selective for specific proteins, organelles or pathogens through the use of adaptors such as Atg32 (yeast) and SQSTM1/p62 (mammals) or non-specifically through bulk loading of cytoplasmic contents (Mizushima et al. 2011). Autophagosomes fuse with lysosomes, resulting in cargo degradation and concomitant release of the metabolic byproducts such as amino acids through permeases in the lysosomal membrane. As a membrane trafficking pathway, autophagy shares several points of convergence with endocytosis, although these converging points, such as lysosomal fusion, can be differentially regulated in autophagy and endocytosis (Ganley et al. 2011). The turnover of long-lived, aggregated, or damaged proteins and organelles by autophagy is essential for maintaining cellular homeostasis. While autophagy occurs at basal levels under normal conditions, it is activated in response to cellular stresses such as nutrient starvation, hypoxia, growth factor withdrawal, ER stress and pathogen infection. Upon these stresses, the cell employs autophagy either to redeploy its resources to tide over the period of stress or to degrade harmful components (such as damaged mitochondria or invading pathogens) via lysosomal degradation (Kroemer et al. 2010). Dysregulation of autophagy has been implicated in a range of diseases including neurodegenerative disorders most typically involving the accumulation of pathogenic proteins, inflammatory disorders such as Crohn’s Disease, and cancer (Kimmelman 2011; Wong et al. 2011).
The study of autophagy has accelerated in the last decade because of the discovery of autophagy related genes (Atg) by yeast genetics (Tsukada and Ohsumi 1993; Harding et al. 1995; Thumm et al. 1994) and subsequently, their mammalian homologues (Mizushima et al. 2011; Yang and Klionsky 2010). The original genetic screens performed by the Ohsumi lab (Tsukada and Ohsumi 1993), Thumm lab (Thumm et al. 1994), and Klionsky lab (Harding et al. 1995) identified multiple complementation groups affecting both autophagosome accumulation (Tsukada and Ohsumi 1993; Thumm et al. 1994) and cytoplasm-to-vacuole targeting (Cvt) (Harding et al. 1995). To date, up to 36 Atg genes have been identified, and the Atg genes making up the core machinery of autophagy can be classified into several major functional units: the Atg1/ULK1 complex, commonly considered as an initiator in the autophagic cascade, the VPS34 phosphatidylinositol 3-kinase (PI3K) complex, two ubiquitin-like conjugation systems (Atg12–Atg5, Atg8/LC3–PE), PI3P effector Atg2–Atg18 complex and the transmembrane protein Atg9.
It appears that most of the Atg proteins cluster at the autophagy-initiating membrane structure, phagophore (Fig. 4.1). The current model suggests the Atg1/ULK1 complex is the most upstream component of the pathway (Itakura and Mizushima 2010a). Its activation and translocation to the phagophore is thought to cause subsequent recruitment of the VPS34 Type III PI3K complex (containing the kinase VPS34 and its regulators p150, Atg6/Beclin 1 and Atg14) to the phagophore. Although the precise mechanism for this process is not clear, it seems the protein kinase activity of Atg1/ULK1 is required (Chan et al. 2009; Hara et al. 2008; Jung et al. 2009). The VPS34 complex catalyzes the conversion of the phospholipid, phosphatidyl inositol (PI), to phosphatidyl inositol-3-phosphate (PI3P). PI3P is a second messenger involved in intracellular membrane trafficking. PI3P presumably recruits its effector proteins, eventually resulting in translocation of the Atg12–Atg5–Atg16 complex to the phagophore. In the Atg5 complex, Atg12 is a ubiquitin-like protein (Ubl) which is conjugated to Atg5 protein through a typical Ubl conjugation reaction (Mizushima et al. 1998). The Atg5 complex can function as the E3 ligase for the other autophagy-essential Ubl reaction, Atg8/LC3–phosphatidyl ethanolamine (PE) conjugation (Ichimura et al. 2000; Fujita et al. 2008; Hanada et al. 2007). Atg8/LC3–PE conjugation in turn is required for autophagosome formation, and probably autophagosomal membrane tethering (Nakatogawa et al. 2007) and cargo recognition (Bjorkoy et al. 2005; Pankiv et al. 2007; Komatsu et al. 2007).
ATG protein complexes on the surface of the phagophore. The figure shows the ULK1 complex, VPS34 complex, the two autophagy Ubl systems, and the ATG9 recycling protein on the phagophore. While the epistasis of the upstream ATG complexes has been suggested, the precise mechanism underlying the signaling events for these complexes to communicate with each other is not defined
2 Interplay of Autophagy with Various Cellular Signaling Processes
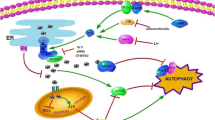
The most extensively studied molecular intersection between the autophagy pathway and other cellular signal transduction pathways is regulation of the Atg1/ULK1 complex by the nutrient-sensing kinase TOR (in yeast) or mTOR (in mammals). Because of the intimate relationship of mTOR with various signaling events including proliferation, protein synthesis, and nutrient metabolism, autophagy is immediately implicated in these processes (Fig. 4.2). Indeed, mounting evidence has confirmed that inducers and inhibitors of these signaling processes can modulate autophagy as expected. On the other hand, autophagy can also impact the biological outcomes of these signaling processes. Molecularly, a clear picture is emerging concerning how autophagy feedbacks to activate mTOR. These contents are discussed in more detail later.
Interplay of autophagy with various cellular signaling pathways. Various signal transduction pathways regulate autophagy and likely employ autophagy to fulfill their eventual biological functions. The cartoon focuses on the connection between mTOR and autophagy, which mutually regulate each other. The interplay of autophagy with multiple cellular signaling events occurs through mTOR, a master regulator of these events. Other crosstalks depicted in the figure include the connection of apoptosis with autophagy, which occurs through the antagonistic interaction between Bcl-2 and the VPS34 component Beclin 1, and regulation of autophagic membrane trafficking and fusion by monomeric GTPases and cytoskeleton. GPCR G protein-coupled receptors, RTK receptor tyrosine kinases
The autophagy pathway also functionally interacts with apoptosis. Beclin 1, the mammalian homolog of yeast Atg6, was originally identified by Levine and colleagues as a Bcl-2 interacting protein (Liang et al. 1998, 1999), thus linking autophagy to apoptotic signaling (Fig. 4.2). The interaction between Beclin 1 and Bcl-2 is thought to be mutually antagonistic, so that Bcl-2 will suppress autophagy through sequestration of Beclin 1 and correspondingly Beclin 1 binding to Bcl-2 will potentiate apoptosis (Pattingre et al. 2005a).
Beyond apoptotic cell death, the role of autophagy in cell death (apoptosis or otherwise) is intriguing and under hot debate. The term “autophagic cell death” originated from the frequent morphological association of autophagy with cell death, especially death with no apparent apoptotic features (Gozuacik and Kimchi 2004; Levine and Yuan 2005). However, under these conditions whether autophagy is a causal reason or inhibitory attempt (or simply a passenger event with no impact on the death process) became testable only after the more recent understanding of the molecular mechanisms of autophagy. Molecular studies indicate that autophagy is more often a pro-survival mechanism (Levine and Yuan 2005; Edinger and Thompson 2004; Berry and Baehrecke 2008), but under certain pathological and physiological conditions, it can be pro-death as well (Gozuacik and Kimchi 2004; Bursch 2001; Yu et al. 2004; Shimizu et al. 2004; Berry and Baehrecke 2007). The apparent opposite roles of autophagy in cell death raises this critical question: how do specific upstream signals instruct autophagy to be pro-death or anti-death, and subsequently how does autophagy contribute to these two opposite processes? And is it possible that excessive autophagy can exhaust the resource required for cell survival, and if so, what would be the signaling that prevents or promotes such “eat-to-death” activity of autophagy?
As a membrane reorganizing and trafficking event, autophagy is apparently subjected to regulation by the Rab subfamily of small G proteins. Therefore, the signaling pathways feeding into Rab function should also regulate autophagy (Klionsky 2005; Gutierrez et al. 2004; Hutagalung and Novick 2011) (Fig. 4.2). Further, signaling dictating cytoskeleton-driven intracellular mobility has also been shown to control autophagy process (Mostowy and Cossart 2011; Di Bartolomeo et al. 2010a; Xie et al. 2011) (Fig. 4.2).
Importantly, autophagy can also interact with many other signal transduction pathways, which are not discussed in this chapter. The rest of this chapter mainly focuses on the function of the Atg1/ULK1 complex, which is considered to be the most upstream player of the core autophagy pathway that directly senses nutrient signals.
3 The Atg1/ULK1 Complex, Signaling Within
3.1 Atg1 and ULK1
In yeast, Atg1 mutants failed to accumulate autophagic bodies in the presence of the protease inhibitor PMSF and died at a faster rate during nutrient starvation, linking autophagy and Atg1 with cellular metabolism (Tsukada and Ohsumi 1993). Atg1 is a serine-threonine kinase and the only protein kinase among the 36 Atg genes described so far.
As with many other Atg proteins, Atg1 also has its mammalian ortholog. But the identification and validation of the mammalian orthologs took a few twists and turns. Atg1 shares strong homology with Caenorhabditis elegans uncoordinated-51 (UNC-51) (Matsuura et al. 1997), which has two mammalian homologs known as Unc-51 like kinase-1 (ULK1) and ULK2. Therefore, the ULK proteins had long been suspected to be mammalian counterparts of Atg1 (Kuroyanagi et al. 1998). The direct experimental evidence came from an RNAi-based screen by Tooze and colleagues to identify protein kinases involved in mammalian autophagy (Chan et al. 2007). In this screen they identified ULK1, but not ULK2, as an essential component for amino acid starvation-induced autophagy in HEK293 cells, and subsequent mechanistic analysis confirmed that ULK1 is indeed the functional equivalent of yeast Atg1 (Chan et al. 2007; Young et al. 2006). Because the sequence homology between ULK1 and ULK2 is significant (55 % identical sequence), it was somewhat a surprise that RNAi knockdown of ULK1 was sufficient to block starvation-induced autophagy in HEK293 cells whereas RNAi of ULK2 had no effect (Chan et al. 2007). Later studies indicate that ULK1 and ULK2 possess redundant roles in autophagy (Lee and Tournier 2011; Yan et al. 1999), and indeed, it takes double knockout of ULK1 and ULK2 to completely block amino acid starvation-induced autophagy in mouse embryonic fibroblasts (MEFs) (Cheong et al. 2011). Mechanistically, both ULK1 and ULK2 have been shown to be recruited to isolation membranes upon autophagy induction, both are able to bind to the regulatory proteins Atg13 and FIP200, and their kinase-dead mutants have dominant negative properties (can block autophagy) when overexpressed in cells (Chan et al. 2009; Hara et al. 2008; Jung et al. 2009). Therefore, both ULK1 and ULK2 are the functional homologs of yeast Atg1, and the initial observation for a predominant autophagic role of ULK1 observed in HEK293 cells might simply be due to the much higher expression of ULK1 protein in these cells compared to that of ULK2. Additionally, it is highly likely that the existence of two Atg1 homologs provides additional mechanisms for differential regulation of autophagy in mammals.
ULK1 and ULK2 share significant homology in their C terminal regions in addition to the N-terminal kinase domains. The role of the C terminal regions of ULK1 and ULK2 in autophagy is intriguing. Domain studies of ULK1 have shown that its C terminal domain (CTD) is required for both interaction with Atg13/FIP200 (two regulatory proteins of ULK1, see later for more detail) and translocation of ULK1 to phagophores (Chan et al. 2007, 2009; Hara et al. 2008). Mammals have three additional protein kinases that share high homology with the kinase domains of ULK1 and ULK2. They are ULK3, ULK4, and STK36 (also known as “fused”). These additional ULKs do not have the conserved C terminal sequence and are not thought to be involved in starvation-induced autophagy. It is, however, possible that they may play a role in other forms of autophagy. In support of this, ULK3 has been recently linked to autophagy induction during senescence (Young et al. 2009).
3.2 The Atg1 Complex and the ULK1 Complex
In yeast, Atg1 has been reported to interact with up to eight other Atg proteins, some of which are specific to autophagy (Atg13, 17, 29, 31), while others are required for the Cvt pathway (Atg11, 20, 24 and vac8) (Mizushima 2010). Both pathways are morphologically distinct although molecularly they share many of the same Atg proteins. It is thought that Atg1 may function as a switch between autophagy and the Cvt pathway through changes in posttranslational modifications or interacting partners. Atg17, 29, and 31 form a stable complex regardless of nutritional status, whereas recruitment of Atg13 and Atg1 to the complex is triggered by nutrient starvation (Fig. 4.3) (Kabeya et al. 2005, 2009; Kawamata et al. 2008). Under nutrient-rich conditions, Atg13 is hyper-phosphorylated by the nutrient-sensing kinase TOR, thus cannot interact with Atg1. Conversely, upon nutrient-starvation, TOR is inactivated, leading to dephosphorylation of Atg13 and subsequent Atg13–Atg1 interaction and autophagy activation. Therefore, the regulated interaction between Atg13 and Atg1 dictates the autophagic function of Atg1 and the Atg1 complex. Further, Atg1 kinase activity was reported to increase during starvation and require Atg13 and Atg17 for maximal activation (Kabeya et al. 2005; Kamada et al. 2000).
Regulation of the Atg1 complex in yeast and the ULK1 complex in mammalian cells. In yeast, TOR phosphorylates Atg13 under nutrient-rich conditions, preventing complex formation (for simplicity, other protein components are not shown here). During starvation, TOR is inactivated, thus the inhibitory phosphorylation on Atg13 is removed, triggering complex formation with Atg1 and Atg17. In mammals, the ULK1 complex is stable even under nutrient-rich conditions. Inhibitory phosphorylation by mTOR on ULK1 and Atg13 prevent complex activation, likely through a specific conformational change of the ULK1 complex
In mammalian cells, there appears to be no equivalent of the yeast Cvt pathway. The main autophagic ULK1 complex consists of ULK1, Atg13, and FIP200 to mediate the signal of nutrient-sensing kinase mTOR, as discovered by the laboratories of Mizushima (Hara et al. 2008), Jiang (Ganley et al. 2009), and Kim (Jung et al. 2009). Mammalian Atg13 possesses mild homology with C. elegans Atg13, but its similarity with yeast Atg13 is rather limited (Chan et al. 2009; Ganley et al. 2009; Meijer et al. 2007); this is perhaps the reason why mammalian Atg13 was a relatively late discovery. FIP200 does not have an obvious sequence homolog in yeast, but functional analysis suggests it is the functional counterpart of yeast Atg17. Complex formation with Atg13 and FIP200 appears to promote the stability of ULK1 as a decrease in ULK1 protein levels were detected in Atg13−/− cells and cells with Atg13 or FIP200 knockdown (Hara et al. 2008; Ganley et al. 2009; Hosokawa et al. 2009a). In yeast, Atg13 is thought to mediate the interaction between Atg1 and Atg17 as Atg1 could interact with Atg13 in Atg17-null cells but not with Atg17 in Atg13-null cells (Kabeya et al. 2005; Cheong et al. 2005). In mammalian cells, Atg13 has been proposed to enhance the interaction between ULK1 and FIP200 as well (Hosokawa et al. 2009a). However, FIP200–ULK1 interaction, as detected by co-immunoprecipitation, was not affected in cells with Atg13 knockdown compared to control-depleted cells. Further, ULK1 could interact with FIP200 or Atg13 in an in vitro binding assay, even in the absence of the other, demonstrating that ULK1 could interact directly with FIP200 in a manner independent of Atg13, and vice versa (Ganley et al. 2009). Additionally, similar to the yeast scenario, both Atg13 and FIP200 can enhance the kinase activity of ULK1 (Jung et al. 2009; Ganley et al. 2009).
Albeit functioning in a highly similar fashion, regulation of the yeast Atg1 complex and that of the mammalian ULK1 complex also has obvious dissimilarity (Fig. 4.3). Perhaps the most striking difference is that under both nutrient-rich and nutrient-starved conditions, the majority of ULK1 exists as part of a macromolecular complex containing Atg13 and FIP200 (Ganley et al. 2009; Hosokawa et al. 2009a). While starvation-induced Atg13 dephosphorylation and subsequent Atg1–Atg13 interaction provide a simple mechanism for controlling the autophagy function of Atg1 in yeast, how does dephosphorylation of mammalian Atg13 turn on ULK1 since the ULK1–Atg13–FIP200 complex formation is constitutive? Can the phosphorylation status of Atg13 influence the conformation of the ULK1 complex, although not affecting complex formation? Is the ULK1 complex subjected to more complex regulation through other posttranslational modifications or interaction with other proteins? Do Atg13 and FIP200 have other functions beyond stabilizing ULK1 and enhancing its kinase activity, such as acting as scaffolds for recruitment of other proteins including the ULK1 substrate(s) whose phosphorylation by ULK1 is required for autophagy? Indeed, several other interacting partners of ULK1, some of which do not have putative homologs in baker’s yeast, have been reported, such as Atg101 (Hosokawa et al. 2009b; Mercer et al. 2009), Ambra1 (Di Bartolomeo et al. 2010b), HSP90-CDC37 (Joo et al. 2011), SynGAP and syntenin (Tomoda et al. 2004), GABARAP (Okazaki et al. 2000), and FEZ1 (McKnight et al. 2012). It is yet to be firmly demonstrated whether and/or how these interactions are relevant to the role of ULK1 in autophagy.
4 Sensing of Upstream Signals by the ULK1 Complex
Previous work in both yeast and mammalian systems suggest that the ULK1 complex senses upstream signals, particularly nutrient and energy signals, mainly via changes in its phosphorylation status. However, this does not appear to be the complete story. Recent studies demonstrate the significance of other regulatory mechanisms, which provide additional versatility for cells to control the intensity and timing of ULK1-mediated autophagy (Fig. 4.4). In this section, we discuss the current knowledge concerning how the ULK1 complex responds upon upstream stimulation. It should be emphasized that in order to monitor the effect of an upstream signal to the ULK1 complex, one should not only measure the eventual autophagy, but also the direct impact on the ULK1 complex per se, such as stimulation of ULK1 kinase activity and translocation of the ULK1 complex to isolation membranes (Ganley et al. 2009).
The ULK1 complex integrates upstream nutrient and energy signals to coordinate the induction of autophagy. The nutrient sensing pathways for growth factors, amino acids and energy converge on ULK1 through unique posttranslational modifications, which regulate the activity of this autophagy-induction complex. mTOR inhibits the induction of autophagy through phosphorylation of ULK1 and Atg13, which undergo a global dephosphorylation upon starvation. The acetyl-transferase TIP60, which is regulated by the growth factor-sensitive kinase GSK3, catalyzes acetylation of ULK1 to bolster its kinase activity. The energy-sensing kinase AMPK can promote autophagy through inhibition of mTOR; however, the role of AMPK phosphorylation on ULK1 is unknown. Dark blue arrows indicate events that promote autophagy while light blue arrows indicate events that are inhibitory to autophagy. The grey arrow between AMPK and ULK1 reflects that it is still unclear if AMPK phosphorylation of Ulk1 activates or inhibits autophagy
4.1 Phosphorylation
In mammalian cells, in addition to Atg13, ULK1 is also hyper-phosphorylated in nutrient-rich conditions and undergoes global dephosphorylation upon starvation. At least 30 phosphorylation sites have been identified on ULK1, although the majority of the responsible kinases and the functions of these phosphorylation events remain to be identified (Mack et al. 2012). Regardless, this suggests that phosphorylation is an important mode of ULK1 regulation.
4.1.1 Phosphorylation by mTOR
As a major nutrient/energy sensor, mTOR has been reported to directly phosphorylate ULK1 and Atg13. The mTOR complex 1 (mTORC1), consisting of mTOR, Raptor and mLST8, is responsive to growth factors and amino acid levels and has been shown to phosphorylate both ULK1 and Atg13 (Jung et al. 2009; Ganley et al. 2009; Hosokawa et al. 2009a). The mTORC1 complex binds to ULK1 directly through Raptor under nutrient-rich conditions and dissociates from ULK1 complex upon starvation (Hosokawa et al. 2009a; Lee et al. 2010). mTOR-driven ULK1 phosphorylation correlates with autophagy inhibition and weaker ULK1 kinase activity (Jung et al. 2009; Ganley et al. 2009; Hosokawa et al. 2009a). It is unclear if mTOR association occurs in the cytosol or on certain autophagosome precursor membrane structures. mTOR phosphorylation events may influence the localization of the ULK1 complex and exert its inhibitory effects by sequestering or physically separating the ULK1 complex from its enzymatic substrates. In yeast, Atg13 phosphorylation by TOR under nutrient-rich conditions inhibits the assembly of Atg1 complex and recruitment to the pre-autophagosomal structure (PAS) (Kamada et al. 2000). Inhibition of TOR by rapamycin also stimulates dimerization/oligomerization of Atg1 in an Atg13-dependent manner, which likely leads to subsequent activation of Atg1 kinase activity (Yeh et al. 2011). Importantly, Ohsumi and colleagues showed that an Atg13 mutant defective in phosphorylation by TOR was sufficient to trigger autophagy in yeast in under nutrient-rich conditions (Kamada et al. 2010). On the other hand, the functional significance of mTOR phosphorylation sites on mammalian Atg13 and ULK1 are less clear as complex formation is not dependent upon mTOR inactivation, and the phosphorylation sites of mammalian Atg13 by mTOR have not been completely mapped, making mutational analysis similar to that performed in yeast impossible at this stage. Furthermore, as ULK1 is also under regulation by mTOR, it is possible that preventing mTOR-catalyzed Atg13 phosphorylation alone will not be sufficient for autophagy activation.
4.1.2 Phosphorylation by PKA
In yeast, protein kinase A (PKA) was also shown to phosphorylate Atg1 and Atg13 (Budovskaya et al. 2005; Stephan et al. 2009). PKA phosphorylation had no effect on Atg1 kinase activity but appeared to regulate its association with the PAS, because phospho-mutants of PKA sites were constitutively located at the PAS (Budovskaya et al. 2005). Inhibition of PKA was sufficient to induce autophagy, indicating that TOR and PKA regulated autophagy through distinct pathways (Stephan et al. 2009). It remains to be seen whether mammalian PKA regulates ULK1. In Arabidopsis thaliana, nutrient starvation induces dephosphorylation and protein turnover of both Atg1 and Atg13. It is likely that this regulation might function as a negative feedback mechanism to tightly connect autophagy with nutritional status in the cell (Suttangkakul et al. 2011).
4.1.3 Phosphorylation by AMPK
It has been known for some time that another important cellular energy sensor, AMPK, can play a role in autophagy induction by phosphorylating TSC2 and Raptor to inactivate mTOR, hence indirectly activating the ULK1 complex. Recently, AMPK has also been shown to directly interact with and phosphorylate ULK1 in a nutrient-dependent manner (Lee et al. 2010; Kim et al. 2011; Shang et al. 2011; Egan et al. 2011). Several phosphorylation sites on ULK1 have been mapped by several groups and in some cases attributed to either mTOR or AMPK, but with very few overlapping sites between the groups (Kim et al. 2011; Shang et al. 2011; Egan et al. 2011; Dorsey et al. 2009). Among the phosphorylation sites reported, Ser555, Ser637 and Ser757 were reported by three or more independent groups. AMPK phosphorylation site Ser555 is thought to recruit phospho-binding protein 14-3-3 to the ULK1 complex (Lee et al. 2010; Egan et al. 2011). mTOR site Ser757 is required for AMPK binding as its mutation disrupts ULK1–AMPK interaction (Kim et al. 2011; Shang et al. 2011). The effect of AMPK phosphorylation on the ULK1 complex and autophagy are not well established, with conflicting reports on whether it leads to stimulation or inhibition of autophagy. The study by Guan and colleagues (Kim et al. 2011) suggests that phosphorylation of AMPK sites on ULK1 are stimulated by glucose starvation, contributing to ULK1 activation. In this case, ULK1 kinase activity (measured by ULK1 autophosphorylation) increased upon glucose starvation and correlated with AMPK activation. However, the study by Shang et al. (2011) identified AMPK sites that were dephosphorylated upon amino acid starvation; phospho-mutants defective in AMPK binding exhibited faster protein degradation upon autophagy stimulation, leading them to the conclusion that AMPK phospho-sites may be inhibitory to autophagy induction. The differences in their observations might reflect the distinct autophagy role of AMPK when sensing different triggers (i.e., glucose starvation versus amino acid starvation); they could also be due to the monitoring of different phosphorylation sites in these two studies. In yet another study by Shaw and colleagues, AMPK regulation of ULK1 was linked to mitophagy (Egan et al. 2011). This study identified four ULK1 sites that were phosphorylated upon treatment with the AMPK activators metformin and phenformin. AMPK-deficient primary mouse hepatocytes accumulated aberrant mitochondria, suggesting a mitophagy defect. ULK1 with compound phospho-deficient mutations at these AMPK sites could not reconstitute starvation-induced p62 degradation in ULK1−/− mouse embryonic fibroblasts (MEFs), suggesting that the AMPK phosphorylation on ULK1 was required for ULK1 function in autophagy. However, the fact that AMPK−/− MEFs can still undergo autophagy, rules out an essential autophagic role for this kinase (at least in response to amino acid starvation) (Mack et al. 2012; Kim et al. 2011). It is possible that AMPK fine-tunes ULK1 activity and the subsequent autophagy outcome in response to various energy requirements.
4.1.4 Feedback Regulation and Dephosphorylation
Intriguingly, there have also been reports on the reverse, i.e., ULK1 regulation of mTOR and AMPK. ULK1 was reported to phosphorylate Raptor in vitro to negatively regulate mTOR activity. In cells, knockdown of ULK1 resulted in increased mTORC1 signaling as assessed by the phosphorylation of mTORC1 substrates (Dunlop et al. 2011; Jung et al. 2011). This could constitute a feed forward mechanism that ensures rapid shutdown of mTOR signaling during autophagy induction and maintenance of mTOR inhibition during nutrient-limiting conditions. Interestingly, FIP200 is also reported to regulate mTOR through its interaction with TSC1 (Gan et al. 2005, 2006). Overexpression of FIP200 alone had no effect on TSC1−/− MEFs, but in wild-type MEFs it caused an increase of S6K phosphorylation which correlated with a mild (5 %) increase in average cell size. It is unclear if ULK1 is involved in the FIP200 regulation of mTOR or vice versa. However, ULK1 and FIP200 appear to have opposite regulatory effects on mTOR and are likely to represent distinct mechanisms, as ULK1 inhibition of mTOR activity could still be observed in TSC2−/− MEFs (Jung et al. 2011). In terms of AMPK, ULK1 has been reported to be able to phosphorylate all three AMPK subunits (Löffler et al. 2011). ULK1 phosphorylation was found to be inhibitory to AMPK activity and was proposed to be a form of negative feedback to dampen autophagy-induction signals.
Recent work identified another form of feedback regulation from autophagy to mTOR. A complete autophagy flux leads to recycling of amino acids from lysosomes to the cytoplasm, which causes reactivation of mTOR (Yu et al. 2010). Reactivation of mTOR is not only able to inhibit excessive autophagy through phosphorylating Atg13 and ULK1 but is also required for reformation of lysosomes (Yu et al. 2010).
Dephosphorylation of ULK1 complex components upon autophagy activation requires more than just suppressing their protein kinases such as mTOR, yet the protein phosphatase(s) involved in ULK1 or Atg13 dephosphorylation is currently unknown, let alone whether these phosphatases are regulated in a coordinated manner. The phosphatase inhibitor okadaic acid has long been described as an inhibitor of autophagy (Blankson et al. 1995; Samari et al. 2005; Bánréti et al. 2012), though its mechanism of action is not known. It may function through inhibiting dephosphorylation of inhibitory phospho-sites, such as the mTOR sites in the ULK1 complex (Shang et al. 2011). Contrary to the reported effects of okadaic acid in eukaryotic cells, a yeast study reported Tap42-PP2A, a target of okadaic acid, as a negative regulator of autophagy, although it did not identify the substrate of the phosphatase (Yorimitsu et al. 2009). Further, a recent screen of PP2A subunit mutant fly strains identified two PP2A complexes required for starvation-induced autophagy in Drosophila larval fat body cells and proposed the dAtg1 complex to be a target of PP2A regulation (Bánréti et al. 2012).
4.2 Acetylation of ULK1 and Other Autophagy Proteins
Aside from phosphorylation, acetylation is another posttranslational modification that has recently been found to play a role in autophagy (McEwan and Dikic 2011; Yi et al. 2012). Acetylation involves the transfer of acetyl groups from acetyl-CoA to the lysine residue of the target protein. The modification regulates diverse processes and has been shown to have an effect on protein–DNA interactions, protein–protein interactions, subcellular localization and protein stability (Deribe et al. 2010). Acetyl-CoA is also a critical building block for cellular metabolism, and as such, protein acetylation/deacetylation has been implicated as a critical regulatory mechanism for metabolism (Wellen and Thompson 2012). Given the catabolic nature of autophagy, regulation of autophagy by acetylation is thus an attractive hypothesis. Protein acetylation/deacetylation was initially implicated in autophagy by studies of Jiang and colleagues using the histone deacetylase (HDAC) inhibitor suberoylanilide hydroxamic acid (SAHA) (Shao et al. 2004; Gammoh et al. 2012). It was found that SAHA induced autophagy in various cell types in a ULK-dependent manner. However, the role of SAHA in maintaining the acetylation status of Atg proteins was not determined. More recently, a systematic genetic analysis of histone acyltransferase (HAT) complexes in yeast, conducted by Yu and colleagues, identified Esa1, the catalytic subunit of HAT complex NuA4, to be required for autophagy. By a process of elimination, Atg3 was deemed to be the substrate of Esa1. Acetylation mutants of Atg3 could not bind to Atg8 and failed to reconstitute Atg3-null cells, indicating that the acetylation event is required for autophagy (Yi et al. 2012). In a separate study by Lin and colleagues, TIP60, the mammalian homologue of Esa1, was reported to acetylate ULK1. Knockdown of TIP60 impaired starvation induced autophagy (Yi et al. 2012; Lin et al. 2012) and acetylation mutants of ULK1 could not rescue LC3 conversion in ULK1−/− MEFs. Further, TIP60 activation was regulated by glycogen synthase kinase-3 (GSK3), a protein whose activity is responsive to growth factor withdrawal through PI3K/AKT/mTOR signaling pathways (Lin et al. 2012), thus directly linking the acetylase TIP60 to nutrient sensing and ULK1-mediated autophagy.
4.3 ULK1 Ubiquitination
Autophagy and proteasome-mediated protein degradation are considered to be the two major mechanisms for cellular protein turnover. Therefore, conceptually it is not a surprise to note that these two processes intimately communicate with each other (Korolchuk et al. 2009a, b, 2010; Gao et al. 2010). Further, both autophagy and proteasome-mediated protein degradation are subjected to a common posttranslational regulatory mechanism, ubiquitination. For example, ubiquitination has been established as a critical event for autophagosomes to recognize and recruit specific cargos, including protein aggregates, mitochondria, and invading pathogens (Korolchuk et al. 2010; Kraft et al. 2010).
Recent evidence suggests that ubiquitin-mediated proteasomal degradation also targets the components of the ULK1 complex. There is evidence of direct ubiquitination of ULK1 in response to nerve growth factor in neurons, although the significance of this with respect to autophagy is unknown (Zhou et al. 2007). Ubiquitination is also alluded to as part of the HSP90-CDC37 regulation of ULK1. Kundu and colleagues reported that treatment with the HSP90 antagonist 17AAG, leads to a decrease in ULK1 protein levels that can be rescued by co-treatment with the proteasomal inhibitor MG132 (Joo et al. 2011). Interestingly, ULK2 is not an HSP90 client, suggesting that although these two proteins are functionally redundant, they may be subjected to distinct regulation. Additionally, Atg13 protein level is stabilized by ULK1, ULK2, and Atg101 (Mercer et al. 2009; Joo et al. 2011). In the absence of Atg101, Atg13 protein levels decrease, and such decrease can be blocked by the proteasomal inhibitor MG132 (Mercer et al. 2009).
Interestingly, it has been observed that upon amino acid starvation, activation of the ULK1 complex is accompanied by a concurrent decrease of ULK1 protein level. The functional relevance of this protein level change is not clear. It is possible that the decrease in ULK1 is a feedback mechanism to ensure that autophagy can be effectively turned off once nutrition becomes available. Further study is needed to determine the biological role of starvation-associated ULK1 decrease and whether it is through ubiquitin-mediated degradation. If so, this will represent an elegant mechanism through which a massive autophagic degradation is controlled by specific proteasomal degradation of an autophagy component.
5 Relaying the Nutrient Signals by the ULK1 Complex
Knockout mice of autophagy essential genes such as Atg3, 5, or 7 do not have gross developmental defects but die within a day after birth, because the animals are unable to survive the neonatal starvation period (Kuma et al. 2004; Komatsu et al. 2005; Sou et al. 2008). This phenotype seems to be the hallmark of autophagy-specific and essential genes. While siRNA knockdown of ULK1 is sufficient to block starvation-induced autophagy in multiple cell types, ULK1−/− mice show a relatively mild autophagy phenotype, with delayed red blood cell maturation (mitophagy) but are viable (Kundu et al. 2008). This has been attributed to the redundancy between ULK1 and the closely related ULK2. Indeed, ULK1−/−Ulk2−/− mice (Cheong et al. 2011), as well as Atg13−/− mice (Shang et al. 2011), have the same neonatal death phenotype as other Atg knockout mice. FIP200−/− mice die between embryonic day 13.5 and 16.5 due to defects in heart and liver development, reflecting its involvement in other processes outside of autophagy (Gan et al. 2006).
The ULK1 kinase complex has been suggested to function in the initial stages of the canonical autophagy pathway. However, the exact functional relationship between the ULK complex and other Atg proteins is still not completely defined and many controversial studies have been reported. Related to this issue, although the kinase activity of Atg1/ULK1 is required for autophagy, the protein substrate(s) of ULK1 that mediates its autophagic function has not been identified; and whether Atg1/ULK1 also has kinase-independent functions in autophagy is not clear. Therefore, to understand how the ULK1 complex relays upstream nutrient signals to the downstream autophagy pathway, these crucial questions need to be solved.
5.1 Signaling Between the ULK1 Complex and Other Upstream Atg Complexes
In addition to the ULK1 complex, other Atg components that function upstream of LC3 conjugation to phosphatidylethanolamine (PE) include the VPS34 complex and the Atg12–Atg5–Atg16 complex. As the ULK1 complex directly senses mTOR activity, if the autophagy “cascade” proceeds in a strictly linear fashion, it is logical to place the ULK1 complex at the most upstream position in the autophagy pathway, at least for amino acid starvation-induced autophagy. While mounting evidence supports this view, such as the recent imaging-based hierarchy study by Mizushima and colleagues (Itakura and Mizushima 2010b), there are also conflicting results suggesting that the autophagy pathway may not be a simple linear process.
5.1.1 Signaling from the ULK1 Complex to VPS34 Complex
VPS34 is a lipid kinase, and its enzymatic product phosphatidylinositol 3-phosphate (PI3P) is required, directly or indirectly, for the recruitment of multiple autophagy components (WIPI-1, DFCP1, Atg5 and LC3) to forming autophagosomes (Matsunaga et al. 2010; Polson et al. 2010). Like the ULK1 complex, the VPS34 complex has been proposed as a major point of regulation for autophagy induction. For example, the autophagic function of the VPS34 complex can be closely controlled through dynamic interaction of the VPS34 component Beclin 1 with its inhibitory binding partners, the Bcl-2 family of proteins Bcl-2, Bcl-xL, and Bim (Liang et al. 1999; Pattingre et al. 2005b; Luo et al. 2012).
As both the ULK1 and VPS34 complexes need to be activated to initiate autophagy, their regulation must be coordinated. It has been suggested that recruitment of the VPS34 complex to autophagic membranes is dependent on ULK1, thus placing the ULK1 complex upstream of the VPS34 complex. The major evidence for this claim is that (1) in FIP200−/− cells, starvation failed to induce the VPS34 complex to form punctate structures; and (2) inhibition of the kinase activity of VPS34 cannot completely prevent starvation-induced ULK1 translocation (Itakura and Mizushima 2010b). Recent work lends mechanistic insights into ULK1–VPS34 communication: in response to nutrient starvation, AMBRA1, a binding partner of the VPS34 complex, is reported to be phosphorylated in a ULK1-dependent manner (Di Bartolomeo et al. 2010b). This phosphorylation releases the AMBRA1–VPS34 complex from dynein and the microtubule network, freeing the complex to translocate to autophagy initiation sites on the endoplasmic reticulum [it should be noted that AMBRA1 is not the only link between ULK1/Atg1 and cytoskeletal motors: in Drosophila, Atg1 could activate myosin II to help drive autophagosome formation; and in mammalian cells this interaction appeared to regulate Atg9 trafficking (Tang et al. 2011)]. Albeit a reasonable explanation for ULK1/Atg1–VPS34 connection, because AMBRA1 does not have a functional counterpart in yeast, it is unlikely to be the universal mediator for these two autophagy complexes.
Although current evidence supports a unidirectional signaling flow from the ULK1 complex to VPS34 complex, this model is likely to be oversimplified, and additional crosstalk should be considered. Particularly, experimental outcomes using the pharmacological agent wortmannin should be interpreted with caution. In addition to VPS34, wortmannin can inhibit other PI3 kinases as well, and thus might impact autophagy and the behavior of the ULK1 complex via multiple mechanisms. Further, wortmannin treatment often results in formation of large amounts of intracellular vesicles. Therefore, what exactly is the nature of ULK1 puncta upon wortmannin treatment? And can knockout of Atg14 or other components of the VPS34 complex recapitulate the effect of wortmannin, i.e., inhibiting starvation-induced LC3 conjugation but not ULK1 puncta formation?
5.1.2 Signaling from the ULK1 Complex to Atg5 Complex
The functional relationship between the ULK1 complex and Atg5 complex is equally complex; for example, FIP200 knockout can block Atg5 puncta formation and conversely Atg5 knockout can also block ULK1 puncta formation (Hara et al. 2008). A somewhat puzzling observation is that starvation could induce ULK1 puncta formation in Atg5−/− cells only in the presence of wortmannin (Itakura and Mizushima 2010b). This observation led to the model that ULK1 is the first complex recruited to the autophagosome formation site, preceding the Atg5 complex; and the requirement for wortmannin is attributed to the potential transient nature of ULK1 localization on autophagic membranes which may be stabilized by blocking the immediate downstream event, the function of the VPS34 complex (Itakura and Mizushima 2010b). On the other hand, alternative possibilities should be considered here: if indeed ULK1 localization on the autophagic membrane is so transient and can only be observed in Atg5−/− cells in the presence of wortmannin, why can it be readily observed in wild-type cells in the absence of wortmannin? And again, what exactly is the nature of ULK1 puncta upon wortmannin treatment?
It is possible that all these puzzling observations of imaging analysis are the outcomes of the limited optical resolution, i.e., even if there is absolute autophagic membrane translocation of a certain Atg protein, when the membrane structure is too small (e.g., at the very early phase of phagophore nucleation), such translocation events will not be able to be monitored microscopically. For the phagophore to grow in size upon starvation, the biochemical conjugation of LC3 to PE is required. Starvation-stimulated LC3 conjugation is dependent on the ULK1 complex, VPS34 complex and Atg5, thus explaining the inter-dependence of the “translocation” of these three complexes in imaging-based analyses. Regardless, given the likely upstream requirement of ULK1 for localization of other autophagy components and the apparent fact that ULK1 is a mediator of multiple upstream kinases, it is clear that the prime function of ULK1 may be as a node to convert incoming signals into autophagosomes.
5.2 The ULK1 Complex and Atg9 Cycling
Atg1/ULK1 has been implicated in Atg9 cycling in both yeast and mammalian cells, as reported independently by Klionsky, Tooze, and Ohsumi (Young et al. 2006; Reggiori et al. 2004; Sekito et al. 2009). In yeast, Atg9 cycles between the PAS and peripheral sites. Atg1 is required for Atg9 cycling and it is thought to be recruited to the PAS through interaction with Atg17 (Sekito et al. 2009). The role of ULK1 in Atg9 cycling in mammalian cells is not as defined. While earlier reports showed that knockdown of ULK1 blocked starvation-induced redistribution of Atg9 (Young et al. 2006), recent work based mostly on confocal imaging suggests that Atg9 localizes to membrane structures adjacent to early autophagy markers such as ULK1, DFCP1, and WIPI2, and is recruited independently of ULK1 (Orsi et al. 2012). In a model for Parkinson’s disease, Atg9-positive structures could still be recruited to damaged mitochondria in FIP200−/− MEFs. Likewise, ULK1-positive punctate structures could be observed in Atg9−/− MEFs, indicating that recruitment of Atg9 and ULK1 to membrane structures were independent of each other (Itakura et al. 2012). Similarly, independent recruitment of Atg9 and ULK1 was observed in Salmonella-induced xenophagy in Atg9−/− and FIP200−/− MEFs (Kageyama et al. 2011). It is noteworthy that GFP-LC3 and GFP-Atg5 but not GFP-WIPI or GFP-ULK1 could still be recruited to salmonella-containing vacuoles in FIP200−/− MEFs. However, the growth of salmonella was not suppressed in FIP200−/− MEFs, indicating a failure to complete Xenophagy to achieve bacterial clearance in these cells.
5.3 Potential Signaling Role of the ULK1 Complex in Downstream Events
While ULK1 is traditionally viewed as orchestrating early events in autophagosome formation, an Atg1–Atg8 interaction was recently reported in two independent yeast studies, suggesting a role for Atg1 in late stages of autophagy as well. The Atg8 interaction targets Atg1 to the vacuole for degradation. Expression of an Atg1 mutant that cannot interact with Atg8 resulted in an autophagy defect and reduced the accumulation of autophagic bodies in vacuolar protease-deficient cells, indicating that the interaction promotes production of fully formed autophagosomes (Nakatogawa et al. 2012; Kraft et al. 2012). In addition, Kraft et al. showed that ULK1 also interacted with mammalian Atg8s and an Atg8-binding mutant of ULK1 showed reduced recruitment to autophagosomes. However, the authors did not show that this interaction resulted in lysosomal turnover of ULK1 or that it was essential for functional autophagy (Kraft et al. 2012). Prior to these studies, mammalian ULK1 was reported to interact with the LC3-related proteins GATE16 and GABARAP, and to a lesser extent LC3 in a yeast two hybrid screen of a human fetal brain cDNA library. While the interaction was validated by co-immunoprecipitation experiments in cells, the functional involvement of this interaction in autophagy was not addressed (Okazaki et al. 2000; Behrends et al. 2010). Taken together, it is possible that ULK1 may have a role in later stages of the autophagy pathway in the mammalian system as well. Technically, to confirm such a role of ULK1 in cells could prove to be difficult, unless distinct mutants of ULK1 can be created to precisely dissect the upstream and downstream functions of ULK1.
5.4 The Kinase Substrate(s) of ULK1
Although the protein kinase activity of Atg1/ULK1 has been demonstrated to be essential for its autophagy function, the relevant substrate(s) has yet to be identified. This substrate should meet the following criteria: its phosphorylation by ULK1 is enhanced upon amino acid starvation; and blocking such phosphorylation (for example by mutating the phosphorylation site) will prevent ULK1-dependent autophagy. The identity of this substrate and the mechanism by which it mediates the autophagy activity of ULK1 might reveal how the ULK1 complex relays the upstream signals to the downstream autophagy pathway, including the puzzling relationship between the ULK1 complex and VPS34 or Atg5.
To date, it has been shown that ULK1 can phosphorylate itself as well as its regulatory proteins Atg13 and FIP200 (Chan et al. 2009; Jung et al. 2009; Ganley et al. 2009). Auto-phosphorylation in the activation loop of Atg1 correlated with enhanced Atg1 kinase activity in yeast (Yeh et al. 2010; Kijanska et al. 2010). Mutation of these sites disrupted starvation-induced autophagy without affecting Atg1 complex formation or localization at the PAS (corresponding sites in ULK1 have not been identified). Therefore, it is most likely that additional substrate(s) of Atg1/ULK1 accounts for its autophagic function. Large scale phospho-proteomic studies (Ptacek et al. 2005) and Atg1 consensus site mapping (Mok et al. 2010) will help the search for this critical component. It is also likely that Atg13 or FIP200 is required for recruiting ULK1 substrate(s) in a phosphorylation (by mTOR and/or ULK1)-regulated manner.
5.5 The Potential Kinase-Independent Autophagic Function of ULK1
Although the kinase activity of Atg1/ULK1 is indispensable for its autophagic function, Atg1/ULK1 might also possess additional kinase-independent functions in autophagy. In yeast, Atg9 cycling is disrupted in Atg1-null mutants but is unaffected by loss of Atg1 kinase activity, suggesting a kinase independent function for the Atg1 complex (Reggiori et al. 2004). Also supporting this possibility is the observation that Atg1-null yeast fail to recruit Atg17 and Atg8 to the PAS, but yeast cells reconstituted with kinase-dead Atg1 mutants show abnormal accumulation of Atg17 and Atg8 at the PAS, suggesting that Atg1 may act as a scaffold to recruit Atg proteins to the PAS, with kinase activity being required only for subsequent steps (Cheong et al. 2008). In mammalian cells, kinase-dead ULK1 cannot rescue starvation-induced autophagy in ULK1−/− cells. A kinase-dead ULK1 mutant can still interact with Atg13 and FIP200 (Chan et al. 2009) but is unable to efficiently recruit downstream complexes such as WIPI and Atg5 (Hara et al. 2008). While the kinase-dead ULK1 complex does not form prominent punctate structures like its wild-type counterpart (Hara et al. 2008), it may still translocate from the cytoplasm onto an autophagosome precursor membrane structure that would be hard to detect if membrane elongation of the autophagosome cannot proceed without ULK1 kinase activity.
5.6 Non-autophagic Functions of the ULK1 Complex Components
5.6.1 ULK1
Before their roles in autophagy were characterized, ULK1 and FIP200 had been studied in other contexts. As its name implies, ULK1 was named due to its homology to the UNC-51 gene in C. elegans (Kuroyanagi et al. 1998; Yan et al. 1998). The C. elegans UNC-51 mutants are mostly paralyzed, reflecting a defect in axonal elongation, and they have aberrant accumulation of enlarged vesicles and other membranous structures in a subset of neuronal cells (Ogura et al. 1994; McIntire et al. 1992). The Drosophila homologue of UNC-51 is also reported to be important for neuronal development through mediation of vesicular transport in axons (Toda et al. 2008; Mochizuki et al. 2011). In mice, a role of mammalian homologues ULK1/2 in neuronal development has been shown for several neuronal populations, mostly in tissue culture systems (Tomoda et al. 1999, 2004; Zhou et al. 2007). ULK1 could be detected in cerebellar granule cells and had a punctate staining pattern along axons and growth cones of dorsal root ganglion neurons, indicating recruitment to membranous structures (Zhou et al. 2007; Tomoda et al. 1999). While it is currently unclear why neuronal defects have not been reported in ULK1−/− mice (possibly due to redundancy between members), the role of ULK1 in neuronal development appears to be highly conserved as murine ULK1 can be used to rescue the UNC-51 defect in C. elegans (Tomoda et al. 1999). In all three model systems, ULK1 kinase activity is required to fulfill its neuronal functions, with the kinase dead (K46R) ULK1 mutant having dominant negative effects on neurite extension (Ogura et al. 1994; Toda et al. 2008; Mochizuki et al. 2011; Tomoda et al. 1999).
There is currently no evidence that autophagy is required for axon elongation during neuronal development; an uncoordinated phenotype has not been reported for knockouts of other autophagy essential genes in C. elegans (Meléndez and Neufeld 2008). Likewise neuronal development is rarely specifically mentioned in Drosophila or mouse genetic knockout models addressing Atg gene function in the context of autophagy. No neuronal functions have been documented for other members of the autophagic ULK1 complex (Atg13, FIP200 or Atg101) either. Certainly autophagy is important in maintaining homeostasis in neurons, as exemplified by the mouse models for neurodegenerative diseases that utilize conditional and neural specific knockout of Atg genes such as Atg5, Atg7 and FIP200 (Liang et al. 2010; Komatsu et al. 2006; Hara et al. 2006). On the other hand, the requirement of autophagy in neural development, if any, remains murky. It is possible that the role of ULK1 in neuronal development represents a specialized function involving novel interacting partners and may be cell type-specific. Further studies are needed to clarify whether this documented neuronal role of ULK1 is because of its autophagy function.
5.6.2 FIP200/RB1CC1
Aside from its role in autophagy, FIP200 (FAK-interacting protein of 200-kDa) has been implicated in a diverse range of cellular processes. The functions of FIP200 have been reviewed elsewhere and are carried out mainly through switching among its numerous interacting partners which include PYK2, FAK, TSC1, ASK1, TRAF2, p53, and E3 ubiquitin ligase Arkadia (Gan and Guan 2008). Most interacting partners of FIP200 were discovered by yeast two-hybrid screens.
FIP200 has been reported to possess nuclear functions. The other alias for FIP200 is RB1CC1 (RB1-inducible coiled-coil 1) due to its ability to regulate the retinoblastoma tumor suppressor protein RB1 as well as p16 through direct transcriptional activation (Ochi et al. 2011; Ikebuchi et al. 2009). As mentioned for ULK1, the other reported functions of FIP200 may give us insights into the molecular mechanisms and role of FIP200 in the ULK1 complex and autophagy. For instance, FIP200 interacts with β-Catenin and can mediate its degradation independent of the classical APC destruction complex (Choi et al. 2012). It was also found to enhance proteasomal degradation of several negative regulators of TGFβ signaling by acting as a cofactor for the E3 ubiquitin ligase Arkadia (Koinuma et al. 2011). FIP200 may thus exert similar scaffolding functions to bind degradation factors for ULK1 which is also degraded during starvation. FIP200 can also bind to protein phosphatase 1 (PP1) (Meiselbach et al. 2006), although the biological function of this interaction has not been studied. One possible function may be to recruit phosphatases to act upon one or all of the ULK1 complex components, which have been shown to be regulated via phosphorylation–dephosphorylation.
6 Sensing ULK1-Independent Signaling by the Autophagy Pathway
LC3 conversion still occurs in Atg13−/−, FIP200−/−, and ULK1−/−ULK2−/− double knockout MEFs (Hara et al. 2008; Cheong et al. 2011; Shang et al. 2011), suggesting that the ULK1 complex is not essential for activation of the LC3 conjugation machinery and that there are other ULK1/2-independent ways for setting off the autophagy cascade (Fig. 4.5). However, it should be noted that LC3 conjugation and delivery to the lysosome is not the sole purvey of autophagosomes, given that LC3 conjugation can occur on non-autophagsosomal structures such as phagosomes and entotic vacuoles (detailed later). Recent work has shown that overnight glucose deprivation-induced LC3 conversion that was dependent on Atg5 yet did not require ULK1 or ULK2 (Cheong et al. 2011). The authors attributed this to the accumulation of ammonia as a result of glutaminolysis (glutamine degradation) within the cells, and they were able to recapitulate the effect by treating cell cultures with ammonium chloride. Prior to and consistent with their discovery, ammonia derived from glutamine degradation was reported to induce autophagy in an mTOR-independent manner (Eng et al. 2010). It should also be noted that glucose deprivation has been reported to stimulate autophagy in a ULK1-dependent manner (Kim et al. 2011). Unlike the ammonium scenario, this is a typical bioenergetic response involving mTOR suppression plus AMPK activation, which subsequently activates the ULK1 complex. Seemingly contradictory with each other, glucose starvation might induce autophagy via both mechanisms, depending on exact biological contexts, such as growth conditions and cell types.
Signaling sensing by the autophagy pathway via ULK1-dependent and independent mechanisms. The role of the ULK1 complex in amino acid starvation-induced autophagy is well established. There are, however, triggers for autophagy that appear to be independent of the ULK1 complex, such as lithium chloride and glucose starvation. This suggests that there are one or more pathways aside from ULK1 that can feed in to the downstream autophagy machinery to trigger the autophagic cascade and LC3 conjugation
Several other mTOR-independent autophagy inducers have been reported in the literature but did not address ULK1 dependency directly. These include amino sugars such as glucosamine and mannosamine, which were reported to induce LC3 conversion without dephosphorylation of mTOR substrates (Shintani et al. 2010). Lithium, which is studied more in the context of neurodegenerative diseases, also induces LC3 conversion and degradation of mutant huntingtin without mTOR inactivation (Sarkar et al. 2005, 2009). Typically these mTOR-independent autophagy inducers require long incubation times before significant LC3 conversion can be detected. If such mTOR-independent autophagy is indeed also independent of the ULK1 complex, it is possible then that a major “purpose” of the ULK1 complex is to trigger a more rapid autophagy response.
In DT40 chicken cells, knockout of Atg13, but not ULK1/2, abrogated autophagy upon amino acid starvation. Furthermore, only Atg13 splice variants that could interact with FIP200 could reconstitute autophagy, suggesting that the vital function of Atg13 in starvation-induced autophagy is independent of ULK1/2 but dependent on FIP200 in chicken cells (Alers et al. 2011). However, in this study and in contrast to mammalian cells, inhibition of TOR with rapamycin or Torin1, failed to induce autophagy in wild-type DT40 cells. Since knockout of ULK1/2 is sufficient to disrupt amino acid starvation-induced autophagy in mammalian cells, the regulation of autophagy might differ even among different vertebrates. Alternatively, it is possible that chicken cells may express another ULK1/2 homolog to complement ULK1/2 deletion.
Strikingly, the autophagy machinery is also utilized in certain cellular processes that are obviously not canonical autophagy (Florey and Overholtzer 2012). One such example is the membrane trafficking process phagocytosis, which, like autophagy, leads to lysosomal degradation of its specific cargo. However, in phagocytosis, extracellular particles in the form of cell debris or pathogens are engulfed to form an internal phagosome which then undergoes maturation and degradation (Sanjuan et al. 2009). Surprisingly, transient recruitment of GFP-LC3 to phagosomes was detected in macrophages when fed with beads coated with toll-like-receptor ligands (Sanjuan et al. 2007). This LC3-associated-phagocytosis (LAP) was dependent on VPS34 complex activity, Atg5, and Atg7. It was later found to be independent of ULK1 as ULK1−/− macrophages displayed similar levels of LC3 lipidation in response to phagocytosis of dead cells (Martinez et al. 2011). Another macro-endocytic process known as entosis is similar to phagocytosis but involves the engulfment of a live epithelial cell by another epithelial cell, resulting in the formation of an entotic vacuole. Morphologically, entosis is characterized by the formation of “cell-in-cell” structures and can be found in human cancers although its significance is currently not known (Overholtzer et al. 2007). The majority of cells that are engulfed by entosis undergo non-apoptotic cell death that is mediated by lysosomal fusion to the entotic vacuole (Overholtzer et al. 2007; Florey et al. 2011). Acidification of the entotic vacuole is preceded by LC3 recruitment that is dependent on VPS34, Atg5 and Atg7 but not FIP200 (Florey et al. 2011). The same study indicates that recruitment of LC3 to micropinosomes was not affected by FIP200 knockdown either, thus representing another process in which the autophagy machinery can be recruited to membranes independent of the ULK1 complex. This suggests that the ULK1 complex may be important in the recruitment of the autophagy machinery in the generation of de novo, double-membrane autophagosomes, but not in the recruitment of the machinery to preformed, single-membrane vesicular structures. On the other hand, these new findings also raise the question of how the canonical Atg proteins are hijacked to exert non-autophagic functions.
7 Concluding Remarks
Mounting evidence indicates that autophagy is regulated by a wide range of signal transduction pathways. Recent studies demonstrate that autophagy can also conversely modulate many of these pathways. However, the precise mechanisms underlying such two-way regulation have not been well defined, and how autophagy contributes to the biological outcomes of these signaling events is also poorly understood. Particularly, as autophagy might play either pro-survival or pro-death role, likely in a context-dependent manner, how do specific upstream signals instruct autophagy machinery to perform these obviously opposite functions? Among the myriad signaling-relevant topics, this chapter focuses on the ULK1 complex, an upstream integrating node in the autophagy pathway. Even for this specific area, many important questions remain to be answered. For example, how does the ULK1 complex sense upstream nutrient signaling from multiple molecules in addition to mTOR, and how does it subsequently relay the upstream signaling to the downstream autophagy machinery? Because of the recently discovered ULK-independent, non-canonical autophagy events, what are the signaling events that trigger autophagy machinery independent of the ULK1 complex, and what specifies or dictates whether autophagy should proceed in a ULK1 complex-dependent or -independent manner? Addressing these questions will provide insights into the molecular mechanisms of signal transduction in autophagy; it could also guide the exploration of the therapeutic potential of targeting autophagy pathway.
References
Alers S, Löffler AS, Paasch F, Dieterle AM, Keppeler H, Lauber K et al (2011) Atg13 and FIP200 act independently of Ulk1 and Ulk2 in autophagy induction. Autophagy 7:1423–1433
Bánréti A, Lukácsovich T, Csikós G, Erdélyi M, Sass M (2012) PP2A regulates autophagy in two alternative ways in Drosophila. Autophagy 8(4):623–636
Behrends C, Sowa ME, Gygi SP, Harper JW (2010) Network organization of the human autophagy system. Nature 466:68–76
Berry DL, Baehrecke EH (2007) Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell 131:1137–1148
Berry DL, Baehrecke EH (2008) Autophagy functions in programmed cell death. Autophagy 4:359–360
Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A et al (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171:603–614
Blankson H, Holen I, Seglen PO (1995) Disruption of the cytokeratin cytoskeleton and inhibition of hepatocytic autophagy by okadaic acid. Exp Cell Res 218:522–530
Budovskaya YV, Stephan JS, Deminoff SJ, Herman PK (2005) An evolutionary proteomics approach identifies substrates of the cAMP-dependent protein kinase. Proc Natl Acad Sci USA 102:13933–13938
Bursch W (2001) The autophagosomal-lysosomal compartment in programmed cell death. Cell Death Differ 8:569–581
Chan E, Kir S, Tooze S (2007) siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem 282:25464–25474
Chan E, Longatti A, McKnight N, Tooze S (2009) Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol Cell Biol 29:157–171
Chen Y, Klionsky DJ (2011) The regulation of autophagy—unanswered questions. J Cell Sci 124:161–170
Cheong H, Yorimitsu T, Reggiori F, Legakis JE, Wang CW, Klionsky DJ (2005) Atg17 regulates the magnitude of the autophagic response. Mol Biol Cell 16:3438–3453
Cheong H, Nair U, Geng J, Klionsky DJ (2008) The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol Biol Cell 19:668–681
Cheong H, Lindsten T, Wu J, Lu C, Thompson CB (2011) Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc Natl Acad Sci USA 108:11121–11126
Choi JD, Ryu M, Ae Park M, Jeong G, Lee JS (2012) FIP200 inhibits β-catenin-mediated transcription by promoting APC-independent β-catenin ubiquitination. Oncogene advance online publication; doi:10.1038/onc.2012.262
Deribe YL, Pawson T, Dikic I (2010) Post-translational modifications in signal integration. Nat Struct Mol Biol 17:666–672
Di Bartolomeo S, Corazzari M, Nazio F, Oliverio S, Lisi G, Antonioli M et al (2010) The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol 191:155–168
Dorsey F, Rose K, Coenen S, Prater S, Cavett V, Cleveland J et al (2009) Mapping the phosphorylation sites of Ulk1. J Proteome Res 8:5253–5263
Dunlop EA, Hunt DK, Acosta-Jaquez HA, Fingar DC, Tee AR (2011) ULK1 inhibits mTORC1 signaling, promotes multisite Raptor phosphorylation and hinders substrate binding. Autophagy 7:737–747
Edinger AL, Thompson CB (2004) Death by design: apoptosis, necrosis and autophagy. Curr Opin Cell Biol 16:663–669
Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W et al (2011) Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331:456–461
Eng CH, Yu K, Lucas J, White E, Abraham RT (2010) Ammonia derived from glutaminolysis is a diffusible regulator of autophagy. Sci Signal 3:ra31
Florey O, Overholtzer M (2012) Autophagy proteins in macroendocytic engulfment. Trends Cell Biol 22:374–380
Florey O, Kim SE, Sandoval CP, Haynes CM, Overholtzer M (2011) Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat Cell Biol 13:1335–1343
Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T (2008) The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell 19:2092–2100
Gammoh N, Lam D, Puente C, Ganley I, Marks PA, Jiang X (2012) Role of autophagy in histone deacetylase inhibitor-induced apoptotic and nonapoptotic cell death. Proc Natl Acad Sci USA 109:6561–6565
Gan B, Guan JL (2008) FIP200, a key signaling node to coordinately regulate various cellular processes. Cell Signal 20:787–794
Gan B, Melkoumian ZK, Wu X, Guan KL, Guan JL (2005) Identification of FIP200 interaction with the TSC1–TSC2 complex and its role in regulation of cell size control. J Cell Biol 170:379–389
Gan B, Peng X, Nagy T, Alcaraz A, Gu H, Guan JL (2006) Role of FIP200 in cardiac and liver development and its regulation of TNFalpha and TSC-mTOR signaling pathways. J Cell Biol 175:121–133
Ganley IG, du Lam H, Wang J, Ding X, Chen S, Jiang X (2009) ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem 284:12297–12305
Ganley IG, Wong PM, Gammoh N, Jiang X (2011) Distinct autophagosomal–lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol Cell 42:731–743
Gao Z, Gammoh N, Wong PM, Erdjument-Bromage H, Tempst P, Jiang X (2010) Processing of autophagic protein LC3 by the 20S proteasome. Autophagy 6:126–137
Gozuacik D, Kimchi A (2004) Autophagy as a cell death and tumor suppressor mechanism. Oncogene 23:2891–2906
Gutierrez MG, Munafo DB, Beron W, Colombo MI (2004) Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci 117:2687–2697
Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T et al (2007) The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem 282:37298–37302
Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R et al (2006) Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441:885–889
Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan J et al (2008) FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 181:497–510
Harding TM, Morano KA, Scott SV, Klionsky DJ (1995) Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol 131:591–602
Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y et al (2009a) Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 20:1981–1991
Hosokawa N, Sasaki T, Iemura S, Natsume T, Hara T, Mizushima N (2009b) Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 5:973–979
Hutagalung AH, Novick PJ (2011) Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev 91:119–149
Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N et al (2000) A ubiquitin-like system mediates protein lipidation. Nature 408:488–492
Ikebuchi K, Chano T, Ochi Y, Tameno H, Shimada T, Hisa Y et al (2009) RB1CC1 activates the promoter and expression of RB1 in human cancer. Int J Cancer 125:861–867
Itakura E, Mizushima N (2010) Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 6:764–776
Itakura E, Kishi-Itakura C, Koyama-Honda I, Mizushima N (2012) Structures containing Atg9A and the ULK1 complex independently target depolarized mitochondria at initial stages of Parkin-mediated mitophagy. J Cell Sci 125:1488–1499
Joo JH, Dorsey FC, Joshi A, Hennessy-Walters KM, Rose KL, McCastlain K et al (2011) Hsp90-Cdc37 chaperone complex regulates Ulk1- and Atg13-mediated mitophagy. Mol Cell 43:572–585
Jung C, Jun C, Ro S, Kim Y, Otto N, Cao J et al (2009) ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 20:1992–2003
Jung CH, Seo M, Otto NM, Kim DH (2011) ULK1 inhibits the kinase activity of mTORC1 and cell proliferation. Autophagy 7:1212–1221
Kabeya Y, Kamada Y, Baba M, Takikawa H, Sasaki M, Ohsumi Y (2005) Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell 16:2544–2553
Kabeya Y, Noda NN, Fujioka Y, Suzuki K, Inagaki F, Ohsumi Y (2009) Characterization of the Atg17–Atg29–Atg31 complex specifically required for starvation-induced autophagy in Saccharomyces cerevisiae. Biochem Biophys Res Commun 389:612–615
Kageyama S, Omori H, Saitoh T, Sone T, Guan JL, Akira S et al (2011) The LC3 recruitment mechanism is separate from Atg9L1-dependent membrane formation in the autophagic response against Salmonella. Mol Biol Cell 22:2290–2300
Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y (2000) Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol 150:1507–1513
Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K et al (2010) Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol 30:1049–1058
Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y (2008) Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell 19:2039–2050
Kijanska M, Dohnal I, Reiter W, Kaspar S, Stoffel I, Ammerer G et al (2010) Activation of Atg1 kinase in autophagy by regulated phosphorylation. Autophagy 6:1168–1178
Kim J, Kundu M, Viollet B, Guan KL (2011) AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 13:132–141
Kimmelman AC (2011) The dynamic nature of autophagy in cancer. Genes Dev 25:1999–2010
Klionsky DJ (2005) The molecular machinery of autophagy: unanswered questions. J Cell Sci 118:7–18
Koinuma D, Shinozaki M, Nagano Y, Ikushima H, Horiguchi K, Goto K et al (2011) RB1CC1 protein positively regulates transforming growth factor-beta signaling through the modulation of Arkadia E3 ubiquitin ligase activity. J Biol Chem 286:32502–32512
Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I et al (2005) Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 169:425–434
Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I et al (2006) Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441:880–884
Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T et al (2007) Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131:1149–1163
Korolchuk VI, Menzies FM, Rubinsztein DC (2009a) A novel link between autophagy and the ubiquitin-proteasome system. Autophagy 5:862–863
Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC (2009b) Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell 33:517–527
Korolchuk VI, Menzies FM, Rubinsztein DC (2010) Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett 584:1393–1398
Kraft C, Peter M, Hofmann K (2010) Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol 12:836–841
Kraft C, Kijanska M, Kalie E, Siergiejuk E, Lee SS, Semplicio G et al (2012) Binding of the Atg1/ULK1 kinase to the ubiquitin-like protein Atg8 regulates autophagy. EMBO J 31(18):3691–3703
Kroemer G, Mariño G, Levine B (2010) Autophagy and the integrated stress response. Mol Cell 40:280–293
Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T et al (2004) The role of autophagy during the early neonatal starvation period. Nature 432:1032–1036
Kundu M, Lindsten T, Yang C, Wu J, Zhao F, Zhang J et al (2008) Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood 112:1493–1502
Kuroyanagi H, Yan J, Seki N, Yamanouchi Y, Suzuki Y, Takano T et al (1998) Human ULK1, a novel serine/threonine kinase related to UNC-51 kinase of Caenorhabditis elegans: cDNA cloning, expression, and chromosomal assignment. Genomics 51:76–85
Lee EJ, Tournier C (2011) The requirement of uncoordinated 51-like kinase 1 (ULK1) and ULK2 in the regulation of autophagy. Autophagy 7:689–695
Lee JW, Park S, Takahashi Y, Wang HG (2010) The association of AMPK with ULK1 regulates autophagy. PLoS One 5:e15394
Levine B, Yuan J (2005) Autophagy in cell death: an innocent convict? J Clin Invest 115:2679–2688
Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G et al (1998) Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol 72:8586–8596
Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H et al (1999) Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402:672–676
Liang CC, Wang C, Peng X, Gan B, Guan JL (2010) Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J Biol Chem 285:3499–3509
Lin SY, Li TY, Liu Q, Zhang C, Li X, Chen Y et al (2012) GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science 336:477–481
Löffler AS, Alers S, Dieterle AM, Keppeler H, Franz-Wachtel M, Kundu M et al (2011) Ulk1-mediated phosphorylation of AMPK constitutes a negative regulatory feedback loop. Autophagy 7:696–706
Luo S, Garcia-Arencibia M, Zhao R, Puri C, Toh PP, Sadiq O et al (2012) Bim inhibits autophagy by recruiting beclin 1 to microtubules. Mol Cell 47:359–370
Mack HID, Zheng B, Asara J, Thomas SM (2012) AMPK-dependent phosphorylation of ULK1 regulates ATG9 localization. Autophagy 8(8):1197–1214
Martinez J, Almendinger J, Oberst A, Ness R, Dillon CP, Fitzgerald P et al (2011) Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci USA 108:17396–17401
Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, Izumi T et al (2010) Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol 190:511–521
Matsuura A, Tsukada M, Wada Y, Ohsumi Y (1997) Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 192:245–250
McEwan DG, Dikic I (2011) The three musketeers of autophagy: phosphorylation, ubiquitylation and acetylation. Trends Cell Biol 21:195–201
McIntire SL, Garriga G, White J, Jacobson D, Horvitz HR (1992) Genes necessary for directed axonal elongation or fasciculation in C. elegans. Neuron 8:307–322
McKnight NC, Jefferies HB, Alemu EA, Saunders RE, Howell M, Johansen T et al (2012) Genome-wide siRNA screen reveals amino acid starvation-induced autophagy requires SCOC and WAC. EMBO J 31:1931–1946
Meijer WH, van der Klei IJ, Veenhuis M, Kiel JA (2007) ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy 3:106–116
Meiselbach H, Sticht H, Enz R (2006) Structural analysis of the protein phosphatase 1 docking motif: molecular description of binding specificities identifies interacting proteins. Chem Biol 13:49–59
Meléndez A, Neufeld TP (2008) The cell biology of autophagy in metazoans: a developing story. Development 135:2347–2360
Mercer C, Kaliappan A, Dennis P (2009) A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy 5:649–662
Mizushima N (2010) The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol 22:132–139
Mizushima N, Komatsu M (2011) Autophagy: renovation of cells and tissues. Cell 147:728–741
Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD et al (1998) A protein conjugation system essential for autophagy. Nature 395:395–398
Mizushima N, Yoshimori T, Ohsumi Y (2011) The role of atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27:107–132
Mochizuki H, Toda H, Ando M, Kurusu M, Tomoda T, Furukubo-Tokunaga K (2011) Unc-51/ATG1 controls axonal and dendritic development via kinesin-mediated vesicle transport in the Drosophila brain. PLoS One 6:e19632
Mok J, Kim PM, Lam HY, Piccirillo S, Zhou X, Jeschke GR et al (2010) Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci Signal 3:ra12
Mostowy S, Cossart P (2011) Autophagy and the cytoskeleton: new links revealed by intracellular pathogens. Autophagy 7:780–782
Nakatogawa H, Ichimura Y, Ohsumi Y (2007) Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130:165–178
Nakatogawa H, Ohbayashi S, Sakoh-Nakatogawa M, Kakuta S, Suzuki SW, Kirisako H et al (2012) The autophagy-related protein kinase Atg1 interacts with the ubiquitin-like protein Atg8 via the Atg8 family interacting motif to facilitate autophagosome formation. J Biol Chem 287(34):28503–28507
Ochi Y, Chano T, Ikebuchi K, Inoue H, Isono T, Arai A et al (2011) RB1CC1 activates the p16 promoter through the interaction with hSNF5. Oncol Rep 26:805–812
Ogura K, Wicky C, Magnenat L, Tobler H, Mori I, Müller F et al (1994) Caenorhabditis elegans unc-51 gene required for axonal elongation encodes a novel serine/threonine kinase. Genes Dev 8:2389–2400
Okazaki N, Yan J, Yuasa S, Ueno T, Kominami E, Masuho Y et al (2000) Interaction of the Unc-51-like kinase and microtubule-associated protein light chain 3 related proteins in the brain: possible role of vesicular transport in axonal elongation. Brain Res Mol Brain Res 85:1–12
Orsi A, Razi M, Dooley H, Robinson D, Weston A, Collinson L et al (2012) Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, is required for autophagy. Mol Biol Cell 23(10):1860–1873
Overholtzer M, Mailleux AA, Mouneimne G, Normand G, Schnitt SJ, King RW et al (2007) A nonapoptotic cell death process, entosis, that occurs by cell-in-cell invasion. Cell 131:966–979
Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H et al (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282:24131–24145
Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N et al (2005) Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122:927–939
Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbé S, Clague MJ et al (2010) Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy 6:506–522
Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J et al (2005) Global analysis of protein phosphorylation in yeast. Nature 438:679–684
Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ (2004) The Atg1–Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell 6:79–90
Samari HR, Møller MT, Holden L, Asmyhr T, Seglen PO (2005) Stimulation of hepatocytic AMP-activated protein kinase by okadaic acid and other autophagy-suppressive toxins. Biochem J 386:237–244
Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S et al (2007) Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450:1253–1257
Sanjuan MA, Milasta S, Green DR (2009) Toll-like receptor signaling in the lysosomal pathways. Immunol Rev 227:203–220
Sarkar S, Floto R, Berger Z, Imarisio S, Cordenier A, Pasco M et al (2005) Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol 170:1101–1111
Sarkar S, Ravikumar B, Floto RA, Rubinsztein DC (2009) Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ 16:46–56
Sekito T, Kawamata T, Ichikawa R, Suzuki K, Ohsumi Y (2009) Atg17 recruits Atg9 to organize the pre-autophagosomal structure. Genes Cells 14:525–538
Shang L, Chen S, Du F, Li S, Zhao L, Wang X (2011) Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc Natl Acad Sci USA 108:4788–4793
Shao Y, Gao Z, Marks PA, Jiang X (2004) Apoptotic and autophagic cell death induced by histone deacetylase inhibitors. Proc Natl Acad Sci USA 101:18030–18035
Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB et al (2004) Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol 6:1221–1228
Shintani T, Yamazaki F, Katoh T, Umekawa M, Matahira Y, Hori S et al (2010) Glucosamine induces autophagy via an mTOR-independent pathway. Biochem Biophys Res Commun 391:1775–1779
Sou YS, Waguri S, Iwata J, Ueno T, Fujimura T, Hara T et al (2008) The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell 19:4762–4775
Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, Herman PK (2009) The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci USA 106:17049–17054
Suttangkakul A, Li F, Chung T, Vierstra RD (2011) The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23:3761–3779
Tang HW, Wang YB, Wang SL, Wu MH, Lin SY, Chen GC (2011) Atg1-mediated myosin II activation regulates autophagosome formation during starvation-induced autophagy. EMBO J 30:636–651
Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M et al (1994) Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett 349:275–280
Toda H, Mochizuki H, Flores R, Josowitz R, Krasieva TB, Lamorte VJ et al (2008) UNC-51/ATG1 kinase regulates axonal transport by mediating motor-cargo assembly. Genes Dev 22:3292–3307
Todde V, Veenhuis M, van der Klei IJ (2009) Autophagy: principles and significance in health and disease. Biochim Biophys Acta 1792:3–13
Tomoda T, Bhatt RS, Kuroyanagi H, Shirasawa T, Hatten ME (1999) A mouse serine/threonine kinase homologous to C. elegans UNC51 functions in parallel fiber formation of cerebellar granule neurons. Neuron 24:833–846
Tomoda T, Kim JH, Zhan C, Hatten ME (2004) Role of Unc51.1 and its binding partners in CNS axon outgrowth. Genes Dev 18:541–558
Tsukada M, Ohsumi Y (1993) Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 333:169–174
Wellen KE, Thompson CB (2012) A two-way street: reciprocal regulation of metabolism and signalling. Nat Rev Mol Cell Biol 13:270–276
Wong AS, Cheung ZH, Ip NY (2011) Molecular machinery of macroautophagy and its deregulation in diseases. Biochim Biophys Acta 1812:1490–1497
Xie R, Nguyen S, McKeehan K, Wang F, McKeehan WL, Liu L (2011) Microtubule-associated protein 1S (MAP1S) bridges autophagic components with microtubules and mitochondria to affect autophagosomal biogenesis and degradation. J Biol Chem 286:10367–10377
Yan J, Kuroyanagi H, Kuroiwa A, Matsuda Y, Tokumitsu H, Tomoda T et al (1998) Identification of mouse ULK1, a novel protein kinase structurally related to C. elegans UNC-51. Biochem Biophys Res Commun 246:222–227
Yan J, Kuroyanagi H, Tomemori T, Okazaki N, Asato K, Matsuda Y et al (1999) Mouse ULK2, a novel member of the UNC-51-like protein kinases: unique features of functional domains. Oncogene 18:5850–5859
Yang Z, Klionsky DJ (2010) Eaten alive: a history of macroautophagy. Nat Cell Biol 12:814–822
Yeh YY, Wrasman K, Herman PK (2010) Autophosphorylation within the Atg1 activation loop is required for both kinase activity and the induction of autophagy in Saccharomyces cerevisiae. Genetics 185:871–882
Yeh YY, Shah KH, Herman PK (2011) An Atg13 protein-mediated self-association of the Atg1 protein kinase is important for the induction of autophagy. J Biol Chem 286:28931–28939
Yi C, Ma M, Ran L, Zheng J, Tong J, Zhu J et al (2012) Function and molecular mechanism of acetylation in autophagy regulation. Science 336:474–477
Yorimitsu T, He C, Wang K, Klionsky DJ (2009) Tap42-associated protein phosphatase type 2A negatively regulates induction of autophagy. Autophagy 5:616–624
Young A, Chan E, Hu X, Köchl R, Crawshaw S, High S et al (2006) Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci 119:3888–3900
Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF et al (2009) Autophagy mediates the mitotic senescence transition. Genes Dev 23:798–803
Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S et al (2004) Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science 304:1500–1502
Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J et al (2010) Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465:942–946
Zhou X, Babu JR, da Silva S, Shu Q, Graef IA, Oliver T et al (2007) Unc-51-like kinase 1/2-mediated endocytic processes regulate filopodia extension and branching of sensory axons. Proc Natl Acad Sci USA 104:5842–5847
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer Science+Business Media New York
About this chapter
Cite this chapter
Wong, PM., Jiang, X. (2013). Signal Transduction Regulation of Autophagy. In: Wang, HG. (eds) Autophagy and Cancer. Current Cancer Research, vol 8. Springer, New York, NY. https://doi.org/10.1007/978-1-4614-6561-4_4
Download citation
DOI: https://doi.org/10.1007/978-1-4614-6561-4_4
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4614-6560-7
Online ISBN: 978-1-4614-6561-4
eBook Packages: MedicineMedicine (R0)