Abstract
The microcellular polymer known as polyHIPE polymer (PHP), with modified physico-chemical characteristics, was developed as a cell matrix for the immobilization of the starch-degrading bacterium, Bacillus subtilis. Suspension of B. subtilis spores was inoculated into a synthesized PHP matrix which is chemically modified, and this PHP matrix was named sulphonated PHPs (SPHPs). These inoculated spores were then activated by supplying continuously well-aerated culture medium (LB medium) and placed in a 37 °C constant temperature room for 24-h incubation. The growth and enzyme productivity data were evaluated and compared. Three different pore and interconnect sizes of SPHPs were evaluated: 42.0 ± 0.61, 36.0 ± 0.50 and 30.0 ± 0.64 μm. The collected samples obtained from the 24-h cultivation were used to determine α-amylase productivity and the loss of cells from the matrices. The morphology, viability and proliferation of the immobilized cells on PHP matrices were observed by scanning electron microscopy (SEM). The SPHP with a pore size of 36.0 μm performed better with respect to the production of α-amylase and the penetration of cells through the whole matrix compared to other SPHPs. The data showed that the total productivity of α-amylase enzyme produced by immobilized cells (on the basis of SPHP volume) was 7.6-fold higher than the batch cell culture. However, if productivity was determined on the basis of used total volume of nutrient medium, that of the immobilized cells was relatively low compared to batch cell culture.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
Introduction
Enzymes are biological products that can act as catalysts. Owing to their specific, effective and versatile features, they tend to exhibit much higher reaction rates as compared to reactions catalysed by chemical catalysts. One of the world’s largest industrial enzyme producers is Novozymes (Denmark), which is involved in the development, production and distribution of microbial enzymes. These enzymes are produced from genetically modified microorganisms, indicating that microbial fermentation is the major route for the production of industrial enzymes. There is a need to explore novel processes that could improve the production systems to meet the anticipated increased demand for industrial enzymes.
The application of immobilized cell technology can improve bioreactor productivity by maintaining high cell concentrations as well as operational stability. Additionally, immobilized cell technology can reduce processing time and down-stream processing as it can act as a biomass separator. A number of previous studies have attempted to investigate the influence of immobilization on cell growth and metabolic activity (Duran-Paramo et al. 2000; Argirakos et al. 1992; Dobreva et al. 1998; Konsoula and Liakopoulou-Kyriakides 2006). The selection of the materials used for the matrices is of the utmost importance because it determines their characteristics. Currently, most of the support matrices used for cell immobilization is derived from natural sources such as alginate, carrageenan, gelatine and chitosan. These natural matrices lack mechanical rigidity and stability and are therefore prone to damage during long processing conditions. This is due to the impact of rapidly growing colonies on the outer surface of the matrix and subsequently leads to the release of cells into the medium. Additionally, the concentration of cells in the interior of these matrices is inconsistent because some treatments allow significant number of cells to escape. This means that more cells are observed growing on the outer surface rather than migrating deeper into the matrix. As a result, the efficiency of immobilization is compromised, affecting the final output of the bioreactor. The main factor that causes these problems is the limited mass transfer rates for nutrients, products or waste to and from the immobilized matrix. Thus, there is a need to improve the characteristics of the immobilized matrices used as cell supports. The matrix of the immobilized cell support must be porous enough to provide easy access to nutrients and waste but retain most or all of the bacteria. In this study, a polymeric material known as polyHIPE polymer (PHP) was used to immobilize the starch-degrading bacterium, Bacillus subtilis. This polyHIPE matrix is synthesized through the high internal phase (HIPE) route and its physico-chemical properties can be modified as desired. This type of polymeric matrix has a highly interconnected porous structure and is mechanically durable in the presence of harsh conditions such as high temperature, pH and toxic chemicals. Therefore, it can be sterilized and used for long operating conditions without significant physical or chemical degradation.
The PHP matrix used in the developed system was designed to improve the production of α-amylase by B. subtilis. The confined microenvironment was designed to promote the growth and metabolic activity of this bacterium. Moreover, the applied forced flow seeding technique yielded a more uniform distribution of cells within the polymeric support, which also helps to improve nutrient transport. This approach is essential in the area of bioprocess development specifically for microbial fermentations. The results of these studies may provide a platform for developing microreactors with sufficient mass transfer capacities, to compete with traditional fermenters.
Methodology
Materials
Phenol, styrene, DVB (divinylbenzene), sorbitanmonooleate (Span 80), potassium persulfate, vinylpyridine, sulphuric acid and isopropanol were purchased from Sigma.
Microorganisms and Growth Medium
B. subtilis 168(pKTH10) strain was obtained from Cell and Molecular Biosciences, Medical School, Newcastle University. The spore suspensions of B. subtilis 168 (pKTH10) were induced in Schaeffer’s sporulation medium as described in Harwood et al. (1990). After overnight incubation, the cell cultures were centrifuged at 10,000 g for 10 min. The pelleted cells were washed five times with sterilized distilled water to remove all cell debris. The supernatant was discarded, and the spores were suspended in sterilized distilled water and stored at −20 °C. Luria–Bertani medium containing 1 % (w/v) Bacto-tryptone, 0.5 % (w/v) Bacto-yeast extract and 1 % (w/v) NaCl was used to cultivate the spore suspension in both immobilization and batch culture.
Preparation of Sulphonated PolyHIPE Polymer Matrix
PHPs were prepared by polymerization of a HIPE emulsion as described by Akay et al. (2005b) the chemical modification of PHPs via sulphonation was carried out using methods developed by Akay et al. (2005a, b).
Preparation of Immobilized Cell Reactor
The PolyHIPE (PHP) discs were mounted into a sealed PTFE block and sterilized in an autoclave at 121 °C for 15 min. Then, this PHP disc was initially wetted with distilled water overnight and then with LB media for an hour before use. Spores of B. subtilis strain 168 (pKTH10) (~2 × 108) were suspended in 2 ml of LB broth and pre-germinated by incubating for 90 min at 37 °C. The pre-germinated spores were force seeded into the PHP by passing the suspension through the matrix at a flow rate of 0.55 ml/min using a syringe pump. The seeded PHP was left for an hour to allow the pre-germinated spore to attach to the surface of the PHP, and then culture medium (LB broth) was pumped continuously through the matrix at a flow rate of 1.0 ml/min. This experimental setup was placed in a 37 °C constant-temperature room. Samples were collected at various time intervals to determine the numbers of cells released from the PHP matrix and α-amylase activity. At the end of experiment, sterilized 0.1-M phosphate-buffered saline (PBS) was pumped through the matrix at the same flow rate to remove the remaining medium. As a control, the same amount of medium was also added into the PHPs discs, without inoculated spores and then incubated overnight at 37 °C. The matrix samples were fixed in 2 % glutaraldehyde/PBS and stored at 4 °C until required.
Determination of Pore and Interconnect Size by Scanning Electron Microscopy
Pore and interconnect sizes of each type of the PHPs were analysed using scanning electron microscopy (SEM, Cambridge s240). Each sample was fractured in order to obtain a fresh internal surface and was then mounted onto aluminium stubs at room temperature using conductive carbon cement (LEIT carbon or glued sticker). Prior to analysis, the samples were then left overnight followed by sputter coated with gold at 20 nm, using a Polaron E5100 sputter coater and stored in silica gel. The analysed micrograph images of each polymer were used to determine the diameter of 50 pores and interconnect using ImageJ software (NIH image).
Quantification of Released Cell Numbers and Cell Dry Weight
The number of cells released from the PHP matrix was determined by plating serial dilutions of the outflowing medium onto LB agar plates containing 1 % starch. The plates were incubated overnight at 37 °C, and the number of colonies was determined. Exposing the surfaces of these plates to iodine vapour revealed clear zones around the colonies able to hydrolyse starch. Cells were recovered from batch culture samples (with volume of 1 ml) by centrifugation (3,000 g, 10 min, 4 °C) and washed twice with 0.9 % NaCl. The resulting pellet was dried at 105 °C for 24 h to constant weight. In the case of cells in the polyHIPE matrix, they were washed by passing distilled water through the matrix to remove culture medium, and the matrix dried to constant weight (by weighing several times) and then weighed. The cell dry weight was determined by subtracting the initial weight of the polyHIPE and its final weight. Weight of blank polyHIPE disc (without cells) was also determined as a control.
Quantification of Protein
1 ml of protein assay reagent (Coomassie Plus (Bradford) Assay reagent, Thermo Scientific) was added to 1 ml of sample and mixed by using vortex. The mixture was incubated for 10 min at room temperature, and the absorbance was determined at 595 nm against distilled water blank. Each sample was treated in duplicate, and the average value was determined. The protein concentration of each sample was determined from the standard curve using the known concentrations of bovine serum albumin (BSA).
Quantification of α-amylase Assays
Phadebas®amylase tablets, a water-insoluble starch substrate, was used to determine α-amylase activity. One tablet of Phadebas was suspended in 5 ml of sterile distilled water. 40 μl of sample was added to 0.8 ml of Phadebas suspension, and the mixture was incubated with frequent mixing at 37 °C in a water bath for 15 min. The reaction was stopped by the addition of 200 μml of 0.5 M NaOH and vortexed immediately. The mixture was centrifuged at 10,000 g for 5 min, and the supernatant was transferred into a cuvette. The absorbance of the supernatant was measured at 620 nm against a LB broth blank subjected to the same assay conditions. Samples with very high α-amylase activity were diluted up to fivefold using distilled water. Each sample was duplicated, and the average was taken. One unit (U) of α-amylase activity is defined as the amount of enzyme catalyzing the hydrolysis of 1-μmol glucosidic linkage per minute at 37 °C.
Preparation of Matrix Samples for SEM Analysis
The samples were dehydrated in an ethanol series (each step for 15 min excepting 100 % ethanol treatment was for 1 h). The dehydrated sample in 100 % ethanol was critical-point dried with liquid CO2. The sample was mounted on aluminium stubs and coated with gold using a Polaron E1500 sputter coater. The sample was examined under SEM (Cambridge s240). SEM analysis for samples without cells was also undertaken. In this case, the samples were directly coated with gold without dehydration and critical drying.
Results and Discussions
Architecture of PolyHIPE Polymer Matrices
In this study, PHPs were synthesized through the water-in-oil HIPE route in order to obtain a good structural stability polyHIPE. The resulting PHPs have high porosities and interconnectivities. In order to enhance its hydrophilicity and diffusivity, the resulting w/o HIPE was subjected to sulphonation using sulphuric acid, and the treated porous, hydrophilic polymeric support is called sulphonated PHP (SPHP), and this chemically modified PHP was then used to immobilize the industrial starch-degrading bacterium, B. subtilis. The microstructure of SPHP was examined by SEM, and representative images (Fig. 1) and their characteristics are shown in Table 1.
As can be seen in Fig. 1a, the pore sizes in sample SPHP-2 (tD5/tM5) were significantly larger than those for samples SPHP-4 and SPHP-6. The pore and interconnect sizes of samples SPHP-4 and SPHP-6 were well distributed and uniform, and this may be due to the additional 5-min dosing time, allowing for the uniform formation of interconnects throughout the structure. The increase in mixing time resulted in the emulsion reaching higher energy state, with a resultant decrease in droplet size. However, since the pore and interconnect sizes of SPHP-6 were only marginally smaller than those of SPHP-4, it appears that the additional 10 min of mixing time had little or no effect on the final structure of the PHP. This was likely to be due to the emulsion having reached a highly viscous and stabile state such that the further break-up of the droplets was not possible without increasing the vigour of the stirring.
α-Amylase Production in Batch and Immobilized Cultures
In this study, a strain of B. subtilis 168 encoding pKTH10 was used for the high-level production of α-amylase. Recombinant plasmid pKTH10, based on the high-copy-number plasmid pUB110, encoded the structural gene for the Bacillus amyloliquefaciens α-amylase AmyQ (Palva 1982). As can be seen from Fig. 2, the cells grow rapidly in exponential phase for the first 8 h. Cell mass then reached a plateau for the next 12 h, before slightly declining for the last 4 h. The specific growth rate, μ (h−1), and doubling time, td (h−1), of the batch culture were determined from the growth curve; the values are 0.41 h−1 and 35 min, respectively. The concentration of α-amylase in the culture medium continued to increase from the exponential phase until the first 5 h of stationary phase.
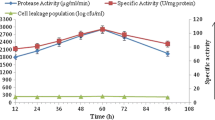
In the case of immobilized cultures, the production of α-amylase by cells of B. subtilis immobilized in sulphonated polyHIPE matrices with different pore sizes (42.0 ± 0.61, 36.0 ± 0.50 and 30.0 ± 0.64 μm) was determined at various times during cultivation (Fig. 3). The data show that SPHP with a pore size 36.0 ± 0.50 μm (SPHP-4) was the most productive—it reached an optimum production rate (~0.5 U/ml) after ~12 h and maintained a similar rate throughout the experiment. This optimum enzyme production rate by SPHP-4 matrix was 20 % more compared to SPHP-6 matrix (~0.4 U/ml). In the case of matrix with a pore size 42.0 ± 0.61 μm (SPHP-2), the production of α-amylase from B. subtilis immobilized on the SPHP-2 matrix reached a maximum production rate (~0.2 U/ml) after 15 h and maintained at 0.25 U/ml towards the end of the experiment. The release of bacterial cells from the SPHP matrices was determined from samples taken every 2 h (Fig. 3). The cells were enumerated by counting the number of colonies on nutrient agar plates after growth overnight at 37 °C. The data show that the SPHP with the smallest pore size (30 μm) released the fewest number of cells. A comparison of performance of sulphonated polyHIPE matrices with batch cell culture is shown in Table 2. The productivity of α-amylase enzyme produced by immobilized cells (on the basis of SPHP volume) with pore size of 36 μm was 7.6-fold higher than the batch cell culture. However, the volumetric productivities of α-amylase per total nutrient medium of immobilized cells on the SPHP matrices were relatively low (<1) compared to that of the batch cell cultures.
The time course of the production of α-amylase and released cells in 106 cells/ml by B. subtilis immobilized in sulphonated polyHIPE with pore sizes of 42.0, 36.0 and 30.0 μm (for 24-h incubation with 1 ml/min of nutrient medium flow rate). The data are the result of independent triplicate experiments
Morphology of Immobilized B. subtilis Cells
After 24 h cultivation at 37 °C with continuous supply of well-aerated LB medium at a flow rate of 1 ml/min, the surfaces of all three sulphonated polyHIPEs were covered by a dense homogeneous layer of vegetative B. subtilis cells that were free of an extensive extracellular matrix (Fig 4d, h, l). Following inoculation and incubation, a large number of vegetative cells were observed at the surface of all sulphonated polymeric matrices: SPHP-2, SPHP-4 and SPHP-6 after 24 h incubation at 37 °C (Fig. 4a–c, e–g, i–k). The germinated spores had successfully migrated throughout the matrix of the polyHIPE supports with pore sizes of 42 μm (Fig. 4b) and 36 μm (Fig. 4f). However, in the case of SPHP-6, a thick layer of cells had accumulated on the surface (Fig. 4k) only migrating up to approximately 3 mm into the support. The pore walls had an almost continuous layer of bacterial cells on the top surfaces, whereas smaller cell numbers were found further deep into the matrices.
Electron micrograph of cross section of the SPHP-2 (ϕ = 42 μm) matrix, showing the appearance of vegetative cells of B. subtilis immobilized on after 24-h cultivation. a Top edge, b further deep (x2000), c bottom edge and d top surface at high magnification. SPHP-4 (ϕ = 36 μm) matrix, showing the appearance of vegetative cells of B. subtilis immobilized on after 24-h cultivation. e Top edge, f in the middle of cross-section matrix x2000, g bottom edge and h top surface at high magnification. SPHP-6 (ϕ = 30 μm) matrix, showing the appearance of vegetative cells of B. subtilis immobilized on after 24-h cultivation. i Top edge, j in the middle of cross-section matrix (x2000), k deep further and l top surface at high magnification
Discussion
One of the design characteristics of matrices used for the immobilization bacterial cells is the need to optimize pore and interconnect size to ensure good mass transport into and out of matrix. In this study, sulphonated polyHIPE matrices with three different pore sizes were evaluated. The results indicated that pore size significantly affected the production of α-amylase by cells immobilized on sulphonated polyHIPE matrices (Fig. 4). SPHP matrices with a pore size of 36 μm (SPHP-4) yielded the highest total α-amylase enzyme and cell dry weight. The volumetric productivity obtained by SPHP-4 was 2.4-fold and 1.3-fold higher than that obtained by the SPHPs with pore sizes of 42 and 30 μm, respectively. As expected, there was a direct relationship between dry cell mass and α-amylase production and entrapping a high proportion of cell in the matrix a high priority. The low level of α-amylase production and low cell mass in the matrix with the smallest pore size (30 μm) are likely to be due to diffusional limitations that restrict the access of nutrients and oxygen to the cells and the release of the secreted enzyme to the medium. It might be also due to limited loading of cells onto this matrix.
Steady-state α-amylase production was achieved after 13 h, indicating that the matrix had reached the limit of its capacity to sustain further increases in biomass. Such a steady state represents the most efficient state for the production of α-amylase since net cell growth and nucleic acid syntheses are repressed while protein synthesis is still active (Kinoshita et al. 1968). This represents a shift from biomass increase to cell maintenance and product formation. Moreover, in B. subtilis, the secretion of macromolecular hydrolases like α-amylase is higher during stationary phase than exponential phase (Antelmann et al. 2001). Bacterial cells were released from all of the SPHP matrices (Fig. 3) although the SPHP matrix with the smallest pore size (30 μm) released the fewest numbers. The release of cells from the SPHP matrices with pore sizes of 36 and 42 μm was similar.
Previous studies have shown that polyHIPE provides a suitable matrix for the immobilization of both bacterial (Akay et al. 2005a, b; Erhan et al. 2004) and eukaryotic (Akay et al. 2004; Bokhari et al. 2005) cells. These studies have shown that B. subtilis can migrate up to 3 mm into such matrices over 30-day cultivation. Multicellular layers of osteoblast were seen on the top surface of polyHIPE, migrating to a maximum depth of 1.4 mm within the matrix. Previous studies used styrene–divinylbenzene polyHIPEs, which are hydrophobic and therefore exhibit reduced permeability to aqueous fluids. In the current study, we used sulphonated polyHIPEs, which are hydrophilic and show increased permeability to aqueous fluids. Sulphonated polyHIPEs (SPHPs) were used to immobilize B. subtilis spores, which were then germinated and cultured in situ. After 24-h continuous culture, a dense layer of B. subtilis cells was seen on the top surface of all the SPHPs matrices, irrespective of pore size. SEM analysis showed that the cells were elongated rods with little evidence of an extracellular matrix or cell lysis. This result indicates that the B. subtilis spores can be efficiently germinated and cultured throughout hydrophilic polymeric supports that provide a permissive microenvironment. A thick homogeneous layer of cells on the top surface of the polymeric supports is likely to be due to the presence of high nutrient and oxygen concentrations in this location. The reduction in cell concentration within the matrix probably reflects the fact that nutrient and oxygen concentrations are likely to decrease as the culture medium passes through the matrix. The production and excretion of extracellular substances, such as polysaccharides, are other parameters that affect the diffusive resistance in a cell mass (Karel et al. 1985). However, in this study, virtually no exopolysaccharide matrix was formed, and there was little or no evidence of cell debris resulting from cell lysis. This indicates that the microporous architecture of the polymeric matrix provided efficient dynamic flow conditions and presumably the continuous removal of dead and degraded cells and toxic materials. The only minor exception was the sulphonated polyHIPE, with pore size of 30 μm (SPHP-6), which was found to contain small amounts of cell debris. The results obtained have shown that the microarchitecture of the matrices is important for the growth, viability and penetration of cells, and was dependent on the pore size and the physico-chemical properties of the matrix. The volumetric productivity of α-amylase of the immobilized cells was significantly higher than that of the batch cell cultures if calculated on the basis of the volume occupied by the SPHP matrix. However, the volumetric productivities of α-amylase per used total nutrient medium of immobilized cells on the SPHP matrices were relatively low (<1) compared to that of the batch cell cultures. Consequently, the overall yield of α-amylase by immobilized cell cultures was relatively low compared to batch cell cultures.
Conclusion
The sulphonated polyHIPE matrix with a pore size of 36 μm was observed to give better cell growth, with cells penetrating throughout the matrix. The volumetric productivity of immobilized cells per volume occupied by SPHP matrix was 7.6-fold higher than the batch cell cultures, but its productivity was relatively low (<1) if determined on the basis of used total volume of medium.
References
Akay, G. (2005). Bioprocess and chemical process intensification. In S. Lee (Ed.), Encyclopedia of chemical processing. New York: Taylor & Francis Group, CRC Press.
Akay, G., Dogru, M., Calkan, B., & Calkan, O. F. (2005a). Flow induced phase inversion phenomenon in process intensification and micro-reactor technology. In Y. Wang & J. D. Holladay (Eds.), Microreactor technology and process intensification (ACS Symposium Series, vol. 914).
Akay, G., Erhan, E., & Keskinler, B. (2005b). Bioprocess intensification in flow-through monolithic microbioreactors with immobilized bacteria. Biotechnology and Bioengineering, 90(2), 180–190.
Antelmann, H., Tjalsma, H., Voigt, B., Ohlmeier, S., Bron, S., & Van Dijl, J. M, et al. (2001). A proteomic view on genome-based signal peptide predictions. Genome Research, 11(9), 1484–1502.
Argirakos, G., Thayanithy, K., & Wase, D. A. J. (1992). Effect of immobilization on the production of alpha- amylase by an industrial strain of Bacillus-Amyloliquefaciens. Journal of Chemical Technology and Biotechnology, 53(1), 33–38.
Bokhari, M. A., Akay, G., Zhang, S., & Birch, M. A. (2005). The enhancement of osteoblast growth and differentiation in vitro on a peptide hydrogel -polyHIPE polymer hybrid material. Journal of Biomaterials, 26, 5198–5208.
Duran-Paramo, E., Garcia-Kirchner, O., Hervagault, J. F., Thomas, D., & Barbotin, J. N. (2000). α-Amylase production by free and immobilized Bacillus subtilis. Applied Biochemistry and Biotechnology—Part A Enzyme Engineering and Biotechnology, 84–86, 479–485.
Dobreva, E., Tonkova, A., Ivanova, V., Stefanova, M., Kabaivanova, L., & Spasova, D. (1998). Immobilization of Bacillus licheniformis cells, producers of thermostable α-amylase, on polymer membranes. Journal of Industrial Microbiology and Biotechnology, 20(3), 166–170.
Erhan, E., Yer, E., Akay, G., Keskinler, B., & Keskinler, D. (2004). Phenol degradation in a fixed-bed bioreactor using micro-cellular polymer-immobilized Pseudomonas syringae. Journal of Chemical Technology and Biotechnology, 79(2), 195–206.
Karel, S. F., Libicki, S. B., & Robertson, C. R. (1985). Immobilization of whole cells: Engineering principles. Chemical Engineering Science, 40(8), 1321–1354.
Kinoshita, S., Okada, H., & Terui, G. (1968). On the nature of α-amylase forming system in Bacillus subtilis. Fermentation Technology, 46, 427–436.
Konsoula, Z., & Liakopoulou-Kyriakides, M. (2006). Thermostable alpha-amylase production by Bacillus subtilis entrapped in calcium alginate gel capsules. Enzyme and Microbial Technology, 39(4), 690–696.
Palva, I. (1982). Molecular cloning of a-amylase gene from Bacillus amyloliquefaciens and its expression in B. subtilis. Gene, 19(1), 81–87.
Acknowledgments
This work was funded by Ministry of Higher Education (MOHE), Malaysia. I would like to thank Tracey Davey and Pauline Carrick for their technical help.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer Science+Business Media New York
About this chapter
Cite this chapter
Jimat, D.N., Harwood, C., Akay, G. (2013). Production of α-Amylase by Immobilized Bacillus Subtilis in Polymeric PolyHIPE Matrix. In: Pogaku, R., Bono, A., Chu, C. (eds) Developments in Sustainable Chemical and Bioprocess Technology. Springer, Boston, MA. https://doi.org/10.1007/978-1-4614-6208-8_21
Download citation
DOI: https://doi.org/10.1007/978-1-4614-6208-8_21
Published:
Publisher Name: Springer, Boston, MA
Print ISBN: 978-1-4614-6207-1
Online ISBN: 978-1-4614-6208-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)