Abstract
H-nuclear magnetic resonance (NMR) spectroscopy was used to analyze pot honey samples of different geographical and entomological origin. An NMR-based metabolomic approach, applied to 67 samples, tested and confirmed the validity of the multivariate statistical analysis in the discrimination. We developed an efficient tool to differentiate the honeys by their geographical origin; additionally, we showed that within limited geographical areas it is possible to distinguish honey samples in terms of the bee species that produced it. Furthermore, the structural identification of a geographical marker compound (in the Brazilian honeys) was achieved, and a specific region in the NMR spectrum was found to be responsible for the entomological separation.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Nuclear Magnetic Resonance
- Nuclear Magnetic Resonance Spectrum
- Nuclear Magnetic Resonance Spectroscopy
- Honey Sample
- Nuclear Magnetic Resonance Experiment
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Nuclear Magnetic Resonance (NMR) is a powerful spectroscopic method, traditionally used as a very important tool in chemistry for structure verification, elucidation and purity analysis. However, driven by the needs of multidisciplinary topics such as biochemistry, medicine, pharmaceutical sciences, food chemistry, and others, NMR has rapidly expanded its applications to many other fields, and recent examples are the analysis of complex mixtures and screening applications (Lindon et al. 2000; Spraul et al. 2009).
NMR is an especially suited detector in the analysis of fluids of biological origin, food materials or drinks. It combines truly quantitative and structural information with high throughput (a 1D spectrum can be measured in a few minutes) and excellent reproducibility, which depends mostly on the minimal sample preparation required and the absence of any derivatization step.
For these reasons, it can be used to detect small molecules to generate global metabolite profiles in metabolomic studies, which aim to categorize or classify samples and to understand the basic underlying principles that contribute to the differences among them (Kang et al. 2008). Pattern recognition is followed by related multivariate statistical approaches to analyze the latent structures in the multivariate data.
Principal Component Analysis (PCA) and Partial Least-Squares Discriminant Analysis (PLS-DA) have often been used to identify sample groups and to relate specific biochemical compounds to the group separation.
1H NMR-based metabolomic studies have been applied also to food science (Cevallos-Cevallos et al. 2009), including for example assessments of green tea (Tarachiwin et al. 2007), rosemary (Xiao et al. 2008), honey (Schievano et al. 2012), and grape wine (Son et al. 2008).
In this chapter, a study of the honey matrix is illustrated, performed using an NMR-based metabolomic approach combined with multivariate analysis.
2 Nuclear Magnetic Resonance
NMR is a branch of spectroscopy which uses radio waves, with a frequency between 20 MHz and 1 GHz on sensitive nuclei. The most common NMR experiments are performed on 1H nuclei, but spectra on many other nuclei (13C, 31P, 19F, and 15N are the most common ones) are frequently acquired. The principles of NMR spectroscopy are well known nowadays; they are available in many textbooks (Günther 1995; Claridge 1999) and they are not discussed in detail here. Some basic principles will be quickly illustrated, to enhance the comprehension of this work. Nuclei with an intrinsic magnetic moment may be oriented by a strong magnetic field; two orientations are possible for 1H nuclei. A consequence is the tendency to absorb and emit energy at a specific resonance frequency. Based on this phenomenon, a very large number of different NMR experiments have been developed, which explore different properties of the material under study. Samples can be analyzed in the solid (CP-MAS NMR), semisolid (HR-MAS NMR), and solution state (HR-NMR): the last one has been utilized in this work.
The 1H 1D spectrum is the simplest NMR experiment: a radio frequency pulse inverts the orientation of some of the 1H nuclei in the magnet; then, relaxation toward the original situation results is an electric signal (free induction decay: FID), which can be processed with a Fourier Transform to give a resonance peak.
Samples must be completely dissolved in a solvent. In the solvents for NMR analyses, protons are normally replaced with deuterium atoms to avoid saturation of the NMR receiver with the solvent protons, which would otherwise hide the signals of the protons of the solute. Each peak in the spectrum is the signal of a particular kind of proton in the mixture and its resonance position, the chemical shift, is measured in ppm units on the x-axis of the spectrum. The y-axis is an intensity scale, relative to the amount of protons. Integration of a peak area is directly proportional to the number of protons resonating at that same frequency.
3 Metabolomic Analysis
Metabolomics is the study of the global metabolic profile in a system (cell, tissue, or organism) under a given set of conditions. Metabolic profiling first appeared in the literature in the 1950s, and developed throughout the following decades (Rochfort 2005). The metabolome is formally defined as a collection of small molecules, including a range of endogenous and exogenous chemical entities such as peptides, amino acids, nucleic acids, carbohydrates, organic acids, vitamins, polyphenols, alkaloids, minerals, and just about any other chemical that can be used, synthesized, or ingested by a given cell or organism. Over the past few years, two schools of thought have emerged for processing and interpreting metabolomic data: the chemometric and the quantitative metabolomics (or targeted profiling) approaches (Wishart 2008).
The chemometric approach (untargeted metabolomics) includes the analysis of multiple samples (for example by NMR) and statistical comparison of the results, without identifying the chemical compounds, but only using the recorded spectral pattern to recognize the relevant spectral features that distinguish sample classes. This method involves unsupervised clustering (PCA) or supervised classification (e.g., PLS-DA). After discovering significant differences, the most informative peaks in the spectra are identified and these molecules can then be used as markers.
In the quantitative metabolomics approach, most compounds in the sample are first identified and quantified, and this information is then used to perform multivariate statistical analyses and to find the most important markers and informative metabolic pathways.
From the perspective of a metabolomics researcher, most foods can essentially be viewed as complex chemical mixtures consisting of various metabolites and chemical additives in a solid, semisolid, or liquid mixture. In food science, metabolomics has become a tool to assess the quality, the processing history, and the safety of raw materials and final products. Recent applications involve geographical or botanical origin, or authenticity, of several foods.
In this work, a chemometric approach to differentiate the geographical and entomological origin of stingless bee honey has been used: 1H NMR spectra provided signals, which were integrated and used as inputs for PCA and PLS-DA studies. Formally, PCA is a clustering technique that reduces the dimensions of a complex data matrix to orthogonal linear combinations (Principal Components visualized as principal axes) which describe variation in the data. These components can be displayed graphically as a score plot, where the separation of the observations is visualized in the space between the two axes.
Unsupervised PCA was initially used to explore variation in the NMR spectra dataset while PLS-DA was subsequently applied to maximize the separation among the samples.
The score plot can be visualized also in 3D corresponding to three principal components. In the loading plot, the most influential variables are highlighted: the farther they are from the center of the graph, the more they influence cluster separation. The Hotelling’s T2 region, shown as an ellipse in score plots of the models, defines the 95% confidence interval of the modeled variation. The quality of the models is described by R 2 x and Q 2 values. R 2 x is defined as the proportion of variance in the data explained by the models and indicates goodness of the fit. Q 2 is defined as the proportion of variance in the data predictable by the model and indicates predictability (Eriksson et al. 2006). Thus, PCA is most commonly used to identify how one sample is different from another, and which variables contribute most to this difference. PLS-DA is based on the same basic principles as PCA, but it uses the labeled set of class identities, enhancing the separation between groups of observations.
4 NMR-Based Metabolomics Applied to Pot-Honey
Because of the complexity and diversity of the metabolites present in a complex food matrix, it is unlikely that one single analytical method would generate information about all the metabolites present and it would probably be necessary to perform a wide range of chemical analyses, which should be both rapid and reproducible. 1H NMR has the potential to detect and identify a large number of compounds; as such, it is emerging as a leading technique in the area of metabolomic studies. An important advantage of the use of NMR spectroscopy in metabolomic studies is that the sample requires hardly any physical or chemical treatment prior to analysis. MS studies usually require separation of the metabolites, and for GC-MS it may be necessary to modify the metabolites to render them volatile. On the other hand, separation via HPLC requires conveniently detectable chromophores or functional groups.
NMR methodologies overcome these problems, and the range of compounds that can be analyzed is not limited by their volatility, presence of chromophores, or polarity, or other properties. Although the detection limit of NMR is still higher than that of other techniques, new pulse sequences have been introduced that lower the detection limit to about 10 μM in the sample solution (Rastrelli et al. 2009). Moreover, NMR spectroscopy simultaneously gives definitive structural information on many different compounds in the sample, maximizing the chance to identify important but unexpected or previously unknown metabolites (Teresa and Fan 1996).
1H NMR has been successfully used, for example, in the area of toxicology, clinical diagnostics, and in the field of plant metabolites; it is frequently applied to food samples that can be directly examined as liquids (Belton et al. 1996), but very simple extraction or sample preparation procedures may also be used (Schievano et al. 2008). In the last decades, specific chemical and physical properties of honey have been used to determine its botanical origin (Anklam 1998; Bogodanov et al. 2004; Arvanitoyannis et al. 2005), and new analytical techniques have been proposed to this aim. An improvement in determination of botanical origin can certainly be achieved by a multivariate analytical approach. Recently, NMR techniques have been proposed also to identify and classify honey of different floral sources (Beretta et al. 2008; Lolli et al. 2008; Schievano et al. 2010) or geographical origin (Donarski et al. 2008; Consonni and Cagliani 2008).
The composition and properties of a particular honey sample depend strongly on the type of bee, on the type of flowers visited by the bees, as well as on the climatic conditions in which the plants grow and on contributions of the beekeeper (Al et al. 2009; Azeredo et al. 2003). In fact, Schievano et al. (2012) have shown that 1H NMR spectra of organic extracts of honey can be used as a fingerprint to differentiate the botanical origin, when coupled with chemometric analysis.
The extraction method is simple and reproducible: a water/chloroform mixture was used as extracting solvent, with the advantage to eliminate the compounds most present in the honey mixture, i.e., the carbohydrates, with the water layer. The aroma compounds and those hydrophobic substances that differ the most in honeys of various sources are retained in the organic solution. Also, the extraction procedure yields a concentrated solution amenable to rapid NMR analysis. In more detail, portions of honey samples (6 g) were weighted in a centrifuge tube and dissolved with 15 ml of deionized water. 15 ml of CHCl3 were added and the mixture was mechanically stirred for 10 min. The biphasic mixture was then centrifuged at 10,000 rpm for 15 min at 4 °C. The lower chloroform phase was collected and the solvent was evaporated under a gentle stream of nitrogen. The solid residue was dissolved in 600 μl of CDCl3 and put in an NMR tube. The scheme of this NMR-based metabolomic approach is shown in Fig. 31.1.
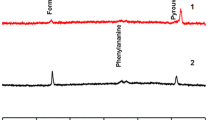
The 1H spectrum provides a fingerprint for each honey type showing many characteristic peaks in all spectral regions. Figure 31.2 shows a representative NMR spectrum from a Melipona fuscopilosa honey sample from the Amazon. Generally, the strongest signals in a honey spectrum are in the aliphatic region (0.0–2.5 ppm) while signals of comparable intensities rise in the other regions. All the regions appear very crowded. Specifically, many peaks are present in the 3.0–3.5 ppm region (–CH 2OH resonances), in the 4.0–4.5 ppm (–CH 2O–CO– signals), in the olefinic proton region (4.5–5.5 ppm), and in the aromatic region (6.5–8.5 ppm); also aldehydic and acidic proton signals are present (9.0–13.0 ppm).
The 1D spectra were acquired at 298 K, with a 600 MHz NMR instrument, using a modified double pulsed field gradient spin echoes (DPFGSE) sequence (Rastrelli et al. 2009). The introduction of a π pulse in the DPFGSE sequence allows the removal of the strongest signals present in the 0–2 ppm region, and this results in improved digitization of the weaker peaks, lower integration errors, and eventually, better quantification of the number of resonant spins. The spectra collection, processing, and analysis require 30 min.
The choice of chloroform as a solvent offers great advantages compared to other solvents previously used in NMR studies of honey. The residual chloroform signal is very sharp, and obscures a very small region at 7.26 ppm, which does not affect the analysis. On the other hand, solvents such as DMSO and MeOH are less suitable since they exhibit large signals in very important areas (around 3.4 ppm for MeOH and around 2.5 ppm for DMSO).
Data were processed using the ACD software (ACD/Specmanager 7.00 software, Advanced Chemistry Development Inc., 90 Adelaide Street West, Toronto, Ont., Canada M5H 3V9). Principal component analysis (PCA) and PLS-DA were conducted using the software SIMCA-P11 (Umetrics, Umea Sweden).
5 Geographical and Entomological Differentiation of Pot-Honey by NMR
The present study was performed on a total of 67 honey samples: 63 were obtained from stingless bees (see Table 31.1), one pot-honey was bought at the Indigenous market of Puerto Ayacucho, Amazonas state, Venezuela, as “erica” honey, one additional pot-honey sample was obtained after sugar feeding M. quadrifasciata bees, São Paulo state, Brazil, and two commercial honeys from Apis mellifera (one from Venezuela and one from Italy).
The pot-honey samples in Table 31.1 are from different entomological and geographical origins: 10 came from Australia, 12 from Brazil, 15 from Venezuela, 13 from Mexico, and 13 from Bolivia. The principal bees are Melipona, Scaptotrigona, Tetragonisca, Tetragonula, and Frieseomelitta nigra. In the sample data set, the differences due to geographical and entomological origin are not easily separable. In fact, the different species of bees generally live in different ecosystems. This consideration suggests that it might be difficult to discriminate the effects of different geographical origin from bee identify.
From the geographical point of view, our data set is composed of five main classes of honeys from five different regions. Furthermore, samples from Venezuela and Brazil can be divided in two other classes of samples collected in different regions of the same nation. Usually, projection methods for classification, such as PLS-DA, are able to produce efficient classification models for not more than four classes of samples.
For this reason, we did not consider the entomological origin of our honey samples at first, and PLS-DA models were obtained from groups of honey samples of different geographical origin, compared three at a time. Figure 31.3 shows the PLS-DA score plots (in 3D, corresponding to PC1/PC2/PC3) derived from the NMR spectra of the honey extracts, and they visualize good separations among these extracts (R 2 x and Q 2 value of 0.70 and 0.80 for the “a” plot, 0.63 and 0.56 for the “b” plot, 0.91 and 0.80 for the “c” plot).
PLS-DA score plots derived from 600 MHz 1H NMR spectra of chloroform honey extracts. (a) PLS-DA on samples from Australia, Brazil, and Venezuela. (b) PLS-DA on samples from Bolivia, Brazil, and Venezuela. (c) PLS-DA on samples from Australia, Brazil, and Mexico. (Filled triangle) Australia, (filled circle) Brazil, (asterisk) Venezuela, (filled diamonds) Bolivia, (open diamonds) Mexico
As a prediction test, we randomly selected two test samples from each region and built the PLS-DA prediction models without them. The approach yielded similar statistical characteristics to those previously obtained using the entire data set and correctly predicted the origins of the ten test samples. These results show that our method could be applicable to discriminate other unknown honey samples on the basis of their geographical origin.
If we apply the same PLS-DA calculations to the classification of the different entomological origins, there can be some ambiguity because some bees are found only in a specific geographical zone (e.g., the Tetragonula carbonaria and the Melipona favosa honeys sampled here are found only in Australia and in the Amazon, respectively). When we considered restricted regions, we were able to achieve good discrimination based on the entomological origin. The best results were obtained with the honeys collected in Venezuela (n° 23–37 of Table 31.1). Within these samples, we have honey of the same geographical origin, but of different entomological origin. A PLS-DA model (Fig. 31.4) is able to discriminate T. clavipes (four samples), M. aff. fuscopilosa (three samples), and M. favosa (eight samples). Specifically, samples from the same ecosystem (the Amazon) are very clearly separated in two groups (R 2 x of 0.88, Q 2 of 0.97) corresponding to honey produced by two different bees (M. aff. fuscopilosa and T. clavipes). The honey sample bought at the local indigenous market in Puerto Ayacucho (State Amazonas) as “erica” M. favosa honey was used to test the predictive capability of our model. In Fig. 31.4, PLS-DA assigns it to the Tetragona group, not to M. favosa as claimed.
PLS-DA on Venezuelan pot-honey samples. M. fuscopilosa (=Melipona aff. fuscopilosa = Melipona (Michmelia) sp. 1, see table in Pedro chapter 4, this book)
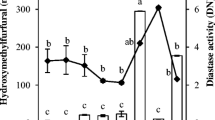
PCA of Mexican honeys (Fig. 31.5a) readily separated the groups of the most numerous samples from Melipona and Scaptotrigona. The remaining samples, produced by different bees, are in different regions of the plot. Pot-honey N° 48 is known to be produced by M. beecheii; however, it is found in a different area, and the most probable reason for that is the presence of a high content of hydroxymethylfurfural (HMF), which indicates lack of freshness or bad storage conditions and substances from fermentation. The sample produced by M. solani is different from the other Melipona honeys.
PC1 and PC2 scores on pot-honey from (a) Mexico (b) Brazil. See honey numbers in Table 31.1
When a PCA on Brazil samples (n° 11–22 of Table 31.1) was performed (Fig. 31.5b), a clear differentiation, by the first PCA component, was seen between M. fasciculata and M. subnitida. The three samples outside the ellipses originated from different species. Again, samples produced by different bees are in different regions of the plot.
Our NMR-based metabolomic approach, even if applied to a limited number of samples, confirmed the validity of the multivariate statistical analysis in discrimination. We developed an efficient tool to differentiate the honeys by their geographical origin; additionally, to highlight the entomological origin, we understood that the field of investigation must be restricted to a smaller geographical region.
The following step was the identification of chemical shift resonances indicating specific marker molecules, responsible for the separation of origins. This was achieved by analyzing the loading plots of PCA, which explain the influence of the selected variables on the PCA model. As an example, we show here the assignment of a chemical compound in the Brazilian honeys. The loading plot is shown in Fig. 31.6a (the corresponding score plot is reported in Fig. 31.5b).
Assignment of abscisic acid. (a) Loading plot of PCA on Brazilian honey samples (the corresponding score plot is shown in Fig. 31.5b on the right). (b) Chemical structures of trans and cis abscisic acid. Asterisks and ellipsoids indicate protons and the corresponding resonances. (c) Expanded region of 1H spectra of three samples of M. subnitida (dotted line) and of three samples of M. fasciculata (continuous line) where the same resonances of abscisic acid are found
M. subnitida honeys from Maranhão are characterized by the following NMR resonances: 5.79, 5.96, 5.89, 6.16, 6.29, 6.44, and 2.28 ppm (see continuous line spectra in Fig. 31.6c, in comparison with the dotted line from Rio Grande du Norte honey). Characteristic peaks for M. fasciculata honey resonate at 5.32, 5.36, 4.24, and 4.12 ppm (see dotted line spectra in Fig. 31.6c).
In the case of M. subnitida honeys, the resonances were assigned and attributed to the cis and trans isomers of abscisic acid, which is present in large amounts (in comparison with the other compounds) in these samples. Unequivocal structural identification of this marker compound was obtained using homo- and hetero-nuclear correlation 2D-NMR experiments, and MS analysis. In Fig. 31.6b, the molecular structure of abscisic acid is reported, with its resonance assignment.
Concerning the Amazon honey samples, PCA led to a clear discrimination of the different bees present in the same ecosystem (M. aff. fuscopilosa and T. clavipes, as evident also in the PLS-DA of Fig. 31.4). According to the loading plot analysis, the discriminating region of the 1H NMR spectrum is between 2.3 and 5.4 ppm. Assuming that in the same territory bees visit the same kind of plants and flowers, these data suggest that signals in this region of the spectrum come from organic compounds secreted by the specific bees.
To find the contribution of the bees and cerumen pots on the honey composition, a blank-trial probe was prepared, in which M. quadrifasciata were fed a sucrose syrup, to obtain a “sucrose honey”. The 1H NMR spectrum was acquired (see Fig. 31.7a,b) and compared with a typical pot-honey spectrum (as Scaptotrigona mexicana in Fig. 31.7c). The expanded aromatic region (6–8 ppm) of the sucrose honey (Fig. 31.7b) is poor of signals, in contrast to floral honey (Fig. 31.7c). However, most of the peaks in the aliphatic region (0–5.5 ppm), at frequencies typical of the fatty acid protons, are present in both sugar and floral honeys. It is evident that these aliphatic compounds must be part of the endogenous metabolism of the bee, rather than of the floral, exogenous resources. Therefore, discriminating signals that differentiate bee species are expected in the aliphatic region, whereas the foraging variation was observed in the aromatic region of the spectrum.
To substantiate this point, a comparison between the 2 – 5.5 ppm regions of the 1H NMR spectra of different species of meliponine honeys was conducted and is shown in Fig. 31.8. This region of the spectra shows the typical resonances of the free or bound fatty acids and many other signals of the glycerol esters. In particular, the very high similarity between A. mellifera from Venezuela and from Italy confirms that these resonances are not geographical but entomological markers, clearly characterizing honeys produced by A. mellifera.
1H NMR spectra (region 2–5.5 ppm) of the chloroform extract of honey produced by A. mellifera, and stingless bees. M. fuscopilosa (= Melipona aff. fuscopilosa= Melipona (Michmelia) sp. 1, see table in Pedro chapter 4, this book)
6 Conclusions
In this work, 65 honey samples from Meliponini (63 in Table 31.1, one from the local market, one obtained from a sugar fed colony) and two from A. mellifera, were analyzed by our NMR-based metabolomic approach. The 1H NMR spectrum of the chloroform honey extract represents a mixture profile containing both endogenous bee metabolites and exogenous compounds coming from plants and flowers visited by the bees. For this reason, the data set is particularly suitable for a multivariate statistical analysis to distinguish both geographical and entomological origin. Moreover, as a preliminary work, the number of the analyzed samples was sufficient a higher number would allow us to have a test set to perform a prediction analysis to confirm these first results. Using an NMR-based metabolomic approach, we showed that:
-
1.
Considering the entire data set samples, the stingless bee pot-honeys were well differentiated by their geographical origin.
-
2.
The structural identification of abscisic acid, as an example of geographical marker compound for the Brazilian honeys, was achieved using 1D and 2D NMR spectroscopy.
-
3.
If the analysis is restricted to a smaller region, it is possible to group honeys according to their entomological origin, because the entomological discriminant character becomes stronger than the geographical differences.
-
4.
The application of NMR to authenticate the entomological origin of pot-honey (i.e., the market honey sold as “erica” was not a honey produced by M. favosa, but by T. clavipes), is demonstrated for the first time.
-
5.
Sugar-fed M. quadrifasciata produced a honey with an unusual NMR profile, very poor in signals in the aromatic region (6–8 ppm), compared to natural floral honey. Therefore, the specific region in the NMR spectrum responsible for entomological separation seems to be the 0–5 ppm aliphatic region, where the protons from endogenous fatty chains resonate.
-
6.
Discriminating signals to differentiate stingless bee species are expected in the aliphatic region of the NMR spectrum of honey.
References
Al ML, Daniel D, Moise A, Bobis O, Laslo L, Bogdanov S. 2009. Physico-chemical and bioactive properties of different floral origin honeys from Romania. Food Chemistry 112(4):863–867.
Anklam E. 1998. A review of the analytical methods to determine the geographical and botanical origin of honey. Food Chemistry 63:549–562.
Arvanitoyannis I S, Chalhoub C, Gotsiou P, Lydakis-Simantiris N, Kefalas P. 2005. Novel quality control methods in conjunction with chemometrics (multivariate analysis) for detecting honey authenticity. Critical Reviews in Food Science and Nutrition 45:193–203.
Azeredo LdaC, Azeredo MAA, De Souza SR, Dutra VML. 2003. Protein contents and physicochemical properties in honey samples of Apis mellifera of different floral origins. Food Chemistry 80:249–254.
Belton PS, Delgadillo I, Gil AM, Holmes E, Nicholls A, Nicholson JK, Spraul M. 1996. Use of high-field 1H NMR spectroscopy for the analysis of liquid foods. Journal of Agricultural and Food Chemistry 44:1483–1487.
Beretta G, Caneva E, Regazoni L, Golbamaki Bakhtyari N, Maffei Facino R. 2008. A solid-phase extraction procedure coupled to 1H NMR, with chemometric analysis, to seek reliable markers of the botanical origin of honey. Analytica Chimica Acta 620:176–182.
Bogodanov S, Ruoff K, Persano Oddo L. 2004. Physico-chemical methods for the characterisation of unifloral honeys: a review. Apidologie 35:S4-S17.
Cevallos-Cevallos JM, Reyes-De-Corcuera JI, Etxeberria E, Danyluk MD, Rodrick GE. 2009. Metabolomic analysis in food science: a review. Trends in Food Science & Technology 20:557–566.
Claridge TDW. 1999. High-resolution NMR techniques in organic chemistry. PERGAMON, Elsevier Science Ltd; Oxford, UK. 382 pp.
Consonni R, Cagliani L. 2008. Geographical characterization of polyfloral and acacia honeys by nuclear magnetic resonance and chemometrics. Journal of Agricultural and Food Chemistry 56:6873–6880.
Donarski JA, Jones SA, Charlton AJ. 2008. Application of cryoprobe 1H nuclear magnetic resonance spectroscopy and multivariate analysis for the verification of Corsican honey. Journal of Agricultural and Food Chemistry 56:5451–5456.
Eriksson L, Johansson E, Kettaneh-Wold N, Trygg J, Wikström C, Wold S. 2006. UMETRICS AB; Umeå, Sweden. 425 pp.
Günther H. 1995. NMR spectroscopy: basic principles, concepts, and applications in chemistry. WILEY; Guildford, UK. 581 pp.
Kang J, Choi MY, Kang S, Kwon HN, Wen H, Lee CH, Park M, Wiklund S, Kim HJ, Kwon SW, Park S. 2008. Application of a 1H nuclear magnetic resonance (NMR) metabolomics approach combined with orthogonal projections to latent structure-discriminant analysis as an efficient tool for discriminating between Korean and Chinese herbal medicines. Journal of Agricultural and Food Chemistry 56:11589–11595.
Lindon JC, Nicholson JK, Holmes E, Everett JR. 2000. Metabonomics: metabolic processes studied by NMR spectroscopy of biofluids. Concepts in Magnetic Resonance 12(5):289–320.
Lolli M, Bertelli D, Plessi M, Sabatini AG., Restani C. 2008. Classification of Italian honeys by 2D HR-NMR. Journal of Agricultural and Food Chemistry 56:1298–1304.
Rastrelli F, Schievano E, Bagno A, Mammi S. 2009. NMR Quantification of trace components in complex matrices by band-selective excitation with adiabatic pulses. Magnetic Resonance in Chemistry 47:868–872.
Rochfort S. 2005. Metabolomics reviewed: a new “omics” platform technology for systems biology and implications for natural products research. Journal of Natural Products 68:1813–1820.
Schievano E, Pasini G, Cozzi G, Mammi S. 2008. Identification of the production chain of asiago d’allevo cheese by nuclear magnetic resonance spectroscopy and principal component analysis. Journal of Agricultural and Food Chemistry 56:7208–7214.
Schievano E, Peggion E, Mammi S. 2010. 1H nuclear magnetic resonance spectra of chloroform extracts of honey for chemometric determination of its botanical origin. Journal of Agricultural and Food Chemistry. 58:57–65.
Schievano E, Stocchero M, Morelato E, Facchin C, Mammi S. 2012. An NMR-based metabolomic approach to identify the botanical origin of honey. Metabolomics 8:679–690.
Son HS, Kim KM, Van De berg F, Hwang GS, Park WM, Lee CH, Hong YS. 2008. H-1 nuclear magnetic resonance-based metabolomic characterization of wines by grape vaieties and production areas. Journal of Agricultural and Food Chemistry 56:8007–8016.
Spraul M, Schütz B, Humpfer E, Mörtter M, Schäfer H, Koswig S, Rinke P. 2009. Mixture analysis by NMR as applied to fruit juice quality control. Magnetic Resonance in Chemistry 47:S130-S137
Tarachiwin L, Ute K, Kobayashi A, Fukusakii E. 2007. 1H NMR based metabolomic profiling in the evaluation of Japanese green tea quality. Journal of Agricultural and Food Chemistry 55:9339–9336.
Teresa W, Fan M. 1996. Metabolite profiling by one- and two-dimensional NMR analysis of complex mixtures. Progress in Nuclear Magnetic Resonance Spectroscopy 28:161–219.
Wishart DS. 2008. Metabolomics: applications to food science and nutrition research. Trends in Food Science & Technologies 19:482–493.
Xiao CN, Dai H, Liu HB, Wang YL, Tang HR. 2008. Revealing the metabolomic variation of rosemary extracts using 1H NMR spectroscopy and multivariate data analysis. Journal of Agricultural and Food Chemistry 56:10142–10153.
Acknowledgments
See the chapter on anticancer activity by Vit et al. in this book for the pot-honey samples received for this study. Prof. Paulo Nogueira-Neto provided the sucrose pot-honey of M. quadrifasciata, from São Paulo, Brazil. The Apis mellifera honeys were provided by Rigoni S.p.A. within the Veneto Region, Italy, UNIMIELE project 2008, and Miel La Encantada, Venezuela. We acknowledge Prof. Vit’s proposal to initiate this research in our lab.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer Science+Business Media New York
About this chapter
Cite this chapter
Schievano, E., Mammi, S., Menegazzo, I. (2013). Nuclear Magnetic Resonance as a Method to Predict the Geographical and Entomological Origin of Pot-Honey. In: Vit, P., Pedro, S., Roubik, D. (eds) Pot-Honey. Springer, New York, NY. https://doi.org/10.1007/978-1-4614-4960-7_31
Download citation
DOI: https://doi.org/10.1007/978-1-4614-4960-7_31
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4614-4959-1
Online ISBN: 978-1-4614-4960-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)