Abstract
Various vectors have been developed in the field of gene transfer. However, important unresolved problems persist such as secondary toxicity or low gene transfer efficiency of viral as well as nonviral vectors. Therefore, an efficient and safe method of DNA delivery needs to be found. DNA electrotransfer is a physical method that consists of the local application of electric pulses after the introduction of DNA into the extracellular medium. Electrotransfer has proven to be one of the most efficient and simple nonviral methods of delivery. Moreover, it may provide an important alternative technique in the field of cell and gene therapies. The present chapter focuses on questions related to the mechanism of DNA electrotransfer, i.e., the basic processes responsible for membrane electropermeabilization, interaction of plasmid DNA with the plasma membrane, and its transport through the cytoplasm and into the nucleus.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Electroporation
- Plasmid DNA
- Plasma membrane
- Intracellular trafficking
- Nuclear envelope
- DNA targeting sequence
Introduction
Gene therapy involves the delivery of therapeutic genes into the target cells in order to prevent, treat, or cure genetic and acquired human diseases. The administration of naked plasmid DNA into target cells and tissues can be considered as the simplest and safest method for gene delivery [1]. Historically, naked plasmid DNA has not been the choice vector for gene therapy because of the variable and relatively low transfection efficiencies compared to those achieved with viral vectors [2]. Unfortunately, viral proteins can induce specific immune responses that can limit the ability to readminister the viral vectors [3]. Moreover, viral vectors, like retrovirus or lentivirus, can evoke insertional mutations during their integration into the host genome [4, 5]. As a result, these adverse events may pose a danger for the patient. In contrast, naked plasmid DNA is composed entirely of covalently closed circles of double-stranded DNA with no associated protein [6]. Moreover, naked plasmid DNA is simple to manipulate and can easily be produced in large pharmaceutical grade quantities [7]. These properties highlight why plasmid DNA is an attractive molecule for human clinical applications. However, the current challenge consists of developing an efficient and safe method for gene delivery.
The controlled use of electric pulses as a safe tool to deliver therapeutic molecules to tissues and organs has been developed over the last decade [8, 9]. This method refers to the transient increase in the permeability of the cell membrane when exposed to electric field pulses. This process is commonly known as electropermeabilization or electroporation [10, 11]. Nowadays, electropermeabilization represents one of the most widespread techniques to transfer genetic material. In vivo gene electrotransfer is of special interest since it is the most efficient nonviral strategy for gene delivery and also because of its low cost, ease of realization, and safety [11]. Moreover, gene delivery is limited to the volume of tissue localized between the electrodes, where the electric field pulses are applied. In contrast to viral vectors, this method allows the delivery of large plasmid DNA (range from 3 to 5 kbp), which greatly expands research and clinical applications. As a result, gene electrotransfer has now been applied to a wide variety of tissues including muscle [12, 13], skin [14, 15], liver [16], lung [17, 18], kidney [19], eyes [20, 21], brain [22], joints [23], and tumors [24, 25]. This strategy is not only promising for the treatment of genetic and acquired diseases, but also for the systemic secretion of therapeutic proteins [26]. Genetic vaccination and cancer gene therapy are also additional fields of application [27, 28]. Therefore, electrogenetherapy is relevant in a variety of research and clinical settings including cancer therapy, modulation of pathogenic immune responses, and treatment of infectious diseases [29]. A phase I dose escalation trial of electroporation of a plasmid expressing interleukin-12 (IL-12) has been carried out in patients with metastatic melanoma. Biopsies showed plasmid dose proportional increases in IL-12 protein levels as well as marked tumor necrosis and lymphocytic infiltrate, indicating this modality to be safe, effective, and titratable [30].
However, the mechanisms underlying DNA electrotransfer are not completely known. The comprehension of these mechanisms is necessary for the rational use (i.e., efficiency and safety) of the method, in vitro and in vivo [31]. Successful electrotransfer of plasmid DNA into target cells depends on the ways the cell membrane has been permeabilized, as well as how the DNA molecules interact with and are transported from the plasma membrane towards the nuclear envelope. The focus of this review is to describe the different aspects of these processes.
Membrane Electropermeabilization
Basic Aspects: Modulation of Membrane Potential Difference
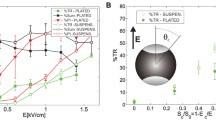
The key effect of an electric field on cells is a position-dependent change in the resting transmembrane potential difference, ΔΨ0, of their plasma membrane. If the cell membrane is modeled as a thin spherical dielectric shell, the electrically induced potential difference, ΔΨ E , which is the difference between the potential inside the cell, Ψin, and the potential outside the cell, Ψout, at a point M on the cell surface, is given by:
where t is the time after the onset of the electric pulse, g depends on the conductivities λ of the membrane, of the cytoplasm, and of the extracellular medium, r is the radius of the cell, E the field strength, θ M is the angle between the normal to the membrane at the position M and the direction of the field, and τ is the membrane charging time (Fig. 13.1a) [32]. The field-induced potential difference adds to the resting potential ΔΨο [33].
Physical principle of electropermeabilization. (a) The plasma membrane is the site of native transmembrane potential difference, Δψ0 (blue arrow). If we consider the cell like a sphere and the plasma membrane like a dielectric spherical shell, when we apply an electric pulse, an induced transmembrane voltage, Δψ E , is created (red arrow). Being dependent on the angular parameter θ, the Δψ E effect is position-dependent on the cell surface; therefore, the side of the cell facing the anode is hyperpolarized, while the side of the cell facing the cathode is depolarized. (b) The localization and asymmetry of electropermeabilization can be detected by propidium iodide uptake in a CHO cell submitted to a train of 10 pulses, 5 ms, 1 Hz at 0.7 kV/cm. Propidium iodide uptake is visualized by fluorescence microscopy. Pseudocolor image and 3D graph are obtained by Image J software
Being dependent on the angular parameter θ, the field effect is position-dependent on the cell surface. Therefore, the side of the cell facing the anode is going to be hyperpolarized, while the side of the cell facing the cathode is depolarized (Fig. 13.1b). This theoretical prediction has been experimentally validated by using a voltage-sensitive fluorescent dye [34]. Therefore, the transmembrane potential difference of a cell exposed to an electric field defines the sites (location, extend) where molecule uptake can take place.
Scaling of Electropermeabilization
Therefore, membrane permeabilization occurs only on the part of the cell membrane where the potential difference has been brought to its critical value [35]. This value has been evaluated to be on the order of 200–300 mV regardless of the cell type [36]. Field intensities, E, larger than a critical value, E p, must be applied where E p is dependent on the size of the target cells. This value ranges from 200 V/cm in the case of large cells such as mice myotubes to 1–2 kV/cm in the case of bacteria. Therefore, electric field values have to be adapted to each cell line. Membrane permeabilization is controlled by the field strength (Fig. 13.2), which is the trigger of permeabilization: when E is larger than E p, it controls the area of the cell surface which is affected [37]. Membrane permeabilization only occurs for electric field values, E, higher than the threshold value, E p, regardless of the number and duration of electric pulses. Increasing E above E p leads to an increase in the area where permeabilization takes place and, in that particular area, the extent of permeabilization is determined by the number and duration of electric pulses (Fig. 13.2). As a whole, these data led to the concept of membrane domains, where macrodomains are regions where permeabilization can take place and in which an area is determined by the pulse intensity according to:
Effect of the electric field parameters on membrane permeabilization. The electric field E leads to the induction of a transmembrane potential difference, Δψ E , which sur-imposes to the resting Δψ0. Cells are hyperpolarized at the anode-facing side, while depolarized at the cathode side. The pink area represents the area where the resulting potential is higher than the threshold value, so where permeabilization is present when E > E p. Increasing E above E p leads to an increase in this area. At a constant E value, increasing the pulse number N or the pulse duration T does not lead to any increase of that area, but leads to a increase in permeabilization efficiency as shown here by a red color. Blue squares represent the electrodes
where A is the cell surface. Within macrodomains, microdomains (defects, pores, permeant structures, etc.) exist as areas where permeabilization actually takes place and in which density depends on the number of pulses and on the pulse duration [37]. However, the molecular characteristics of these domains in terms of composition, structures, and dynamics remain an open question. Theoretical models have been proposed to explain the mechanism of this reversible membrane electropermeabilization. Nevertheless, the molecular definition of the transient permeable structures is not yet known.
Electropermeabilization, A Fast and Localized Process
The use of video microscopy allows visualization of the permeabilization phenomenon at the single cell level. Propidium iodide uptake in the cytoplasm is a fast process that can be detected seconds after the application of electric pulses (Fig. 13.1b). Exchange across the permeabilized membrane is not homogeneous on the whole cell membrane. It occurs at the sides of the cells facing the electrodes in an asymmetrical way where it is more pronounced at the anode-facing side of the cells than at the cathode, i.e., in the hyperpolarized area than in the depolarized area [38], which is in agreement with the above theoretical considerations.
Electropermeabilization can be described as a three-step process in respect with electric field: (1) Before electropulsation, the plasma membrane acts as a physical barrier that prevents the free exchange of hydrophilic molecules between the cell cytoplasm and external medium; (2) during electropulsation, the transmembrane potential increases which induces the formation of transient permeable structures facing the electrodes and allows the exchange of molecules; (3) after electropulsation, membrane resealing occurs.
Gene Electrotransfer
Basic Aspects: Scaling of Gene Expression
Membrane permeabilization is the first step of gene electrotransfer. However, this step, although necessary, is not sufficient to obtain gene expression. Millisecond pulses are generally required to obtain efficient gene expression by preserving cell viability and limiting the electric field intensities required when short pulses are used [39, 40]. Nevertheless, gene electrotransfer can be obtained with short strong pulses [41]. The transfection efficiency obeys the following equation:
where K is a constant, N the number of pulses, and T is the pulse duration. The dependence on the plasmid concentration f(DNA) is rather complex, and it is observed that high levels of plasmids are toxic [42]. The effect of pulse duration also appears to be essential as it is a key parameter for efficient gene expression in target cells and tissues [41].
Additionally, the polarity of the electric field has a direct effect on transfection. This dependence of the transfection efficiency on the direction of the field might be due to the involvement of the electrophoretic force in the translocation of the negatively charged plasmid DNA [43]. While cell permeabilization is only slightly affected by reversing the polarity of the electric pulses or by changing the orientation of pulses, increased transfection levels are observed where these last effects are due to an increase in the cell membrane area where plasmid DNA interacts [44].
Gene Electrotransfer: A Multistep and Localized Process
Fluorescent plasmids allow monitoring of the interaction of DNA molecules with permeabilized cells [45]. DNA molecules, which are negatively charged, migrate electrophoretically when subjected to the electric field. Under electric fields, which are too small to permeabilize the membrane, the DNA simply flows around the membrane in the direction of the anode. However, beyond a critical field value, above which cell permeabilization occurs, the DNA interacts with the plasma membrane. This interaction only occurs at the pole of the cell opposite the cathode and demonstrates the importance of electrophoretic forces in the initial phase of the DNA/membrane interaction. When the DNA/membrane interaction occurs, one observes the formation of microdomains whose dimensions lie between 0.1 and 0.5 μm. Also seen are clusters or aggregates of DNA, which grow during the application of the field. However, once the field is cut, the growth of these clusters stops. DNA electrotransfer can be described as a multistep process with respect to electropulsation. First, the negatively charged DNA migrates electrophoretically towards the plasma membrane on the cathode side, where it accumulates. For electric field values above a certain threshold, the plasma membrane is permeabilized, thereby allowing the accumulated plasmid DNA to be inserted into it. This interaction, which is observed for several minutes, lasts much longer than the duration of the electric field pulse. Translocation of the plasmid from the plasma membrane to the cytoplasm and its subsequent passage towards the nuclear envelope takes place with kinetics ranging from minutes to hours. When the plasmid has reached the nucleus, gene expression can take place and can be detected for up to several weeks afterwards. Due to the good correlation between visualization of DNA/membrane interaction and gene expression, these results are consistent with a multistep process of DNA electrotransfer: (1) Before electropulsation: plasma membrane acts as a physical barrier that prevents the entrance of plasmid DNA into the target cells (Fig. 13.3a); (2) during electropulsation, the plasma membrane is permeabilized on the cell sides facing the electrodes. Negatively charged plasmid DNA migrates electrophoretically towards the permeabilized membrane on the cathode side, where it is inserted in membrane competent sites (Fig. 13.3a). Nevertheless, DNA/membrane interaction can be obtained on the whole cell membrane perimeter by changing the polarity of electric pulses [44]. (3) After electropulsation, plasmid translocation through the cytoplasm to the nucleus takes place, eventually leading to gene expression. This final step, including plasmid DNA trafficking in the cytosol and its passage through the nuclear pore, can limit gene expression. The challenge is to overcome these limiting steps. An alternative approach comes from nanosecond pulsed electric fields (nPEFs). New findings indeed indicate that very short (10–300 ns) but high (up to 300 kV/cm) pulses extend classical electropermeabilization to include events that primarily affect intracellular structures and functions. As the pulse duration is decreased below the plasma membrane charging time constant (see equation 13.1 [12]), plasma membrane effects decrease and intracellular effects predominate [46, 47]. When used in conjugation with classical electropermeabilization, nanopulses can increase gene expression. The idea is to perform classical membrane permeabilization and allow plasmid DNA electrotransfer and then, 30 min later, permeabilize the nuclear envelope by using short nPEFs. Thus, it is possible to not only electropermeabilize cells, but also electromanipulate them.
The different steps involved in DNA electrotransfer. (a) During the application of the electric field, DNA molecules (labeled with a fluorescent marker, TOTO-1) migrate towards the electropermeabilized cell plasma membrane where they interact with and can become inserted in the membrane. This interaction proceeds from a few ms to a few minutes. (b) After the electric field application, transport of DNA in the cytoplasm takes place with kinetics ranging from minutes to hours, finally leading to (c) gene expression lasting several days to weeks
Cytoplasmic Trafficking
Once the membrane has been destabilized allowing for DNA entry, plasmids are confronted by a number of barriers that must be overcome in order for gene expression to occur, none of which depend on the electrical field. Studies with fluorescently labeled plasmids show that the DNA crosses the plasma membrane, but remains near the inner surface of the membrane for a period of time. Since cortical actin forms a meshwork beneath the plasma membrane in order to provide structure to the cell and connections between surface proteins and intracellular networks, it is likely that the entering DNA becomes “trapped” within this area for a brief amount of time. Similar findings have been reported for the entry of a number of viral particles, including HIV and Semliki Forest virus [48, 49]. How the DNA escapes this region is unclear, but the meshwork is not uniform and areas with a larger mesh size may allow for localized diffusion of the DNA out of this region and toward the interior.
How DNA, or any molecule for that matter, moves through the cytoplasm to the nucleus has long been assumed to be by means of diffusion. However, a number of studies have elegantly shown that the cytoplasm is relatively stiff and does not allow for a great deal of free diffusion of large molecules. Using a combination of cell-free extracts, microinjected cells, and fluorescence recovery after photobleaching experiments, Verkman and colleagues demonstrated that DNA fragments longer that 2,000 bp do not diffuse to any significant degree within a biologically relevant time frame [50]. Indeed, while 1,000 bp DNA fragments diffuse relatively freely in cytoplasm, 2,000 bp fragments show greatly reduced movement, and 6,000 bp fragments are immobile. The reason for this limited diffusion is that the cytoplasm is not simply an aqueous environment, but rather a complex system with visco-elastic properties which is composed of multiple cytoskeletal elements, including microfilaments, microtubules, and intermediate filaments. These elements are organized into a complex, crowded latticework that is constantly remodeling itself in response to a variety of internal and external stimuli. This lattice is a barrier to the movement of DNA toward the nucleus, which is evident from the fact that disruption of the actin cytoskeleton using latrunculin B or cytochalasin D resulted in greatly enhanced diffusion of large DNA fragments within the cytoplasm of microinjected cells [51].
If, then, DNA cannot diffuse to any extent through the cytoplasm, how does it reach the nucleus? It has been demonstrated that, like many viruses and cellular proteins, plasmids move through the cytoplasm along the microtubule network using dynein as a microtubule motor protein [52, 53]. Unlike many proteins that are trafficked along microtubules, DNA does not bind directly to either dynein or the microtubules themselves. Rather, it is thought to interact with the motor proteins through adapter proteins that bind to the DNA once it enters the cytoplasm [52, 53]. While it is “naked” DNA that is electroporated into the cell, any plasmid or DNA fragment very quickly becomes associated with a number of cations, peptides, and proteins that interact electrostatically and/or sequence-specifically with the DNA. The resulting protein–DNA complex is the form of DNA that traffics through the cell. Although the exact nature of the protein complement that binds to the DNA to mediate interactions with the microtubule motors to facilitate movement is unknown, several recent studies have used a proteomics approach to begin to address this question [54, 55]. Taking an in vitro approach to identify proteins in cytoplasmic extract, which bind to plasmids that traffic and enter the nuclei of either all cells or specific cell types, a number of proteins have been identified. These include a number of proteins that bind directly, but nonspecifically, to DNA as well as a number of general and cell-specific transcription factors that recognize unique sequences within the plasmids. While both studies focused on the ability of these plasmids to enter the nuclei of nondividing cells, all DNA that enters the nucleus must also be able to move through the cytoplasm, so it is likely that many of the proteins identified may also play a role in cytoplasmic trafficking as well as nuclear import. Since many proteins that bind DNA are nuclear proteins, when they bind to the DNA, the DNA becomes partially coated with nuclear localization signals (NLSs), the small amino acid postal code that directs proteins to the nucleus. Both studies found the small GTPase RAN and several importin β family members that bind to the DNA, supporting previous studies showing that these proteins are necessary for nuclear import of plasmids [56]. Further, coprecipitation assays of cells that had been electroporated with these plasmids demonstrated that both proteins formed complexes in transfected cells following electroporation. RNA interference-mediated reduction of these proteins confirmed that importin β was indeed needed for cytoplasmic movement and nuclear import in living cells. Taken together with several studies demonstrating that importin β can bind to NLSs on proteins destined for the nucleus and at the same time bind to dynein for movement along microtubules [52, 57–59], these findings support the model for trafficking of plasmids (Fig. 13.4).
Intracellular trafficking of plasmids. Following electropermeabilization of the plasma membrane and translocation through the cortical actin layer, plasmids associate with a number of DNA-binding proteins present in the cytoplasm to form DNA–protein complexes. At least some of the NLSs present on these DNA-binding proteins are accessible to the importin machinery and are bound by importin β or transportin, which then interacts with the microtubule motor protein dynein to move the DNA through the cytoplasm toward the nucleus. The motor–cargo complex then falls apart, allowing the DNA to enter the nucleus in a DNA-sequence- and importin-dependent process
Additionally, the speed with which plasmids move to the nucleus has not been directly measured, but in static cells, once DNA is free in the cytoplasm, nuclear DNA can be detected within 1 h in many cell types [60, 61]. Further, in cells electroporated with plasmid, gene expression can be detected within several hours [61], suggesting that movement can be relatively rapid, although much slower than the millisecond timescale needed for transport across the plasma membrane. The rates of plasmid movement through the cytoplasm also can be increased by manipulation of the microtubule network. In initial studies designed to look at how stretching the basement membrane of lung epithelial cells in a cyclic manner to represent tidal breathing in the lung affected the intracellular trafficking of plasmids, it was found that cyclic stretch increased microtubule acetylation and that cells with elevated levels of acetylated microtubules showed greater and more rapid gene expression following transfection [62, 63]. This effect was mediated through the major tubulin deacetylase, HDAC6, whose activity was inhibited by cyclic stretch. When the level of acetylated microtubules increased by the application of cyclic stretch, drug inhibition of HDAC6, or siRNA-mediated silencing of HDAC6, plasmids trafficked along microtubules to the nucleus much faster than in untreated cells, resulting in up to an almost 30-fold increase in transfection efficiency by 24 h postelectroporation [61].
Nuclear Import
Plasmid movement through the cytoplasm toward the nuclear envelope does not guarantee that the plasmid will be translocated across the nuclear envelope into the nucleus for gene expression. Indeed, it has been shown that in the absence of cell division, nuclear import of plasmid DNA is a sequence-specific process that requires binding of specific transcription factors and other proteins to the DNA in order for nuclear translocation to occur. In most laboratory transfections, actively dividing cells are used. Since one of the hallmarks of mitosis is nuclear envelope breakdown, any DNA that has entered the cytoplasm prior to mitosis would gain access to the nuclear compartment once cells enter the M phase. Indeed, it has been repeatedly shown that nonviral transfections, and transductions by numerous viruses, are cell cycle dependent [64–68]. Although it has been shown that mitosis-associated nuclear envelope breakdown greatly enhances nuclear localization of plasmids and transfection efficiency, it is not required.
Numerous groups have demonstrated that plasmids can enter the nuclei through nuclear pore complexes (NPCs) in the absence of cell division, although the efficiency of such transfection is usually much lower than in dividing cells [69–71]. Moreover, certain DNA sequences can increase this nuclear targeting of plasmids prior to mitosis. This nuclear import of plasmid DNA through the NPCs is a sequence-specific process, mediated by specific eukaryotic sequence elements [69]. When delivered side-by-side, plasmids containing as little as 72 bp of the SV40 enhancer target the nucleus of most cells within several hours, whereas an isogenic plasmid lacking this 72 bp sequence remains cytoplasmic until cell division (or indefinitely if the cell is nondividing) [69, 72]. This sequence, termed the SV40 DNA nuclear targeting sequence (DTS), has been shown to mediate plasmid nuclear import in all primary cells and cell lines tested, as well as in tissues of living animals [69, 71–77]. Apart from this SV40 sequence, several other DTSs have been identified and shown to mediate plasmid nuclear import in either all or specific cell types [71, 78–80]. For example, Reich and colleagues have shown that plasmids containing multiple NF-κB binding sites had increased levels of gene transfer compared to identical plasmids lacking NF-kB binding sites, due to more efficient transfer across the nuclear envelope [52, 81]. Further, several DTSs have been shown to act only in specific cell types due to the fact that they bind to transcription factors expressed only in those cells. Two such sequences are the smooth-muscle-cell-specific SMGA DTS, whose activity is dependent on the smooth-muscle-specific transcription factors SRF and Nkx3.1 [71, 76, 78, 79], and the alveolar epithelial type II cell-specific SPC DTS, whose activity requires several transcription factors expressed together in alveolar epithelial type II cells [80]. In all cases, inclusion of the DTS on a plasmid greatly increases nuclear import and gene expression in cells.
The defining feature of the DTS is that it contains binding sites for a number of transcription factors. Because transcription factors act in the nucleus, they contain NLSs that target them to the nucleus through interactions with receptor proteins. Although transcription factors function in the nucleus, they spend a fair amount of time in the cytoplasm, either after their translation or as part of their normal regulation (many factors, such as NF-kB, are sequestered in the cytoplasm as a way of controlling their access to genes in the nucleus). When plasmids carrying a DTS are delivered into the cytoplasm by any method, some of these transcription factors can bind to the DTS, thereby coating a region of the plasmid with NLSs, at least some of which are oriented away from the DNA itself. These DNA-bound NLSs can be recognized by importin β or transportin and transported into the nucleus through the NPC [54, 56, 69, 70, 82, 83].
Although the function of the DTS is mediated by binding of NLS-containing transcription factors, it would seem that any eukaryotic regulatory sequence could function similarly for DNA nuclear import. Surprisingly, this is not the case and although half a dozen or so DNA nuclear targeting sequences have been identified, most promoters and enhancers, including the CMV immediate-early promoter/enhancer, the Herpes TK promoter, and the RSV LTR, have no import activity [72]. The likely explanation for this is that the transcription factors bound to these other promoters may not present their NLSs in an orientation that is accessible to the importins, as demonstrated by studies with GAL4 DNA-importin binding [84]. Further, while the dependence on the DTS for plasmid nuclear import appears almost absolute in cultured cells, a number of studies have shown that in many tissues, especially skeletal muscle, robust gene transfer and expression can be obtained using plasmids lacking any nuclear import sequence. It is likely that when the cytoplasm becomes filled with large concentrations of plasmids, at least some of the plasmids can randomly make their way to the nuclear envelope and be imported into the nucleus independent of any DTS. Indeed, when linear DNA is brought close enough to the nuclear envelope using laser tweezers, it is pulled in [85]. Further, it has been shown that when DTS-lacking plasmids are delivered to the cytoplasm of a mouse myotube in vivo, no gene expression is observed until 1,000,000 plasmids are injected, suggesting that mass action could account for the nuclear localization [86].
Conclusion
All in all, in addition to its potential use in gene therapy, gene electrotransfer is, because of its simplicity, a powerful laboratory tool to study gene expression and function in a given cell or a tissue. The processes by which molecules translocate across the electropermeabilized membrane are dependent on the size of the molecules. Small molecules can freely cross the permeabilized cell membrane, but plasmid DNA transfer involves complex steps including interaction with the permeabilized membrane. New directions of research are needed to characterize membrane competent sites involved in gene electrotransfer in terms of composition, structure, and dynamics. If the effects of the electric field parameters are about to be elucidated (electric pulse strength higher than a threshold value, millisecond pulse duration for efficient gene expression), the associated membrane destabilization, which is a stress for the cells and may affect the cell viability, has still to be clearly described. Moreover, it becomes evident that extracellular barriers (e.g., extracellular matrix and exogenous nucleases) and intracellular barriers (e.g., cytoplasm crowding, endogenous nucleases, nuclear envelope) compromise the transfection efficiency. Furthermore, studies will also be necessary to understand the cascade of events triggered by electropermeabilization at the tissue level where new constraints coming from tissue organization are present, such as the inhomogeneity of the electric field strength and the intracellular distribution of plasmid DNA [38, 87].
References
Wolff JA, Budker V. The mechanism of naked DNA uptake and expression. Adv Genet. 2005;54:3–20.
Wolff JA et al. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–8.
Couzin J, Kaiser J. Gene therapy. As Gelsinger case ends, gene therapy suffers another blow. Science. 2005;307(5712):1028.
Hacein-Bey-Abina S et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302(5644):415–19.
Hacein-Bey-Abina S et al. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N Engl J Med. 2002;346(16):1185–93.
Wells DJ. Gene therapy progress and prospects: electroporation and other physical methods. Gene Ther. 2004;11(18):1363–9.
Bower DM, Prather KL. Engineering of bacterial strains and vectors for the production of plasmid DNA. Appl Microbiol Biotechnol. 2009;82(5):805–13.
Neumann E, Sowers AE, Jordan CA. Electroporation and electrofusion in cell biology. New York: Plenum; 1989.
Weaver JC. Electroporation theory. Concepts and mechanisms. Methods Mol Biol. 1995;55:3–28.
Teissie J, Golzio M, Rols MP. Mechanisms of cell membrane electropermeabilization: a minireview of our present (lack of?) knowledge. Biochim Biophys Acta. 2005;1724(3):270–80.
Escoffre JM et al. Membrane perturbation by an external electric field: a mechanism to permit molecular uptake. Eur Biophys J. 2007;36(8):973–83.
Aihara H, Miyazaki J. Gene transfer into muscle by electroporation in vivo. Nat Biotechnol. 1998;16(9):867–70.
Mir LM et al. High-efficiency gene transfer into skeletal muscle mediated by electric pulses. Proc Natl Acad Sci USA. 1999;96:4262–7.
Titomirov AV, Sukharev S, Kistanova E. In vivo electroporation and stable transformation of skin cells of newborn mice by plasmid DNA. Biochim Biophys Acta. 1991;1088(1):131–4.
Vandermeulen G et al. Optimisation of intradermal DNA electrotransfer for immunisation. J Control Release. 2007;124(1–2):81–7.
Heller R et al. In vivo gene elctroinjection and expression in rat liver. FEBS Lett. 1996;389:225–8.
Dean DA et al. Electroporation as a method for high-level non-viral gene transfer to the lung. Gene Ther. 2003;10(18):1608–15.
Zhou R et al. Electroporation-mediated transfer of plasmids to the lung results in reduced TLR9 signaling and inflammation. Gene Ther. 2007;14(9):775–80.
Isaka Y et al. Electroporation-mediated HGF gene transfection protected the kidney against graft injury. Gene Ther. 2005;12(10):815–20.
Blair-Parks K, Weston BC, Dean DA. Gene delivery to the cornea by plasmid injection and electroporation. J Gene Med. 2002;4:92–100.
Bloquel C et al. Plasmid electrotransfer of eye ciliary muscle: principles and therapeutic efficacy using hTNF-alpha soluble receptor in uveitis. FASEB J. 2006;20(2):389–91.
Judkewitz B et al. Targeted single-cell electroporation of mammalian neurons in vivo. Nat Protoc. 2009;4(6):862–9.
Khoury M et al. A comparative study on intra-articular versus systemic gene electrotransfer in experimental arthritis. J Gene Med. 2006;8(8):1027–36.
Rols MP et al. In vivo electrically mediated protein and gene transfer in murine melanoma. Nat Biotechnol. 1998;16(2):168–71.
Cemazar M et al. Control by pulse parameters of DNA electrotransfer into solid tumors in mice. Gene Ther. 2009;16(5):635–44.
Bloquel C et al. Plasmid DNA electrotransfer for intracellular and secreted proteins expression: new methodological developments and applications. J Gene Med. 2004;6 Suppl 1:S11–23.
Hirao LA et al. Combined effects of IL-12 and electroporation enhances the potency of DNA vaccination in macaques. Vaccine. 2008;26(25):3112–20.
Rice J, Ottensmeier CH, Stevenson FK. DNA vaccines: precision tools for activating effective immunity against cancer. Nat Rev Cancer. 2008;8(2):108–20.
Gehl J. Electroporation for drug and gene delivery in the clinic: doctors go electric. Methods Mol Biol. 2008;423:351–9.
Daud AI et al. Phase I trial of interleukin-12 plasmid electroporation in patients with metastatic melanoma. J Clin Oncol. 2008;26(36):5896–903.
Escoffre JM et al. What is (still not) known of the mechanism by which electroporation mediates gene transfer and expression in cells and tissues. Mol Biotechnol. 2009;41(3):286–95.
Bernhardt J, Pauly H. On the generation of potential differences across the membranes of ellipsoidal cells in an alternating electrical field. Biophysik. 1973;10(3):89–98.
Kotnik T et al. Evaluation of cell membrane electropermeabilization by means of a nonpermeant cytotoxic agent. Biotechniques. 2000;28(5):921–6.
Hibino M, Itoh H, Kinosita Jr K. Time courses of cell electroporation as revealed by submicrosecond imaging of transmembrane potential. Biophys J. 1993;64(6):1789–800.
Kotnik T, Miklavcic D. Analytical description of transmembrane voltage induced by electric fields on spheroidal cells. Biophys J. 2000;79(2):670–9.
Teissie J, Rols MP. An experimental evaluation of the critical potential difference inducing cell membrane electropermeabilization. Biophys J. 1993;65(1):409–13.
Rols MP, Teissie J. Electropermeabilization of mammalian cells. Quantitative analysis of the phenomenon. Biophys J. 1990;58(5):1089–98.
Bureau MF et al. Intramuscular plasmid DNA electrotransfer: biodistribution and degradation. Biochim Biophys Acta. 2004;1676(2):138–48.
Rols MP, Teissie J. Electropermeabilization of mammalian cells to macromolecules: control by pulse duration. Biophys J. 1998;75(3):1415–23.
Kubiniec RT, Liang H, Hui SW. Effects of pulse length and pulse strength on transfection by electroporation. Biotechniques. 1990;8(1):16–20.
Neumann E et al. Gene transfer into mouse lyoma cells by electroporation in high electric fields. EMBO J. 1982;1(7):841–5.
Rols MP, Coulet D, Teissie J. Highly efficient transfection of mammalian cells by electric field pulses. Application to large volumes of cell culture by using a flow system. Eur J Biochem. 1992;206(1):115–21.
Klenchin VA et al. Electrically induced DNA uptake by cells is a fast process involving DNA electrophoresis. Biophys J. 1991;60(4):804–11.
Faurie C et al. Effect of electric field vectoriality on electrically mediated gene delivery in mammalian cells. Biochim Biophys Acta. 2004;1665(1–2):92–100.
Golzio M, Teissie J, Rols MP. Direct visualization at the single-cell level of electrically mediated gene delivery. Proc Natl Acad Sci USA. 2002;99(3):1292–7.
Schoenbach KH, Beebe SJ, Buescher ES. Intracellular effect of ultrashort electrical pulses. Bioelectromagnetics. 2001;22(6):440–8.
Beebe SJ et al. Diverse effects of nanosecond pulsed electric fields on cells and tissues. DNA Cell Biol. 2003;22(12):785–96.
Gruenheid S, Finlay BB. Microbial pathogenesis and cytoskeletal function. Nature. 2003;422(6933):775–81.
Marsh M, Bron R. SFV infection in CHO cells: cell-type specific restrictions to productive virus entry at the cell surface. J Cell Sci. 1997;110(Pt 1):95–103.
Lukacs GL et al. Size-dependent DNA mobility in cytoplasm and nucleus. J Biol Chem. 2000;275(3):1625–9.
Dauty E, Verkman AS. Actin cytoskeleton as the principal determinant of size-dependent DNA mobility in cytoplasm: a new barrier for non-viral gene delivery. J Biol Chem. 2005;280(9):7823–8.
Mesika A et al. Enhanced intracellular mobility and nuclear accumulation of DNA plasmids associated with a karyophilic protein. Hum Gene Ther. 2005;16(2):200–8.
Vaughan EE, Dean DA. Intracellular trafficking of plasmids during transfection is mediated by microtubules. Mol Ther. 2006;13(2):422–8.
Miller AM et al. Identification of protein cofactors necessary for sequence-specific plasmid DNA nuclear import. Mol Ther. 2009;17:1897–903.
Munkonge FM et al. Identification and functional characterisation of cytoplasmic determinants of plasmid DNA nuclear import. J Biol Chem. 2009;284:26978–87.
Wilson GL et al. Nuclear import of plasmid DNA in digitonin-permeabilized cells requires both cytoplasmic factors and specific DNA sequences. J Biol Chem. 1999;274:22025–32.
Roth DM et al. A microtubule-facilitated nuclear import pathway for cancer regulatory proteins. Traffic. 2007;8(6):673–86.
Salman H et al. Nuclear localization signal peptides induce molecular delivery along microtubules. Biophys J. 2005;89(3):2134–45.
Perlson E et al. Vimentin-dependent spatial translocation of an activated MAP kinase in injured nerve. Neuron. 2005;45(5):715–26.
Dean DA. Improving gene delivery and expression of GFP by cyclic stretch. In: Spector DL, Goldman RD, editors. Live cell imaging: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2004. p. 51–66.
Vaughan EE et al. Microtubule acetylation through HDAC6 inhibition results in increased transfection efficiency. Mol Ther. 2008;16(11):1841–7.
Geiger RC et al. Cyclic stretch-induced reorganization of the cytoskeleton and its role in enhanced gene transfer. Gene Ther. 2006;13(8):725–31.
Taylor W et al. Effects of cyclic stretch on gene transfer in alveolar epithelial cells. Mol Ther. 2003;7(4):542–9.
Fasbender A et al. A low rate of cell proliferation and reduced DNA uptake limit cationic lipid-mediated gene transfer to primary cultures of ciliated human airway epithelia. Gene Ther. 1997;4:1173–80.
Brunner S et al. Cell cycle dependence of gene transfer by lipoplex, polyplex and recombinant adenovirus. Gene Ther. 2000;7:401–7.
Brunner S et al. Overcoming the nuclear barrier: cell cycle independent nonviral gene transfer with linear polyethylenimine or electroporation. Mol Ther. 2002;5(1):80–6.
Escriou V et al. Critical assessment of the nuclear import of plasmid during cationic lipid-mediated gene transfer. J Gene Med. 2001;3(2):179–87.
Tseng WC, Haselton FR, Giorgio TD. Mitosis enhances transgene expression of plasmid delivered by cationic liposomes. Biochim Biophys Acta. 1999;1445(1):53–64.
Dean DA. Import of plasmid DNA into the nucleus is sequence specific. Exp Cell Res. 1997;230:293–302.
Dowty ME et al. Plasmid DNA entry into postmitotic nuclei of primary rat myotubes. Proc Natl Acad Sci USA. 1995;92:4572–6.
Vacik J et al. Cell-specific nuclear import of plasmid DNA. Gene Ther. 1999;6:1006–14.
Dean DA et al. Sequence requirements for plasmid nuclear entry. Exp Cell Res. 1999;253:713–22.
Blomberg P et al. Electroporation in combination with a plasmid vector containing SV40 enhancer elements results in increased and persistent gene expression in mouse muscle. Biochem Biophys Res Commun. 2002;298(4):505–10.
Li S et al. Muscle-specific enhancement of gene expression by incorporation of the SV40 enhancer in the expression plasmid. Gene Ther. 2001;8:494–7.
Young JL, Benoit JN, Dean DA. Effect of a DNA nuclear targeting sequence on gene transfer and expression of plasmids in the intact vasculature. Gene Ther. 2003;10(17):1465–70.
Young JL, Zimmer WE, Dean DA. Smooth muscle-specific gene delivery in the vasculature based on restriction of DNA nuclear import. Exp Biol Med. 2008;233(7):840–8.
Zhou R, Dean DA. Gene transfer of interleukin 10 to the murine cornea using electroporation. Exp Biol Med (Maywood). 2007;232(3):362–9.
Miller AM, Dean DA. Cell-specific nuclear import of plasmid DNA in smooth muscle requires tissue-specific transcription factors and DNA sequences. Gene Ther. 2008;15(15):1107–15.
Miller AM, Dean DA. Tissue-specific and transcription factor-mediated nuclear entry of DNA. Adv Drug Deliv Rev. 2009;61(7–8):603–13.
DeGiulio JV, Kaufman CD, Dean DA. Alveolar epithelial cell-specific plasmid nuclear import. Gene Ther. 2010;17(4):541–9.
Mesika A et al. A regulated NFkappaB-assisted import of plasmid DNA into mammalian cell nuclei. Mol Ther. 2001;3(5 Pt 1):653–7.
Sebestyén MG et al. DNA vector chemistry: the covalent attachment of signal peptides to plasmid DNA. Nat Biotechnol. 1998;16:80–5.
Lachish-Zalait A et al. Transportin mediates nuclear entry of DNA in vertebrate systems. Traffic. 2009;10(10):1414–28.
Chan CK et al. Mutual exclusivity of DNA binding and nuclear localization signal recognition by the yeast transcription factor GAL4: implications for nonviral DNA delivery. Gene Ther. 1998;5(9):1204–12.
Salman H et al. Kinetics and mechanism of DNA uptake into the cell nucleus. Proc Natl Acad Sci USA. 2001;98:7247–52.
Utvik JK, Nja A, Gundersen K. DNA injection into single cells of intact mice. Hum Gene Ther. 1999;10(2):291–300.
Wasungu L et al. A 3D in vitro spheroid model as a way to study the mechanisms of electroporation. Int J Pharm. 2009;379(2):278–84.
Acknowledgments
Many thanks for the financial support from the CNRS, the Association Française sur les Myopathies, the Région Midi-Pyrénées, and NIH.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2011 Springer Science+Business Media, LLC
About this chapter
Cite this chapter
Escoffre, JM., Rols, MP., Dean, D.A. (2011). Electrotransfer of Plasmid DNA. In: Kee, S., Gehl, J., Lee, E. (eds) Clinical Aspects of Electroporation. Springer, New York, NY. https://doi.org/10.1007/978-1-4419-8363-3_13
Download citation
DOI: https://doi.org/10.1007/978-1-4419-8363-3_13
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4419-8362-6
Online ISBN: 978-1-4419-8363-3
eBook Packages: MedicineMedicine (R0)