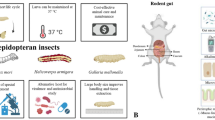
Abstract
The use of invertebrate model hosts has increased in popularity due to numerous advantages of invertebrates over mammalian models, including ethical, logistical and budgetary features. This review provides an introduction to three model hosts, the nematode Caenorhabditis elegans, the fruit fly Drosophila melanogaster and the larvae of Galleria mellonella, the greater wax moth. It highlights principal experimental advantages of each model, for C. elegans the ability to run high-throughput assays, for D. melanogaster the evolutionarily conserved innate immune response, and for G. mellonella the ability to conduct experiments at 37°C and easily inoculate a precise quantity of pathogen. It additionally discusses recent research that has been conducted with each host to identify pathogen virulence factors, study the immune response, and evaluate potential antimicrobial compounds, focusing principally on fungal pathogens.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
Introduction
The study of infectious disease requires model hosts. For obvious ethical reasons it is impossible to conduct human in-vivo primary experimentation to identify pathogen virulence factors, study the immune response to pathogenic infection, or evaluate potential antimicrobial compounds for toxicity and effectiveness. The murine model Mus musculus has long been a favored model host, as it provides a similarity of human and mouse anatomy, immune response, and in some cases pathogen susceptibility. Yet there are many drawbacks to the mouse model. First, there are ethical concerns with mammalian experimentation. Second, there are logistical obstacles, including lengthy reproduction time and the difficulty and expense associated with obtaining and maintaining sufficient numbers of mice to conduct experimentation.
Fortunately, evolutionary conservation extends from humans to distantly related metazoans, permitting the use of invertebrates as model hosts for pathogenesis studies. Invertebrates have numerous advantages over mammalian models. Invertebrates can be inexpensively obtained, easily maintained, and their small size and short lifespan facilitates experimentation in the laboratory setting. For these reasons, the use of invertebrate model hosts has been increasing in popularity. This review covers three invertebrate model hosts, describing principal experimental advantages and highlighting recent research. The soil-living nematode Caenorhabditis elegans is an ideal model for high-throughput screening due to its small size, transparent body, rapid reproductive cycle producing genetically identical progeny, and fully sequenced genome with available genomic tools. The fruit fly Drosophila melanogaster mounts a sophisticated innate immune response that is homologous to the mammalian innate immune response and, like C. elegans, has a fully sequenced genome and available genomic tools. Lastly, the larvae of Galleria mellonella, the greater wax moth, can survive at 37°C (thus providing an analog to humans when studying temperature-sensitive pathogen virulence) and, due to its relatively large size, can easily be inoculated with a precise quantity of pathogen using a syringe.
Caenorhabditis elegans
In the 1960s, Sydney Brenner established the soil-living nematode Caenorhabditis elegans as a genetic model host with tremendous potential for studying cell biology and genetics in-vivo (Brenner 1974). Its small size (adults are ∼1 mm), rapid life cycle (∼3.5 days at 20°C), and transparent body make the organism well suited to experimental observation. Other important features of C. elegans include its ability to produce genetically identical progeny as a self-fertilizing hermaphrodite, its small and fully sequenced genome, and its physiological and anatomical simplicity (∼1,000 fully-mapped cells, including ∼300 neurons) (Riddle et al. 1997). In addition, as discovered by Fire and Mello, delivering double-stranded RNA-mediated interference (RNAi) by feeding is a means of genetic disruption in C. elegans (Fire et al. 1998). Also, C. elegans can be maintained indefinitely in liquid nitrogen, a fact that led to the creation of libraries of thousands of easily and inexpensively obtainable mutant strains (Bazopoulou and Tavernarakis 2009). Most importantly, C. elegans is uniquely well suited to study infectious agents because it naturally feeds on microorganisms and because it is susceptible to many of the same bacterial and fungal pathogens that can kill mammals and humans (O’Callaghan and Vergunst 2010). In laboratory, C. elegans is usually fed a lawn of non-pathogenic Escherichia coli, which can be substituted with pathogenic bacteria or fungi to generate an infection and test genetic virulence factors or screen potential antimicrobial compounds. Broadly, in-vivo screening in the microbiology field using C. elegans can be utilized in three ways to: (1) Identify the genetic basis of pathogen virulence; (2) Gain insight into the immune response; and (3) Identify potential antimicrobial compounds.
One method of discovering the genetic basis of bacterial virulence is screening libraries of various bacterial mutants to identify those with increased or decreased virulence. The gram-negative bacterium Pseudomonas aeruginosa is a free-living opportunistic pathogen capable of causing mortality in immunocompromised patients, and a leading cause of hospital-acquired and ventilator-associated pneumonia, with a high degree of intrinsic virulence and multi-drug resistance (Diaz et al. 2008). P. aeruginosa transposon mutation libraries have been screened for mutant clones that exhibit a reduced ability to kill C. elegans, and many of the mutants identified in this way are also less virulent in murine models of infection (Mahajan-Miklos et al. 1999; Tan et al. 1999). In a separate study, a high throughput screen of 2,200 P. aeruginosa mutants using a liquid infection assay showed attenuated virulence associated with a mutation in the cheB2 gene. This finding was subsequently confirmed in a murine lung infection model, illustrating the applicability of bacterial virulence findings in C. elegans to mammals (Garvis et al. 2009). Similar work has been done with Serratia marcescens, another gram-negative bacterium frequently associated with hospital-acquired urinary tract infections, and with Staphylococcus aureus, a gram-positive bacterium that has increasing antibiotic resistance, to identify the genetic mutations associated with attenuated virulence in C. elegans (Kurz et al. 2003; Begun et al. 2005).
Furthermore, several studies have been done with C. elegans assays to identify virulence factors associated with two of the most prevalent fungal pathogens, Candida albicans and Cryptococcus neoformans. C. albicans is an opportunistic fungal pathogen commonly carried in the human gastrointestinal tract with harmful effects in immunocompromised patients. It is the fourth most common cause of bloodstream infection, with costly treatment and high mortality rates (35%) (Douglas 2003). C. albicans has the ability to undergo morphological change from a yeast form to a hyphal form. Using a C. elegans assay, mutant C. albicans strains with diminished hyphal formation capability were found to have attenuated virulence, and several genes were identified that are important for hyphal formation in vivo (Pukkila-Worley et al. 2009). C. neoformans is the third most common cause of invasive fungal infections in solid organ transplant recipients and can be life threatening in immunocompromised patients (Mueller and Fishman 2003). Using a C. elegans screen, 350 C. neoformans mutants were tested for reduced virulence. Among seven mutants identified, one contained an insertion in a gene encoding a serine/threonine protein kinase (KIN1), which is also an important virulence factor in murine models of infection (Mylonakis et al. 2004).
Increasing microbial resistance to many antibiotics and antifungal agents has created a need to identify new compounds for therapeutic use. C. elegans provides an ideal screening model to identify potential antimicrobial candidates in vivo. Because of its small size C. elegans is particularly well-suited to high throughput screening in standard 348 well plates of thousands of compounds to identify those with potential novel antimicrobial activities (Spring 2005). In vitro high throughput screens against specific microbial targets have produced high attrition rates due to an inability to forecast preclinical and clinical development barriers, including toxicity of the compound under the study (Lindsay 2003). In addition, traditional antibiotic screens for compounds that block pathogen growth in vitro cannot identify compounds that stimulate immune responses or decrease pathogen virulence during infection (often referred to immunomodulatory and antivirulence compounds, respectively). In contrast, in vivo whole animal screens for compounds that cure an infection immediately identify toxic compounds and other potential clinical obstacles, and can identify compounds that may enhance host immunity (Moy et al. 2009). The small size and transparent body of C. elegans also facilitates the use of robots to dispense a precise number of live, C. elegans into microwells and facilitates automatic image analysis to identify the number of surviving organisms during the experiment (Moy et al. 2009).
An automated C. elegans assay has been used to identify compounds that cure infections caused by the bacterium Enterococcus faecalis or the yeast Candida albicans. First, in a screen of 372,000 compounds with E. faecalis, a gram-positive bacterium that is increasingly acquiring resistance to antibiotics, 28 compounds were identified that were not previously reported to have antimicrobial properties, including at least 6 which affected the growth of the pathogen in vivo, but not in vitro, suggesting they act by mechanisms distinct from antibiotics currently in use (Moy et al. 2009). In the case of C. albicans, a screen of 2,560 natural compounds resulted in the identification of 12 saponins, which significantly enhanced survival of the worms, suggesting that saponins have the potential to form a foundation for a new generation of antifungal compounds (Coleman et al. 2010). Another screen of 3,228 bioactive compounds yielded 19 compounds that resulted in an increase in C. elegans survival after infection with C. albicans. Of these, 12 were not primarily used as antifungal agents, including 3 immunosuppressive drugs (Okoli et al. 2009). An earlier screen of 1,266 compounds for antifungal activity identified 15 compounds that prolonged survival of C. elegans after infection with C. albicans and inhibited hyphal formation. Two of these compounds, caffeic acid phenethyl ester, a major active component of honeybee propolis, and the fluoroquinolone enoxacin, also attenuated C. albicans virulence in a murine model (Breger et al. 2007).
Although C. elegans has numerous advantages, the organism does pose some limitations as a model host. In the case of compound screens, it can be difficult to predict mammalian bioactivity given the anatomical simplicity of C. elegans relative to mammals, and challenging to forecast effective concentrations of identified compounds, as the nematode’s thick cuticle blocks absorption and its small size makes it impossible to measure the concentration of compound that has been absorbed (Giacomotto and Ségalat 2010). Additionally, some diseases and immune responses cannot be recreated because the nematode lacks a variety of mammalian anatomical structures. To circumvent these limitations, other models must be used.
Drosophila melanogaster
The fruit fly Drosophila melanogaster has many of the same advantages as C. elegans, including small size, short generation time, a fully sequenced genome, and pre-existing libraries of genetic mutants. In addition, D. melanogaster is an excellent model host because it mounts an extensively studied innate immune response, with genes and pathways similar to those found in mammals (Hoffmann 2003). In particular, the Toll and Imd (immune deficiency) pathways in D. melanogaster are useful models for mammalian study, given the similarity of the Toll receptor to mammalian Toll-like receptors (TLR) and interleukin-1 (IL-1) receptors, and the similarity of the Imd pathway to the mammalian tumor necrosis factor signaling pathway (Sekiya et al. 2008).
The innate immune response in D. melanogaster is comprised of both cellular and humoral components. The cellular response involves specialized hemocytes (blood cells), which engage in phagocytosis and encapsulation of foreign microbes (Rizki and Rizki 1984). The humoral response involves the production of antimicrobial peptides (AMPs) in the fat body, the equivalent of the mammalian liver, which are then secreted into the haemolymph (Lemaitre 2004). Approximately 20 AMPs that have been discovered can be classified into seven groups, with differential effectiveness against fungi (Drosomycin and Metchnikowin), gram-positive bacteria (Defensin), and gram-negative bacteria (Diptericin, Drosocin, Attacin and Cecropin) (Lemaitre and Hoffmann 2007). The regulation of the genes encoding AMP production occurs via the Toll and Imd signaling pathways. The Toll pathway is activated primarily in response to fungal and some gram-positive bacterial infections, whereas the Imd pathway is activated predominantly in response to gram-negative and some gram-positive bacterial infections (Lemaitre et al. 1997).
Utilizing D. melanogaster in studies with fungal pathogens, including C. albicans, Cryptococcus neoformans and Aspergillus fumigatus has provided insights into the innate immune response and mechanisms of pathogenic virulence (Chamilos et al. 2006). These studies have also aided in establishing D. melanogaster as a potential model for testing antimicrobial compounds efficacy. D. melanogaster mutants devoid of functioning Toll receptors are analogous in many ways to immunocompromised mammals that are at risk for infection by opportunistic fungi. For example, a study of C. albicans mutant strains revealed the same rank order of virulence in Toll-deficient D. melanogaster as in mammalian infection models, and moreover showed that increased virulence in Drosophila was associated with the ability of the mutant C. albicans strain to form hyphae, the same virulence mechanism as is demonstrated in mammals (Alarco et al. 2004). Similarly, D. melanogaster has proven to be an important model host in studies of the pathogenic fungi C. neoformans. In mammals, a significant aspect of C. neoformans virulence is its ability to avoid phagocytosis and the immune response via polysaccharide capsule formation. In a study of mutant strains of C. neoformans in D. melanogaster, genes associated with pathogenesis in mammals caused enhanced killing of D. melanogaster, but C. neoformans with a mutated gene essential for capsule formation (cap59) exhibited only a minor decrease in virulence, suggesting factors other than capsular formation contribute to C. neoformans virulence in D. melanogaster (Apidianakis et al. 2004). Additional research has established D. melanogaster as a potential model host for in vivo assessment of antifungal compound efficacy. Although D. melanogaster cannot be grown in liquid medium, which precludes its use in robotic high throughput screening, screening of potential compounds can be done by hand (Giacomotto and Ségalat 2010). Two studies in particular evaluated the response of D. melanogaster infected with A. fumigatus and C. albicans, finding D. melanogaster a reliable model of testing antifungal compounds currently in use, and a potential model for testing combinations of antifungal drugs (Lionakis et al. 2005; Chamilos et al. 2006).
Galleria mellonella
The larval stage of the greater wax moth Galleria mellonella presents unique advantages as a model host. Chief among these advantages is its ability to survive at 37°C, thus providing an analog to humans when studying pathogenic temperature-sensitive virulence. Changes in temperature, however, can provoke the G. mellonella immune response (Mowlds and Kavanagh 2008). G. mellonella caterpillars can be stored at room temperature in the lab, and are easily and inexpensively obtained in sizes large enough (1.5–2.5 cm) to be inoculated by hand with a syringe, permitting the delivery of a precise amount of pathogen (Kavanagh and Fallon 2010). Significant limitations of the G. mellonella model are that its genome has not yet been sequenced and well-established methods of generating mutants have not been developed. Yet, unlike wild-type D. melanogaster, which can survive large inocula of pathogen (often necessitating the use of Toll mutants to study pathogenesis), G. mellonella is susceptible to infection by numerous pathogens (Mylonakis 2008). In particular, G. mellonella serves as an ideal model host for studying pathogen virulence mechanisms and the efficacy of potential antimicrobial compounds.
G. mellonella larvae can be killed by infection with Candida, and the hierarchy of virulence of different Candida species in G. mellonella is consistent with virulence observed in mammalian models, with C. albicans demonstrating the greatest pathogenicity (Cotter et al. 2000). C. albicans virulence is associated with the ability to form hyphae, and a good correlation exists between C. albicans mutants with decreased hyphal formation and decreased virulence in both G. mellonella and mice (Brennan et al. 2002). Recent work with G. mellonella, however, has demonstrated that hyphal formation alone is insufficient to kill G. mellonella, as mutant C. albicans strains exist with the ability to form filaments with impaired virulence (Fuchs et al. 2010).
G. mellonella larvae can also be employed to evaluate the effectiveness of antifungal compounds and treatment protocols. For example, following infection with C. neoformans, G. mellonella was inoculated in one study with amphotericin B, flucytosine, and fluconazole. Treatment guidelines for severe cryptococcal infection in humans call for the combination of amphotericin B plus flucytosine, which was also associated with greatest G. mellonella survival, suggesting G. mellonella may be a valuable model host for future C. neoformans antifungal compound discovery (Mylonakis et al. 2005). Similarly, G. mellonella larvae were used to assess the efficacy of silver (I) and 1,10-phenanthroline after infection with C. albicans, and demonstrated significantly increased survival with both compounds (Rowan et al. 2009). Finally, an infection assay in G. mellonella with C. albicans and A. fumigatus revealed that co-inoculation of an Hsp90 inhibitor enhanced the efficacy of existing antifungal drugs (Cowen et al. 2009).
Conclusion
It is becoming increasingly critical to understand pathogenic virulence factors and identify novel therapeutic options as the population of immunocompromised patients grows and new pathogenic resistance to conventional antimicrobial therapies emerges. Invertebrate model hosts will continue to play important roles in the development of new lifesaving treatments and in finding virulence traits that could be potential targets for new antimicrobials. In particular, further robotic-assisted high throughput assays with C. elegans will be critical in identifying new antimicrobial compounds, our understanding of the innate immune system will increase with further experimentation with D. melanogaster and, as G. mellonella becomes a more established model, genomic tools will emerge that will further increase its usefulness. In addition, new model hosts will continue to be discovered that will further our understanding and assist in the development of novel lifesaving therapies.
References
Alarco AM et al (2004) Immune-deficient Drosophila melanogaster: a model for the innate immune response to human fungal pathogens. J Immunol 172:5622–5628
Apidianakis Y et al (2004) Challenge of Drosophila melanogaster with Cryptococcus neoformans and role of the innate immune response. Eukaryot Cell 3:413–419
Bazopoulou D, Tavernarakis N (2009) The NemaGENETAG initiative: large scale transposon insertion gene-tagging in Caenorhabditis elegans. Genetica 137:39–46
Begun J, Sifri CD, Goldman S, Calderwood SB, Ausubel FM (2005) Staphylococcus aureus virulence factors identified by using a high-throughput Caenorhabditis elegans-killing model. Infect Immun 73:872–877
Breger J et al (2007) Antifungal chemical compounds identified using a C. elegans pathogenicity assay. PLoS Pathog 3:e18
Brennan M, Thomas DY, Whiteway M, Kavanagh K (2002) Correlation between virulence of Candida albicans mutants in mice and Galleria mellonella larvae. FEMS Immunol Med Microbiol 34:153–157
Brenner S (1974) The genetics of Caenorhabditis elegans. Genetics 77:71–94
Chamilos G et al (2006) Drosophila melanogaster as a facile model for large-scale studies of virulence mechanisms and antifungal drug efficacy in Candida species. J Infect Dis 193:1014–1022
Coleman JJ et al (2010) Characterization of plant-derived saponin natural products against Candida albicans. ACS Chem Biol 5:321–332
Cotter G, Doyle S, Kavanagh K (2000) Development of an insect model for the in vivo pathogenicity testing of yeasts. FEMS Immunol Med Microbiol 27:163–169
Cowen LE et al (2009) Harnessing Hsp90 function as a powerful, broadly effective therapeutic strategy for fungal infectious disease. Proc Natl Acad Sci USA 106:2818–2823
Diaz MH et al (2008) Pseudomonas aeruginosa induces localized immunosuppression during pneumonia. Infect Immun 76:4414–4421
Douglas LJ (2003) Candida biofilms and their role in infection. Trends Microbiol 11:30–36
Fire A et al (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811
Fuchs BB et al (2010) Role of filamentation in Galleria mellonella killing by Candida albicans. Microbes Infect 12:488–496
Garvis S et al (2009) Caenorhabditis elegans semi-automated liquid screen reveals a specialized role for the chemotaxis gene cheB2 in Pseudomonas aeruginosa virulence. PLoS Pathog 5:e1000540
Giacomotto J, Ségalat L (2010) High-throughput screening and small animal models, where are we? Br J Pharmacol 160:204–216
Hoffmann J (2003) The immune response of Drosophila. Nature 426:33–38
Kavanagh K, Fallon JP (2010) Galleria mellonella larvae as models for studying fungal virulence. Fungal Biol Rev 24:79–83
Kurz CL et al (2003) Virulence factors of the human opportunistic pathogen Serratia marcescens identified by in vivo screening. EMBO J 22:1451–1460
Lemaitre B (2004) The road to toll. Nat Rev Immunol 4:521–527
Lemaitre B, Hoffmann J (2007) The host defense of Drosophila melanogaster. Annu Rev Immunol 25:697–743
Lemaitre B, Reichhart JM, Hoffmann JA (1997) Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc Natl Acad Sci USA 23:14614–14619
Lindsay MA (2003) Target discovery. Nat Rev Drug Discov 2:831–838
Lionakis MS et al (2005) Toll-deficient Drosophila flies as a fast, high-throughput model for the study of antifungal drug efficacy against invasive aspergillosis and Aspergillus virulence. J Infect Dis 191:1188–1195
Mahajan-Miklos S, Tan MW, Rahme LG, Ausubel FM (1999) Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96:47–56
Mowlds P, Kavanagh K (2008) Effect of pre-incubation temperature on susceptibility of Galleria mellonella larvae to infection by Candida albicans. Mycopathologia 165:5–12
Moy T et al (2009) High-throughput screen for novel antimicrobials using a whole animal infection model. ACS Chem Biol 4:527–533
Mueller NJ, Fishman JA (2003) Asymptomatic pulmonary cryptococcosis in solid organ transplantation: report of four cases and review of the literature. Transpl Infect Dis 5:140–143
Mylonakis E (2008) Galleria mellonella and the study of fungal pathogenesis: making the case for another genetically tractable model host. Mycopathologia 165:1–3
Mylonakis E et al (2004) Cryptococcus neoformans Kin1 protein kinase homologue, identified through a Caenorhabditis elegans screen, promotes virulence in mammals. Mol Microbiol 54:407–419
Mylonakis E et al (2005) Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect Immun 73:3842–3850
O’Callaghan D, Vergunst A (2010) Non-mammalian animal models to study infectious disease: worms or fly fishing? Curr Opin Microbiol 13:79–85
Okoli I et al (2009) Identification of antifungal compounds active against Candida albicans using an improved high-throughput Caenorhabditis elegans assay. PLoS One 4:e7025
Pukkila-Worley R, Peleg A, Tampakakis E, Mylonakis E (2009) Candida albicans hyphal formation and virulence assessed using a Caenorhabditis elegans infection model. Eukaryot Cell 8:1750–1758
Riddle DL, Blumenthal T, Meyer BG, Priess JR (1997) C. elegans II, 2nd edn. Cold Spring Harbor Laboratory Press, Plainview
Rizki RM, Rizki TM (1984) Selective destruction of a host blood cell type by a parasitoid wasp. Proc Natl Acad Sci USA 81:6154–6158
Rowan R, Moran C, McCann M, Kavanagh K (2009) Use of Galleria mellonella larvae to evaluate the in vivo anti-fungal activity of [Ag2(mal)(phen)3]. Biometals 22:461–467
Sekiya M et al (2008) A cyclopentanediol analogue selectively suppresses the conserved innate immunity pathways, Drosophila IMD and TNF-alpha pathways. Biochem Pharmacol 75:2165–2174
Spring DR (2005) Chemical genetics to chemical genomics: small molecules offer big insights. Chem Soc Rev 34:472–482
Tan MW, Rahme LG, Sternberg JA, Tompkins RG, Ausubel FM (1999) Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc Natl Acad Sci USA 96:2408–2413
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2012 Springer Science+Business Media, LLC
About this paper
Cite this paper
Glavis-Bloom, J., Muhammed, M., Mylonakis, E. (2012). Of Model Hosts and Man: Using Caenorhabditis elegans, Drosophila melanogaster and Galleria mellonella as Model Hosts for Infectious Disease Research. In: Mylonakis, E., Ausubel, F., Gilmore, M., Casadevall, A. (eds) Recent Advances on Model Hosts. Advances in Experimental Medicine and Biology, vol 710. Springer, New York, NY. https://doi.org/10.1007/978-1-4419-5638-5_2
Download citation
DOI: https://doi.org/10.1007/978-1-4419-5638-5_2
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4419-5637-8
Online ISBN: 978-1-4419-5638-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)