Abstract
Proteomics technologies are improving at a great pace with the ultimate goal to allow the sensitive and comprehensive analysis of proteins in tissues and body fluids. The comparison of proteomes from two or more sources is still a challenging task but has already made significant inroads into biomarker discovery. This chapter describes strategies and platforms for biomarker discovery in mouse models and human specimens. We submit that the discovery of proteins that are differentially expressed will ultimately provide valuable information about the pathways involved in the pathobiology of psychiatric disorders.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Psychiatric Disorder
- Experimental Autoimmune Encephalomyelitis Mouse
- Quantitative Mass Spectrometry
- Oxidative Stress Mechanism
- Anxiety Phenotype
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
3.1 3.1 Introduction
Unlike other major debilitating diseases including cancer and ischaemic heart disease, where we have seen a significant decrease in mortality over the past 30 years, a similar trend is not observed for psychiatric disorders. Also, the prevalence for psychiatric illness has not shown any appreciable decline. It therefore does not come as a surprise that the “World Health Organization 2002 Report” has labeled psychiatric disorders the major cause for disability in the Western world today (Murray and Lopez, 1997; Kessler et al., 2003).
Genome-wide association studies strongly suggest that a combination of several genes accounts for the various psychiatric disorder phenotypes, with each gene contributing to a small extent. As a consequence, it is now widely accepted that only gene interaction analysis will make possible a comprehensive understanding of the genetic contribution to psychiatric disorders. Furthermore, because of the low effect sizes of the candidate genes that have resulted from poorly powered studies and low rates of replication, a validation by meta-analyses is mandatory in order to validate candidate genes (Sullivan et al., 2000; Camp and Cannon-Albright, 2005). Further complicating the interpretation of genotyping results is the fact that many single nucleotide polymorphisms (SNPs) do not reside inside genes, where they could give rise to altered protein expression, structure, and function, but are found in intergenic regions, the so-called dark matter of the genome, where their impact on the phenotype remains obscure. Finally, epigenetic factors caused by the environment, especially those with effects on critical aspects of developmental experience and stressful life events, seem to greatly contribute to psychiatric disorder pathobiology (Mill and Petronis, 2007). Specific environmental factors will ultimately trigger a genetic risk background to develop into a psychiatric disorder and different patient phenotypes for each genotype. A quotation from Leo Tolstoy's novel Anna Karenina “all happy families resemble one another, but each unhappy family is unhappy in its own way” sums up the heterogeneous presentation of psychiatric disorders that results from the complex interplay between genes and the environment.
Unlike other diseases, psychiatric disorders do not reveal themselves through a lesion and in all likelihood originate from an abnormal processing and/or activity in neural networks that involve several brain areas (Insel and Quirion, 2005). The characterization of these altered networks and neural circuits will be critical to get a better understanding of psychiatric disorders and in turn make possible the definition of targets for the development of more specific medicines. Since current medications act on targets that are in all likelihood quite remote from pathways that are relevant for the pathobiology of psychiatric disorders, they are characterized by limited efficiency and a number of side effects (Hyman, 2007).
Another complicating factor in the area of psychiatric disorders is the imprecise diagnosis that is not based on a molecular pathophysiology but instead relies on clinical observations of symptom clusters (Hyman, 2007). Clinical criteria for the diagnosis of psychiatric disorders are based on the Diagnostic and Statistical Manual of Mental Disorders IV (DSM-IV) and the International Classification of Diseases (ICD-10) and to a large extent include verbal communication between psychiatrist and patient, which renders them very subjective. In addition, an overlap of the general symptoms makes it particularly difficult to deal with the great diversity of psychiatric disorders. In this regard it is also increasingly questioned whether the traditional boundary between psychiatric and neurological disorders is still appropriate despite the fact that the latter are characterized by well-defined lesions (Insel and Quirion, 2005). Even though psychiatric disorders in all likelihood involve dysfunctional neural circuits, there is increasing evidence that certain aspects of their pathobiology are related to neurological diseases, a finding that is also supported by the often-observed comorbidity of neurological and psychiatric disorders. Owing to the late detection of psychiatric disorder onset, their prevention is not strategic and treatment is by and large based on trial and error (Hyman, 2007). New drug candidates are therefore needed, making the obligatory response studies as important as disease susceptibility investigations.
Here is where the field of biomarker discovery can make important contributions. For both types of investigation, drug response and disease susceptibility, biomarkers are needed to move the area of psychiatric disorders into the rest of medicine (Hyman, 2007). Only through a combined interrogation of genetic variations and biomarkers will it be possible to achieve this goal. The obscure etiology and pathogenesis of psychiatric disorders, combined with the fact that today's treatments are empirical and at best symptomatic, also provide a great incentive for psychiatric disorder biomarker discovery efforts. Ultimately, it is hoped that biomarkers will assist in stratifying patient groups with similar clinical features and at the same time help in the identification of neural circuitries that are responsible for disease etiology. This will enable the complementation of the presently applied DSM-IV and ICD-10 criteria with specific sets of biomarkers and result in a more precise nosological framework for psychiatric disorders (Hyman, 2007). In addition, biomarkers will make possible a predictive pharmacogenomics approach for newly developed medicines with new target sites that will result in more specific medicines with fewer side effects.
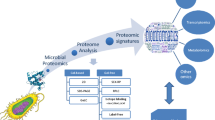
A number of different assays and methods can produce biomarkers for psychiatric disorders (Fig. 3.1 ). In this chapter we will focus on the identification of proteins, the functional molecules in cells and organisms, and their roles in biomarker discovery.
A biomarker is any characteristic that can be objectively measured to reflect physiological, pharmacological, or disease processes in animals or humans (De Gruttola et al., 2001). Various platforms for different types of biomarkers can be used for psychiatric disorders. (a) RNA microarray, (b) Protein expression analysis by gel electrophoresis, (c) Mass spectrometry-based protein and metabolite profiling, (d) Neuroendocrine assay, (e) Brain MRI scan
3.2 3.2 Strategies
The Proteomics and Biomarkers research group at the Max Planck Institute of Psychiatry is engaged in the identification of biomarkers for psychiatric and neurological disorders. Several global proteomics approaches with human and rodent tissue and body fluid specimens are used for this purpose (Turck et al., 2005). In addition, the examination of autoantibodies in patient cerebrospinal fluid (CSF) constitutes an exploratory project that is based on the hypothesis of a dysfunctional immune response in the pathobiology of psychiatric disorders. Finally, we also attempt to exploit the great wealth of information on psychiatric disorders in the public domain through an in silico interrogation of experimental data deposited in various databases and article texts (Fig. 3.2 ).
Roads to protein biomarker discovery. On one hand, brains from animal models that reflect certain aspects of the disease phenotype are investigated by the proteomics methods. Once candidate markers have been identified from the animal model, they need to be validated in patient specimens. Alternatively, clinical specimens are used directly for biomarker discovery, validated in a great number of samples, and ultimately used for establishing a diagnostic assay platform
A particular focus of our biomarker discovery efforts is the analysis of mouse models that represent features relevant for psychiatric disorders. The rationale behind the use of animal models is that the analysis of patient specimens is plagued by several limiting factors. Foremost, as discussed in the Introduction, psychiatric disorders present themselves in a rather heterogeneous manner owing to diverse sets of disease-causing genes and different environmental influences varying from patient to patient. Also, since each gene contributes to the disease to a rather small extent one can expect only minor effects on particular biochemical pathways, resulting in a low disease marker signal-to-noise ratio. Other interindividual differences that are not related to the disease phenotype further complicate this issue. Finally, human specimens relevant for the proteomic analysis of psychiatric disorders are available in limited amounts and their retrieval is difficult to control in a consistent manner. This is true for both types of patient specimen that are commonly used for proteomic analyses of psychiatric disorders: postmortem brain tissue and CSF. In the case of postmortem brain tissue, prolonged times between death and tissue preservation can lead to protein degradation and/or modification, which will produce artifacts picked up during proteomics analysis (Sköld et al., 2007). Similar effects are also seen in the case of CSF specimens obtained by lumbar puncture, a procedure that is difficult to control with regard to timing and patient constitution. Patient studies are also often compromised by medication and other treatments which will affect the proteome constituents.
Despite these challenges, we have not excluded human specimens from our biomarker discovery studies (Turck, 2005). Experienced psychiatrists in our clinic ensure that CSF specimens are obtained from carefully phenotyped patient populations in a consistent manner and include detailed clinical and neuroendocrine data, which make the grouping of samples to be used for proteomic analysis more viable.
In addition to a global proteome analysis of CSF, we are also using a semitargeted approach for the identification of novel disease markers that exploits the great specificity of the body's immune system. For this purpose we are interrogating the antibody pool that is present in CSF. The basis for this approach stands on the hypothesis that several factors may trigger an autoimmune response within the central nervous system (CNS) especially in individuals with psychiatric disorders that are dispositioned to immune system dysfunction and impaired blood—brain barriers. We therefore hypothesize that the presence of markers for psychiatric diseases is reflected by the appearance of autoantibodies in CSF. These autoantibodies can be used for the identification of their respective autoantigen targets.
The mining of already existing data in the public domain is a recent approach that we have begun to explore. It is based on the assumption that a great amount of information pertinent to biomarker discovery has already been acquired in the numerous genetic, proteomic, and clinical studies and deposited in databases or text documents where it awaits exploitation with the right bioinformatic tools. Obviously, not all the publicly deposited data is relevant and of sufficient quality, but the sheer volume of the data warrants a thorough interrogation with regard to their significance as biomarkers.
3.3 3.3 Trait Anxiety Mouse Model
The utilization of animal models for psychiatric disorders comes with the realization that these models represent only certain aspects of the disorder and not the disorder itself (Insel, 2007). Even if one had the knowledge of most, if not all, genes causing a particular psychiatric disorder, it is at present not technically feasible to create models by manipulating a great number of genes at a time in the same animal. The generation of animal models based on the manipulation of single genes, on the other hand, has a poor chance of achieving a penetrating level that is sufficient to reflect the disease phenotype observed in patients.
The symptom anxiety is the normal response to danger but becomes abnormal when the response is unproportional and/or outlasts the danger. Clinical and epidemiological data have shown that the symptom anxiety is often found in other psychiatric disorders including depression, obsessive-compulsive disorders, and post-traumatic stress disorder (Gross and Hen, 2004).
A number of animal models reflecting the anxiety phenotype have been generated through either genetic manipulation, exposure to trauma or social stress, or maternal separation in early developmental stages. Alternatively, inbred mouse strains that inherently differ in their natural anxiety levels are used to study the phenotype in greater detail (Finn et al., 2003; Cryan and Mombereau, 2004; Gordon and Hen, 2004).
Dr. Rainer Landgraf, head of the Behavioral Neuroendocrinology research group at the Max Planck Institute of Psychiatry, has established a robust mouse model of trait anxiety using a selective breeding protocol, based on the animal's behavior in a commonly used assay for anxiety, the elevated plus maze (Krömer et al., 2005). This model does not have the disadvantage inherent to studies dealing with a comparison of unselected inbred or outbred mouse lines, which in addition to anxiety also differ in other phenotypes. The intrastrain breeding approach has the benefit that it centers on only a limited set of traits related to anxiety. Consequently, studies with this type of mouse model will increase the likelihood of identifying trait-relevant parameters no matter whether they are genetic, proteomic, metabolomic, or otherwise in nature. On the basis of a bidirectional breeding approach, mouse lines of hyperanxious (HAB) and hypoanxious (LAB) phenotypes were established and validated. These lines were derived from CD1 mice, which, due to their outbred nature, differ in their anxiety-related behavior using the elevated plus maze and other behavioral assays that reveal symptoms indicative of psychiatric disorders.
Our biomarker discovery efforts with the above mouse lines initially used a classical proteomics platform, two-dimensional polyacrylamide gel electrophoresis (2DPAGE) (Klose, 1975; O'Farrell, 1975), for differential protein expression analysis (Krömer et al., 2005). Two proteins were identified that showed quantitative and qualitative differences, respectively, between HAB and LAB mice. The quantitative difference was identified as glyoxalase 1 (Glx1), a protein expressed in the cytosol of cells and tissues of many organisms (Hayes et al., 1989; Thornalley, 1993). The enzyme plays a major role in the detoxification of methylglyoxal, which is a potent cytotoxic metabolite. Glx1 catalyzes the transformation of methylglyoxal and glutathione to S-lactoylglutathione, which is then converted to D-lactic acid by glyoxalase 2. Owing to its ubiquitous expression, Glx1 is believed to be of fundamental importance for cellular metabolism. The fact that the enzyme uses glutathione as a cosubstrate points to a functional role of Glx1 in oxidative stress mechanisms. Studies have implicated Glx1 in several brain disorders including Alzheimer's disease (Chen et al., 2004) and autism (Junaid et al., 2004). A possible connection between Glx1 and unipolar affective disease has been found in a linkage study (Tanna et al., 1989). Interestingly, in an analysis using different inbred strains of mice it was also found that Glx1 and glutathione reductase 1, another enzyme with a function in oxidative stress mechanisms, play a causal role in anxiety (Hovatta et al., 2005). Still, the question whether Glx1 represents a risk marker or a risk factor for the anxiety-related phenotype in mice remains unknown at the present time (Thornalley, 2006).
In the HAB/LAB animals, Glx1 is present in many cell types in which its expression level reflects the one found in the brain and makes its determination in blood cells feasible. On the basis of this finding we have used a western blot assay specific for Glx1 to screen red blood cell specimens from patients afflicted with anxiety disorders and depression (Ditzen et al., 2006). These studies are presently expanded to a great number of samples to find out whether Glx1 can be used as a biomarker for anxiety in the clinical laboratory.
The other difference found during proteome analysis of HAB and LAB brains also represents an enzyme, enolase phosphatase (EP), which is expressed as different isoforms in the two lines (Ditzen et al., 2006). Caused by two SNPs that result in amino acid changes, the protein isoforms migrate at different positions during 2DPAGE. Apart from its altered mobility in SDS gels, the HAB EP isoform has also a lower enzymatic activity compared to the LAB/NAB isoform. This activity difference probably affects the methionine salvage pathway, of which EP is a member. The metabolic pathway is of interest in the area of psychiatric disorders as it includes a metabolite, S-adenosylmethionine (SAM), which is reported to be a natural mood stabilizer. Furthermore, antidepressant activities of SAM have been demonstrated in clinical trials (Bressa, 1994; Silveri et al., 2003). Aside from SAM, the methionine salvage pathway has an interesting connection to another pathway that is relevant for psychiatric disorders. The polyamine pathway includes putrescine, spermidine, and spermin, which have been shown to affect neurotransmission caused by their ability to modulate ion channels such as the N-methyl-d-aspartate-type excitatory amino acid receptor (NMDAR) (Bernstein and Müller, 1999). The latter is involved in glutamatergic neurotransmission and associated with long-term potentiation, neuronal development and neuronal plasticity (Williams, 1997), as well as affective disorder pathobiology (Skolnik, 1999). Furthermore, animal models for depression have shown that levels of the three polyamines are altered in specific brain areas compared to control animals and that SAM administration has the ability to modulate polyamine levels (Genedani et al., 2001). Our analyses indicate elevated levels of spermidine and spermin in HAB relative to LAB mice. This could be a result of the reduced EP activity in the methionine salvage pathway and ultimately leads to differential modulatory effects on the NMDAR by the polyamines (Skolnik, 1999). The proteomic analysis of a robust and valid animal model for trait anxiety has resulted in two marker candidates that are part of metabolic pathways pertinent to the disease phenotype. Although not necessarily causative for the anxiety phenotype, these proteins provide valuable information with regard to the pathways that are involved. As a consequence, we submit that genetic and proteomic differences that are identified in animal models are not only useful as biomarkers themselves but at the same time open the gate for an extended interrogation of metabolites that are part of the affected pathways and may represent biomarkers in their own right. The elucidated pathways can thus provide valuable information for metabolic assays of specimens from patients afflicted with anxiety and affective disorders. Studies are in progress that will assess metabolites from the methionine salvage and polyamine pathways to examine their potential use in clinical assays for anxiety (Fig. 3.3 ).
From protein biomarkers to metabolic pathways to neural circuits. Phenotype-related protein expression differences provide information on metabolic pathways that are affected in psychiatric disorders. A combination of altered metabolic pathways may ultimately precipitate disease by distressing critical neural circuits
Despite our success in biomarker discovery using the 2DPAGE method, we seek to extend the list of trait anxiety markers using more sophisticated proteomic methods. For complex diseases such as psychiatric disorders, a great number of markers will be needed for a reliable diagnosis characterized by high specificity and sensitivity. To achieve this goal, we have established a more sensitive, comprehensive, and consistent proteomics platform that omits the inherent limitations of the 2DPAGE method and allows quantitative mass spectrometry with great precision and sensitivity. This method involves metabolic labeling of mouse models with stable isotopes. Using this method, tissues and body fluids from metabolically labeled case and nonlabeled control mice can be combined before any sample work-up steps such as lysis, protein fractionation, and digestion are performed. The metabolic labeling procedure therefore avoids the introduction of any artificial differences caused by an inconsistent sample preparation prior to and during the proteomic comparison. The method was initially restricted to lower organisms such as bacteria and yeast, but was later also applied to mammalian cells in culture. More recently, the metabolic labeling approach has been used in mammals when rats were differentially grown on 15N-enriched and -depleted diets (Wu et al., 2004).
To achieve optimal sensitivity, mice are labeled with the 15N isotope-enriched diet in utero with continued feeding after birth. Mice are then sacrificed after assessing their phenotype with several behavioral assays, and brain tissue and blood is quick-frozen in liquid nitrogen. Applying the method to the HAB/LAB mouse model, we have found that 15N-enrichment in brain tissue and blood is 93 and 95%, respectively, which is a sufficiently high incorporation rate for sensitive protein analysis and quantitation by tandem mass spectrometry. For a thorough interrogation of the mouse proteomes, we are using brain sections and organellar fractions (cytosolic, membrane, nuclear). Peptide ratios from tryptic digests are determined on the basis of the ion current ratios of each light (14N) and heavy (15N) peptide pair. With the help of a mass spectrometry data quantitation software that we have developed, changes in protein expression can be estimated by using multiple peptide pairs for each protein (Fig. 3.4 ).
3.4 3.4 Cerebrospinal Fluid
Because of the close proximity and perfusion of the brain, CSF contains mediators that reflect metabolic processes in the CNS. CSF therefore represents the most appropriate biomarker source for patients afflicted with psychiatric disorders. Proteins that are secreted or shed from brain cells are present in CSF and, owing to an exchange, also found in blood, albeit at reduced levels. This is an important aspect for a biomarker as, owing to its easier availability, blood will ultimately be the body fluid of choice for a routine clinical diagnostic assay for psychiatric disorders (LaBaer, 2005). We therefore propose to use CSF for biomarker discovery and blood for eventual routine screening in the clinical laboratory.
The proteomic analysis of body fluids such as CSF remains a challenge (Anderson and Anderson, 1998). An important prerequisite for biomarker discovery is that retrieval of CSF by lumbar puncture is carried out in a controlled fashion to minimize variability. The limiting amounts of starting material and the large dynamic range of protein concentrations in CSF (up to 12 orders of magnitude) between the highest and lowest expressed proteins make any proteomic-based analysis difficult (Anderson and Anderson, 1998). As current technologies are limited by the amount of protein they can reliably detect and the large dynamic range, only a fraction of the CSF proteome is interrogated. In addition, owing to a leaky blood—brain barrier that is especially pronounced in patients with brain disorders, many serum proteins can infiltrate CSF. This makes it difficult to know in many cases whether a CSF protein is derived from the brain or serum.
To improve the depth of CSF proteome coverage, abundant proteins are first depleted and the remaining less abundant proteins further fractionated prior to mass spectrometry analysis. As is always the case in protein analysis, the more one fractionates, the more proteins can be identified. However, because of limited sample amounts, this can be done only to a certain extent.
In the first set of studies, we have carried out proteome mining experiments to get a feel of the complexity of the CSF protein constituents (Maccarrone et al., 2004a, b). For this purpose we employed the shotgun mass spectrometry approach after depletion of the abundant proteins followed by an extensive fractionation at the protein or tryptic peptide level. This was achieved by protein anion exchange chromatography, SDS-PAGE and tryptic peptide isoelectric focusing. We have been able to identify over 1,000 CSF proteins that belong to several groups with regard to molecular function, biological process, and cellular compartment distribution. On the basis of a comparison with two human serum protein databases, approximately half of the proteins are derived from brain tissue (Anderson et al., 2004; Chan et al., 2004).
The logical next step in our CSF proteomics studies is to move from a mining to a scoring mode. In other words, we compare CSF proteomes from different patient groups to identify biomarkers for specific psychiatric phenotypes. These endeavors have been met with only limited success. In retrospect, we were probably overly optimistic in assuming that the identification of biomarkers with a small number of CSF specimens was feasible. The problems associated with the proteomic analysis of human specimens have been discussed in the Introduction. In addition, it is now widely accepted in the area of genome-wide association studies of complex disorders, in which individual genes have a rather small effect on the phenotype, that a great number of samples have to be interrogated in order to come up with significant hits. This situation in all likelihood is not much different for protein biomarker discovery efforts. Comparative proteomic analyses that we have attempted with samples from patients with unipolar depression (UPD) and normal controls, using either 2DPAGE or the iTRAQ stable isotope labeling method (Ross et al., 2004) in combination with quantitative mass spectrometry, have resulted in a limited set of differences that were not consistently found for all patients. This is, of course, not surprising in light of the fact that not every patient is predicted to have the same set of biomarkers. Instead, it is more likely that each patient will have different sets of markers with a certain degree of overlap between patients.
Analysis and comparison by 2DPAGE of CSF from UPD patients and controls revealed interesting differences in protein isoform expression. One example of such an isoform variation in UPD patients is the pigment epithelium derived protein (PEDF) (Fig. 3.5 ). PEDF is a glycoprotein with neuroprotective effects by inducing prosurvival genes in neurons (Yabe et al., 2005). If and how these isoform discrepancies contribute to the pathomechanism of depressive disorders is unclear at this point. Since PEDF was also found as a potential marker for early diagnosis of Alzheimer's disease (Yamagishi et al., 2004), it might be indicative of a possible compensatory mechanism of the brain to fight against neuronal cell injury.
The other difference of interest observed by 2DPAGE is in the Dickkopf-3 related protein (Dkk3), which is expressed at lower levels in CSF of UPD patients. Dkk3 is a secreted glycoprotein that suppresses the neurodevelopmental wingless cascade and is critical for embryonic head development and synaptic function in the adult brain. A decrease in Dkk3 mRNA in postmortem brains of schizophrenic individuals has been reported (Ftouh et al., 2005). We are presently assessing to what extent the two protein differences can contribute to a biomarker list for UPD.
On the basis of the CSF protein mining results (Maccarrone et al., 2004a), we are now establishing a different type of proteomic screen that involves an antibody array platform. A project proposal to the Human Proteome Resource in Sweden (Uhlen, 2007) was approved and will provide us with 200 antibodies specific for proteins that we have previously identified in human CSF. This method has also the advantage that it consumes a significantly smaller amount of CSF sample and therefore will permit screening with a greater number of CSF specimens. As explained above, we expect to find different but overlapping sets of biomarkers between patients of the same disease group. It remains to be seen whether different patient groups will also result in such an overlap, which is not inconceivable given the fact that different psychiatric disorders also show a genetic overlap.
3.5 3.5 Autoantibodies
Autoantibodies against brain proteins in serum and CSF of patients afflicted with psychiatric disorders have been reported (Wang et al., 2003; Hornig et al., 1999). In one such study, it was found that an increased immunoglobulin G titer was present in CSF and serum during a state of depression (Hornig et al., 1999). Besides an immune dysfunction, there is also evidence for an impaired blood—brain barrier in patients with psychiatric disorder, which results in an increased risk for an autoimmune response (Wang et al., 2003). The autoimmune response targets, the autoantigens, may represent a valuable class of biomarkers for psychiatric disorders. Similar approaches have already provided important information in other disease areas. These include Type I autoimmune diabetes mellitus, rheumatoid arthritis, and multiple sclerosis (Mathey et al., 2007).
In preliminary studies we were able to demonstrate, by western blot immunoassays, that autoantibodies against brain proteins are indeed present in CSF from patients afflicted with bipolar disorder (BPD). In these experiments, human brain protein extracts were fractionated by SDS gel electrophoresis and transferred to a membrane. The brain protein blot was then probed with different patient CSF specimens, which after blot development resulted in the detection of discrete bands that represent brain proteins specifically recognized by CSF autoantibodies. The limited amount of CSF specimen available prompted us to explore sensitive phage display and protein array screens for the identification of the detected brain autoantigens. Unlike the western blot, these methods allow not only the detection but also the subsequent identification of protein antigens with the small amounts of autoantibodies that are present in CSF.
Phage display screens with CSF from BPD patients resulted in 64 positive clones, with some derived from the same protein family or sharing a common metabolic pathway. After in silico interrogation by pathway analysis and text mining, 20 candidate biomarkers were selected for further validation studies. In a complementary approach, we used a protein array in order to monitor the CSF autoantibody pool. The protein array consisted of over 27,000 proteins derived from expression-verified, full-length, as well as shorter cDNA clones. Proteins printed in duplicate onto 22 × 22 cm membranes were overlaid with CSF and developed, which resulted in a number of positive hits that were also found during the phage display screens (Fig. 3.6 ).
3.6 3.6 Validation
With the great flood of data resulting from today's “omics” technologies, criteria need to be developed for the subsequent validation of the most promising set of candidates before translating them into the clinical laboratory. As mentioned previously, we believe that the protein markers identified with the various experimental platforms may not necessarily be the best biomarkers themselves. However, they may have great potential in identifying disease-pertinent pathways. Other proteins or metabolites that are part of these pathways may be more applicable as biomarkers or could even serve as drug targets. For this reason, we are using pathway programs and protein interaction databases as well as text mining software to establish a list of marker targets by starting with the proteins identified during the proteomic interrogation of mouse model and patient specimens.
As a first step in the validation of a biomarker candidate, we are typically using immunoassay-based screens with the same material that was used as a source in the proteomics discovery process. This assay eliminates any candidate markers that result from artifacts. In the second step of validation, patient body fluid specimens, including serum and CSF, are used. Here it is of paramount importance that the patients are carefully phenotyped by the physicians and grouped according to the results of the clinical phenotyping data. Only then can one expect meaningful results from the validation studies. As explained in the “Introduction”, it should also be kept in mind that not every patient of a group will have the same set of biomarkers. A thorough statistical analysis for biomarker patterns is therefore critical and in all likelihood will result in biomarker overlaps between patients. We take advantage of a large CSF specimen bank that has been established at our institute through the associated Psychiatry and Neurology Clinics. This bank now contains over 1,000 samples that have been carefully prepared and stored and are derived from groups of patients who have been thoroughly characterized by physicians in our clinic. As is the case for all polygenic diseases, we do not anticipate that a single marker will be able to unequivocally distinguish different clinical phenotypes. Only through a combination of a great number of markers will it be possible to gain statistical significance to differentiate complex traits and establish a sensitive and selective diagnostic assay.
3.7 3.7 Conclusion
From the above discussion of our biomarker discovery strategies, it is clear that only a combination of technologies will result in a valid list of markers that can ultimately be used for a clinical assay. Once this goal has been achieved, patient stratification and diagnosis of psychiatric disorders will greatly improve. On the basis of our own data, there is a good chance that a number of different brain disorders including neurological diseases such as Alzheimer's disease, Parkinson disease and multiple sclerosis will share certain biomarkers with psychiatric disorders. Data that we have obtained from biomarker analyses of the Experimental Autoimmune Encephalomyelitis mouse model for multiple sclerosis indicate that this is indeed the case (Jastorff et al., submitted).
Looking ahead, biomarker information will not only be a critical requirement for the establishment of more reliable clinical diagnostic assays but at the same time reveal pathologic mechanisms for psychiatric disorders, which in all likelihood involve dysfunctional neural pathways. A prerequisite, however, will be the consolidation and integration of a number of different data sets resulting from disciplines such as epidemiology, statistical genetics, proteomics, metabolomics, and others (Fig. 3.7 ).
Abbreviations
- BPD:
-
Bipolar disorder
- CSF:
-
Cerebrospinal fluid
- Dkk3:
-
Dickkopf-3 related protein
- DSM-IV:
-
Diagnostic and Statistical Manual of Mental Disorders IV
- EP:
-
Enolase phosphatase
- Glx1:
-
Glyoxalase 1
- HAB:
-
High anxiety-related behavior
- ICD:
-
International Classification of Diseases
- iTRAQ:
-
Isobaric tags for relative and absolute quantitation
- LAB:
-
Low anxiety-related behavior
- NAB:
-
Normal anxiety-related behavior
- NMDAR:
-
N-methyl-d-aspartate-type excitatory amino acid receptor
- PEDF:
-
Pigment epithelium-derived protein
- SAM:
-
S-adenosylmethionine
- SDS:
-
Sodium dodecyl sulfate
- SNP:
-
Single nucleotide polymorphism
- UPD:
-
Unipolar depression
- 2DPAGE:
-
Two-dimensional polyacrylamide gel electrophoresis
References
American Psychiatric Association (2000) Diagnostic and Statistical Manual of Mental Disorders 4th edn, Text Revision, American Psychiatric Association, Washington DC
Anderson NL, Anderson NG (1998) Proteome and proteomics: new technologies, new concepts, and new words. Electrophoresis 19:1853–1861
Anderson NL, Polanski M, Pieper R et al (2004) The human plasma proteome: a nonredundant list developed by combination of four separate sources. Mol Cell Proteomics 3:311–326
Bernstein H-G, Müller M (1999) The cellular localization of the L-ornithine decarboxylase/polyamine system in normal and diseased central nervous systems. Prog Neurobiol 57:485–505
Bressa, GM (1994) S-adenosyl-l-methionine (SAMe) as antidepressant: meta-analysis of clinical studies. Acta Neurol Scand 154 (Suppl):7–14
Camp NJ, Cannon-Albright LA (2005) Dissecting the genetic aetiology of major depressive disorder using linkage analysis. Trends Mol Med 11:138–144
Chan KC, Lucas DA, Hise D et al (2004) Analysis of the human serum proteome. Clin Proteomics 1:101–225
Chen F, Wollmer MA, Hoerndli F et al (2004) Role for glyoxalase I in Alzheimer's disease. Proc Natl Acad Sci U S A 101:7687–7692
Cryan JF, Mombereau C (2004) In search of a depressed mouse: utility of models for studying depression-related behavior in genetically modified mice. Mol Psychiatr 9:326–357
De Gruttola VG, Clax P, DeMets DL et al (2001) Considerations in the evaluation of surrogate endpoints in clinical trials. Summary of a National Institutes of Health workshop. Control Clin Trials 22:485–502
Ditzen C, Jastorff AM, Kessler MS et al (2006) Protein biomarkers in a mouse model of extremes in trait anxiety. Mol Cell Proteomics 5:1914–1920
Finn DA, Rutledge-Gorman MT, Crabbe JC (2003) Genetic animal models of anxiety. Neurogenetics 4:109–135
Ftouh S, Akbar MT, Hirsch SR, de Belleroche JS (2005) Down-regulation of Dickkopf 3, a regulator of the Wnt signalling pathway, in elderly schizophrenic subjects. J Neurochem 94:520–530
Genedani S, Saltini S, Benelli A et al (2001) Influence of SAMe on the modifications of brain polyamine levels in an animal model of depression. Neuroreport 12:3939–3942
Gordon JA, Hen R (2004) Genetic approaches to the study of anxiety. Annu Rev Neurosci 27:193–222
Gross C, Hen R (2004) The developmental origins of anxiety. Nat Rev Neurosci 5:545–552
Hayes JD, Milner SW, Walker SW (1989) Expression of glyoxalase, gluthatione peroxidase and gluthatione S-transferase isoenzymes in different bovine tissues. Biochim Biophys Acta 994:21–29
Hornig M, Amsterdam JD, Kamoun M, Goodman DBP (1999) Autoantibody disturbances in affective disorders: a function of age and gender? J Affect Disord 55:29–37
Hovatta I, Tennant RS, Helton R et al (2005) Glyoxalase 1 and glutathione reductase 1 regulate anxiety in mice. Nature 438:662–666
Hyman SE (2007) Can neuroscience be integrated into the DSM-V? Nat Rev Neurosci 8:725–732
Insel TR (2007) From animal models to model animals. Biol Psychiatry 62:1337–1339
Insel TR, Quirion R (2005) J Am Med Assoc 294:2221–2224
Jastorff AM et al. (submitted)
Junaid MA, Kowal D, Barua M et al (2004) Proteomic studies identified a single nucleotide polymorphism in glyoxalase I as autism susceptibility factor. Am J Med Genet 131:11–17
Kessler RC, Berglund P, Demler O et al (2003) The epidemiology of major depressive disorder: results from the national comorbidity survey replication (NCS-R). JAMA 289:3095–3105
Klose J (1975) Protein mapping by combined isoelectric focusing and electrophoresis of mouse tissues. A novel approach to testing for induced point mutations in mammals. Humangenetik 26:231–243
Krömer SA, Kessler MS, Milfay D et al (2005) Identification of glyoxalase-I as a protein marker in a mouse model of extremes in trait anxiety. J Neurosci 25:4375–4384
LaBaer J (2005) So, you want to look for biomarkers. J Proteome Res 4:1053–1059
Maccarrone G, Birg I, Malisch E et al (2004a) In-depth analysis of the human CSF proteome using protein prefractionation. Clin Proteomics J 1:333–364
Maccarrone G, Milfay D, Birg I (2004b) Mining the human CSF proteome by immunodepletion and shotgun mass spectrometry. Electrophoresis 25:2402–2412
Mathey EK, Derfuss T, Storch MK et al (2007) Neurofascin as a novel target for autoantibody-mediated axonal injury. J Exp Med 204:2363–2372
Mill J, Petronis A (2007) Molecular studies of major depressive disorder: the epigenetic perspective. Mol Psychiatr 12:799–814
Murray CJ, Lopez AD (1997) Global mortality, disability, and the contribution of risk factors: global burden of disease study. Lancet 349:1436–1442
O'Farrell PH (1975) High resolution two-dimensional electrophoresis of proteins. J Biol Chem 250:4007–4021
Ross PL, Huang YN, Marchese JN et al (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 3:1154–1169
Silveri MM, Parow AM, Villafuerte RA et al (2003) S-adenosyl-L-methionine: effects on brain bioenergetic status and transverse relaxation time in healthy subjects. Biol Psychiatr 54:833–839
Sköld K, Svensson M, Norrman M et al (2007) The significance of biochemical and molecular sample integrity in brain proteomics and peptidomics: Stathmin 2–20 and peptides as sample quality indicators. Proteomics 7:4445–4456
Skolnik P (1999) Antidepressants for the new millenium. Eur J Pharmacol 375:31–40
Sullivan PF, Neale MC, Kendler KS (2000) Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatr 157:1552–1562
Tanna VL, Wilson AF, Winokur G, Elston RC (1989) Linkage analysis of pure depressive disease. J Psychiatr Res 23:99–107
Thornalley PJ (1993) The glyoxalase system in health and disease. Mol Aspects Med 14:287–371
Thornalley PJ (2006) Unease on the role of glyoxalase 1 in high-anxiety-related behaviour. Trends Mol Med 12:195–199
Turck CW (2005) Understanding and treating psychiatric and neurological disorders: the benefits of cerebrospinal fluid analysis. Eur Biopharm Rev 46–49
Turck CW, Maccarrone G, Sayan-Ayata E et al (2005) The quest for brain disorder biomarkers. J Med Invest 52 (Suppl):231–235
Uhlen M (2007) Mapping the human proteome using antibodies. Mol Cell Proteomics 6:1455–1456
Wang X-F, Wang D, Zhu W et al (2003). Studies characterizing 60 kDa autoantibodies in subjects with schizophrenia. Biol Psychiatr 53:361–375
Williams K (1997) Interaction of polyamines with ion channels. Biochem J 325:289–297
World Health Organization: International Statistical Classification of Diseases, 2nd edn, 10th Revision
Wu CC, MacCoss MJ, Howell KE et al (2004) Metabolic labeling of mammalian organisms with stable isotopes for quantitative proteomic analysis. Anal Chem 76:4951–4959
Yabe T, Kanemitsu K, Sanagi T et al (2005) Pigment epithelium-derived factor induces pro-survival genes through cyclic AMP-responsive element binding protein and nuclear factor kappa B activation in rat cultured cerebellar granule cells: implication for its neuroprotective effect. Neuroscience 133:691–700
Yamagishi S, Inagaki Y, Takeuchi M, Sasaki N (2004) Is pigment epithelium-derived factor level in cerebrospinal fluid a promising biomarker for early diagnosis of Alzheimer's disease? Med Hypotheses 63:115–117
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2008 Springer Science+Business Media, LLC
About this chapter
Cite this chapter
Turck*, C.W., Ditzen, C., Sayan-Ayata, E. (2008). Proteomic Strategies for Biomarker Discovery: From Differential Expression to Isoforms to Pathways. In: Turck, C. (eds) Biomarkers for Psychiatric Disorders. Springer, Boston, MA. https://doi.org/10.1007/978-0-387-79251-4_3
Download citation
DOI: https://doi.org/10.1007/978-0-387-79251-4_3
Published:
Publisher Name: Springer, Boston, MA
Print ISBN: 978-0-387-79250-7
Online ISBN: 978-0-387-79251-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)