Abstract
Agrobacterium populations live in different habitats (bare soil, rhizosphere, host plants), and hence face different environmental constraints. They have evolved the capacity to exploit diverse resources and to escape plant defense and competition from other microbiota. By modifying the genome of their host, Agrobacterium populations exhibit the remarkable ability to construct and exploit the ecological niche of the plant tumors that they incite. This niche is characterized by the accumulation of specific, low molecular weight compounds termed opines that play a critical role in Agrobacterium ’s lifestyle. We present and discuss the functions, advantages, and costs associated with this niche construction and exploitation.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
1 Introduction
Agrobacterium is known among microbiologists, geneticists, and biotechnologists as a robust and versatile tool used to introduce foreign genes into plants or fungi (for reviews, see Vain 2007; Idnurm et al. 2017). However, most members of this genus are primarily plant pathogens that induce galls on dicotyledonous plants. Formerly, the Agrobacterium genus encompassed various species such as A. rubi , A. larrymoorei , A. vitis , and A. tumefaciens . The latter species is now recognized as a complex of several species including A. fabrum to which belongs A. fabrum C58, whose genome was the first sequenced in Agrobacterium (for more on Agrobacterium taxonomy see, e.g., Mousavi et al. 2014; Kuzmanović et al. 2015; De Lajudie and Young 2017). In this chapter, we deal with members of the Agrobacterium genus and related genera, irrespective of their species designation, but the most abundant literature is associated with the A. tumefaciens species complex, and especially with the strain C58. For consistency, we will retain the ancient name A. tumefaciens to designate this strain.
Collectively, agrobacteria belong to the family Rhizobiaceae of the class alpha-proteobacteria, members of which are often found in soils of various origins and appear to be among the most common inhabitants of these environments (e.g., Bouzar and Moore 1987; Nüsslein and Tiedje 1998; Texeira et al. 2010; Inceoglu et al. 2011; Lundgerg et al., 2012; Bulgarelli et al. 2012). Interestingly, agrobacteria isolated from soils, including rhizospheric soils, are most often avirulent (Bouzar and Moore 1987; Burr et al. 1987), i.e., they do not harbor a Ti plasmid, the key replicon that determines virulence, unless the soil has an history of crown gall disease (Bouzar et al. 1993; Krimi et al. 2002). These findings suggest that agrobacteria are soil- and rhizosphere-adapted bacteria. As expected, agrobacteria exhibit several traits to exploit soil and rhizosphere resources and to survive under competition with other micro- and macro-organisms. Aside from these adaptive traits, the acquisition of a Ti plasmid that confers pathogenicity can be considered as a process leading to the construction of a more specific and less competitive ecological niche on plant hosts. Data to support these views on niche exploitation and construction by agrobacteria in the soil and plant habitats are presented below.
2 Agrobacterium: A Soil-Adapted Bacterium
Depending on the soil type, agrobacteria members can be either rare or relatively abundant among cultivatable bacteria, with concentrations ranging from 103 to 107 CFU/g (Bouzar and Moore 1987; Bouzar et al. 1993; Krimi et al. 2002). Agrobacterial traits that favor the adaptation to the soil environment remain largely unidentified, as they do for most soil bacteria. However, analysis of the metabolic properties of the bacteria and recent genomic data revealed several interesting features that may allow Agrobacterium to colonize the highly competitive soil environment.
2.1 Exploiting Soil Resources
Agrobacteria may survive for weeks and months under oligotrophic conditions, including pure water (Iacobellis and Devay 1986). Surface waters and aerosols could therefore contribute to dissemination of Agrobacterium populations. Members of this genus are also resistant to osmotic stress, both by taking up osmoprotectants (Nobile and Deshusses 1986; Boncompagni et al. 1999) or by synthesizing them (Smith et al. 1990).
However, bare soils are rare. Most often they are covered by plants that decompose in fall and winter to form humic acids in which agrobacteria can survive for months (Süle 1978). Plants also release at their root system a mixture of carbon compounds known as rhizodeposits. The rhizodeposits consist mainly of root cell debris and exudates, these later originating from plant photosynthesis and metabolism (for reviews, see Hinsinger et al. 2009; Jones et al. 2009; Sánchez-Cañizares et al. 2017). In possible relation with the supply of diverse carbon sources in the rhizosphere, agrobacteria have evolved a wide metabolic capability. For instance and with some variations from one strain to another, agrobacteria can degrade a large range of oses, polyols, and sugar derivatives often from plant origin, including cellobiose, trehalose, maltitol (Marasco et al. 1995; Ampomah et al. 2013), altritol and galactitol (Wichelecki et al. 2015), xylose and glucosamine (Zhao and Binns 2014), melezibiose, raffinose, gentobiose, turanose, lyxose, tagatose, d- and l-fucose, aldonitol, d- and l-arabitol, dulcitol, inositol, sorbitol, xylitol, gluconic acid, keto-gluconic acid, arbutin, esculin, and salicin (Dessaux, unpublished). Agrobacteria can also utilize a wide range of nitrogen-containing compounds as nitrogen sources such as urea (Riley and Weaver 1977), amino-valerate, amino benzoate, ethanolamine, tryptamine (Dessaux, unpublished), and gamma amino-butyrate (Chevrot et al. 2006). In relation to these potential nutrients, agrobacteria exhibit potent urease (Dessaux et al. 1986a, b) and transaminase activities (Sukanya and Vaidyanathan 1964) and a putative nitrilase that permits the scavenging of nitrogen from the plant glycoside amygdalin (Dessaux et al. 1989) and possibly from other cyanogenic compounds. In agreement with the above catabolites, agrobacteria also encode a large number of diverse transporters likely used to take up various potential nutrients.
2.2 Facing and Sustaining Competition
In the soil, agrobacteria are armed to face microbial competitors. Indeed, agrobacteria benefit from a set of potent siderophores that permit an efficient recovery of iron in iron-deprived environments. Several types of siderophores have been identified. The first of these discovered is agrobactin, a derivative of 2,3-dihydroxybenzoic acid, spermidine, and threonine (Ong et al. 1979). The second one is a hydroxamate (Penyalver et al. 2001). The third one, detected in strain C58, remains unidentified (Rondon et al. 2004) but may be specific for this strain (Baude et al. 2016). In addition, with respect to microbial competitors, agrobacteria appear to be partly resistant to antibiotics such as chloramphenicol (Tennigkeit and Matzura 1991), penicillin, erythromycin, streptomycin, and moderately to tetracycline (Khanaka et al. 1981). Aside from these traits, some agrobacteria also express a type VI secretion system (T6SS; for review, see Ryu 2015) that drives the injection of at least three effectors with enzymatic activities (DNase and putative peptidoglycan amidase) into neighboring, competing bacteria (Wu et al. 2008; Ma et al. 2014).
When Agrobacterium colonizes a plant habitat, it can resist adverse antimicrobial compounds such as phenolics produced by plants upon wounding or biotic stress (reviews: Kefeli et al. 2003; Bhattacharya et al. 2010; Caretto et al. 2015). Phenolics play multiple roles in plant protection. With respect to the microflora, phenolics can be potent growth inhibitors of fungi and antibacterial agents (for reviews, see Cushnie and Lamb 2005; Lattanzio et al. 2008). However, nonpathogenic Agrobacterium strains possess an efflux pump active on a group of phenolics, the isoflavonoids that include medicarpin and coumestrol (Palumbo et al. 1998). Other phenolics such as vanillyl alcohol, vanillin, coniferyl alcohol, coniferyl aldehyde, sinapyl alcohol, sinapinaldehyde, and syringaldehyde can also be degraded by nonpathogenic agrobacteria (Brencic et al. 2004). Recently, ferulic acid was also shown to be degraded by Agrobacterium strain C58 (Baude et al. 2016). In addition, pathogenic agrobacteria can detoxify other phenolics via the products of two Ti plasmid genes, virH1 and virH2, located in the virulence region. The VirH1 and VirH2 proteins share sequence homology with cytochrome P450-like enzymes (Kanemoto et al. 1989), and VirH2 appears to be an O-demethylase that is active on over 15 phenolic substrates such as sinapinic acid and acetosyringone. VirH2 can also convert vanillic acid to protocatechuate, which can be further metabolized via the β-ketoadipate pathway (Brencic et al. 2004). Taken together, these data indicate that pathogenic agrobacteria are more resistant to phenolics than are nonpathogenic ones, a result confirmed by the analysis of a virH2 mutant (Brencic et al. 2004). Remarkably, many of the above-mentioned phenolics are inducers of the virulence genes of Agrobacterium (Bolton et al. 1986; Engström et al. 1987) and a few may also be chemoattractant (Parke et al. 1987), a feature that could allow agrobacteria to move upward the concentration gradient toward the wounded plant cells (for review, see Shaw 1991). The route to the plant is also traced by root exudates that are also chemoattractant for agrobacteria (Hawes and Pueppke 1987; Hawes and Smith 1989).
3 The Plant Tumor: A Niche Extension for Agrobacteria
The above data indicate that Agrobacterium is well-equipped to survive in the soil and the plant rhizosphere. However, these environments remain quite competitive. The ability of Agrobacterium to generate a plant tumor can therefore be seen as a “coup de génie” that permits these bacteria to benefit from a much more private habitat, i.e., a quasi-specific niche (Fig. 1). Agrobacterium takes a triple ecological advantage from tumor-niche construction: (i) an increase of resources supporting its proliferation to a high population level; (ii) a decrease of plant defense response in plant tumor tissues; and, (iii) a decrease of competition with resident microbiota, especially through the exploitation of specific growth substrates known as opines. The first point is still poorly understood but could be hypothesized from the high abundance of organic and mineral nutrients that accumulate in plant tumors (Deeken et al. 2006; Lang et al. 2016), whereas tumor development represents a metabolic sink from the plant host; this process makes diversified and abundant resources available to the pathogen. The second point was revealed by transcriptomic and genetic analyses of plant defense pathways (Gohlke and Deeken 2014). Tumor tissue development not only results in abnormally proliferating cells, but also causes differentiation and serves as a mechanism to balance pathogen defense, thereby contributing to the long-term coexistence of agrobacteria and the host plant. The third point, i.e., the opine contribution to Agrobacterium lifestyle in plant tumors, is detailed below.
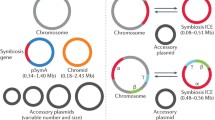
Ecological niches of Agrobacterium. Saprophytic and pathogenic (carrying Ti plasmid) Agrobacterium populations efficiently colonize the rhizosphere of host and non-host plants. Upon permissive conditions (wounding), virulent agrobacteria construct a novel ecological niche that is the plant tumor, as the result of the transfer and expression of the T-DNA in plant host genome. Agrobacterium populations exploit the tumor resources, including opines which confer a selective advantage to Agrobacterium pathogens. Opines also activate quorum-sensing pathways that promote Ti plasmid conjugative transfer, hence contributing to the maintenance and propagation of the virulence genes. The high abundance of virulent Agrobacterium in plant tumor facilitates the dissemination to a new host as well as the maintenance of populations in the rhizosphere and soil
3.1 An Instance of Natural Genetic Engineering
Agrobacterium’s ability to incite a plant tumor, known as crown gall, depends upon the presence in the bacteria of a large plasmid termed the tumor-inducing (Ti) plasmid. During the infection process, a portion of this plasmid, T-DNA, is transferred via a type IV secretion system (T4SST-DNA) as a single-stranded DNA linked with proteins with plant nuclear localization signals. These proteins and T-DNA localize to the nucleus of the plant where T-DNA is eventually integrated into the genome and expressed. These proteins and the T4SST-DNA are encoded by the non-transferred virulence (vir) genes also located on the Ti plasmid (for reviews and more details on the transfer machinery and genetic transformation formation process, see Pitzschke and Hirt 2010; Gelvin 2012; Lacroix and Citovsky 2013; Subramoni et al. 2014; Nester 2015; Christie 2016; Gelvin 2017). Two major sets of genes are borne on T-DNA. The first set, the oncogenes, is responsible for the synthesis of the plant hormones auxin and cytokinin by the transformed host cells, a feature that triggers their proliferation to form a tumor (Ooms et al. 1981; Akiyoshi et al. 1983; Ream et al. 1983). The second set is responsible for the synthesis, of low molecular weight compounds collectively termed opines (see Fig. 2) at the expense of the metabolite pool of the plant. Opines play key ecological roles in the Agrobacterium/plant interaction (for reviews, see Dessaux et al. 1998; Subramoni et al. 2014).
Structural formulas of opines a octopine family, b nopaline family, c agropine family, d agrocinopines family, e cucumopine family, f succinamopine and leucinopine families, g chrysopine family, h ridéopine and heliopine families. Octopine family opines are all synthesized by the enzyme octopine synthase and derive from various proteinous and nonproteinous amino acids, and pyruvate. They include the most recently discovered opine sulfonopine (Flores-Mireles et al. 2012). Nopaline and nopalinic acid synthesized by nopaline synthase derive from alpha-ketoglutarate and, respectively, arginine and ornithine. Succinamopine, leucinopine, cucumopine (and its diastereomer mikimopine) are also alpha-ketoglutarate condensates and exhibit asparagine, leucine, and histidine moieties, respectively. Heliopine (also termed vitopine) is a condensation product of pyruvate and glutamine. The mannityl opines are sugar and glutamate or glutamine-containing compounds as are the closely related opines of the chrysopine family. Other sugar opines include the agrocinopines A and B that are the only phosphorus-containing opines
3.2 The Opine Concept
Opine synthesis by crown gall tumors and their assimilation by agrobacteria represents an archetype of ecological niche construction and exploitation processes by a pathogen. Opines are secreted by transformed plant cells into the intercellular spaces in the tumor, and to a lesser extent the whole plant (Savka and Farrand 1992; Savka et al. 1996). Opines play two major roles in niche construction for agrobacteria. First, they serve as growth substrates for the tumor-inciting strain and, second, they stimulate the conjugative transfer of the Ti plasmid from pathogenic Agrobacterium to other Agrobacterium cells (for a review, see Dessaux et al. 1998). These features are at the origin of the opine concept that describes opines as chemical mediators of parasitism. Synthesis of opines is induced by the pathogen, thus providing an environment favorable to the growth of the bacteria and dissemination of its pathogenicity (Schell et al. 1979; Tempé and Petit 1983).
The opine concept was formulated years before the discovery of plants that naturally harbor in their genomes DNA regions highly homologous to Agrobacterium T-DNA. Among these species are members of the genera Nicotiana, Linaria, and Ipomoea (White et al. 1983, Aoki et al. 1994; Suzuki et al. 2002; Matveeva et al. 2012; Kyndt et al. 2015; Quispe-Huamanquispe et al. 2017). Interestingly, some of these plants produce detectable amounts of opines (Chen and Otten 2017). The opine concept could therefore incorporate both the tumorous temporary niche and the permanent niche that naturally genetically modified plants and their offspring represent. However, no clear demonstration of a stimulation of the community of Opine-degrading bacteria at the root system of these naturally transformed plants has yet been reported.
3.3 Opine Metabolism Genes
Opines are most often synthesized from common molecules such as amino acids, alpha-ketoacids, and sugars. Over 20 opine molecules are known (Fig. 2a–h). They are not all present at the same time in a tumor and some opines are specific for a given agrobacteria species. Indeed, the type opine synthesized by plant cells and degraded by agrobacteria depends upon the type of Ti plasmid, a feature that has been used to classify agrobacterial Ti plasmids (for a review, see Dessaux et al. 1998). The current list of agrobacterial opines is likely to be near complete. Indeed, over the last 15 years no novel opine has been discovered except sulfonopine, a sulfur-containing molecule detected in tumors induced by a single octopine-type Agrobacterium strain (Flores-Mireles et al. 2012).
Genes involved in the biosynthesis and catabolism of opines are known for several opine systems. Generally, opines derived from amino acids and alpha-ketoacids (such as octopine or nopaline; Fig. 2a, b) are synthesized in one step by a protein encoded by a single gene located on T-DNA (De Greve et al. 1982; Koncz et al. 1983). The same is true for phospho-sugar opines of the agrocinopine family (Joos et al. 1983; Fig. 2d). On the contrary, opines derived from condensation of sugars and amino acids, the mannityl opines or the chrysopine family opines (Fig. 2c, g), are synthesized in one, two, or three steps by the corresponding number of enzymes encoded by one, two, or three genes. These are most often located on a T-DNA separate from that which carries the oncogenes (Hood et al. 1986; Palanichelvam et al. 2000).
Opine catabolic genes are generally clustered in operons and regulons in delineated regions of the Ti plasmids, and their expression is inducible by the degraded opines (Klapwik et al. 1977; Klapwik et al. 1978; Chilton and Chilton 1984; Dessaux et al. 1988). Two sets of genes are present in the catabolic region. The first encodes the transport system (e.g., Klapwik et al. 1977; Zanker et al. 1992) that often consists of an ABC transporter and its cognate, high affinity (nM–µM range) periplasmic-binding protein (Lang et al. 2014; El Sahili et al. 2015; Marty et al. 2016; Vigouroux et al. 2017). The second encodes the enzymes involved in the degradation of the opines to molecules that belong to central bacterial metabolism. For example, octopine and nopaline are degraded into arginine, ornithine, and glutamate, and pyruvate or alpha-ketoglutarate, respectively (Montoya et al. 1977; Ellis et al. 1979; Dessaux et al. 1986a, b). Remarkably, for some opines such as the mannityl opines, genes, and functions involved in the synthesis and degradation are closely related, suggesting that duplication events occurred in the course of the evolution of the Ti plasmids (Kim et al. 1996; Hong et al. 1997; Kim and Farrand 1996). A similar duplication also occurred with respect to genes involved in the synthesis and degradation of the phospho-sugar opines agrocinopines A and B (Kim and Farrand 1997).
3.4 Opines as Growth Substrates
The opine concept has been elaborated from the observation that all crown gall tumors, including those initially reported not to contain any opine (i.e., the so-called null type tumors), indeed contain such compounds (Guyon et al. 1980). The opine hypothesis later received experimental validation. The first support for the opine concept came from comparison of the growth of two closely related Agrobacterium strains, one capable of degrading opines, the other not, at the root system of transformed plants producing opines. The experiment revealed that plants producing opines preferentially promote the growth of opine-degrading agrobacteria (Guyon et al. 1993). A second set of experiments involved transformed plants producing opines and two closely related Pseudomonas strains, one engineered—via the introduction of an opine catabolic plasmid—to degrade opines, the other not. The experiment demonstrated that the growth of the opine-degrading pseudomonad was favored at the root and leaf surface of opine-producing plants (Wilson et al. 1995; Savka and Farrand 1997). A recent experiment (Lang et al. 2014) involved the wild-type (WT) Agrobacterium strain C58 and a mutant unable to degrade nopaline, the major opine found in the tumors incited by this strain. When both strains were inoculated separately onto a plant, they multiplied in the tumor to reach a similar bacterial concentration. However, when co-inoculated the WT opine-degrading bacteria outcompeted the mutant. This observation formally demonstrated that the presence of the opine does not increase the carrying capacity of the tumor habitat for Agrobacterium but “selects for those able to assimilate it” (Lang et al. 2014). A similar study extended this paradigm to the octopine-niche (submitted by Vigouroux et al. 2017).
3.5 Opines as Inducers of Ti Plasmid Horizontal Transfer
The discovery of the Ti plasmid as key pathogenic element for Agrobacterium (Van Larebeke et al. 1974; Watson et al. 1975) was rapidly followed by the demonstration that these plasmids can be transferred by conjugation between bacteria; a phenomenon also regulated by opines (Kerr et al. 1977; Genetello et al. 1977). The nature of the opines that induce conjugation varied as a function of the opine-type of the plasmid. Thus, octopine induces the transfer of octopine-type plasmids, whereas agrocinopines A and B induce transfer of nopaline-type plasmids, and agrocinopines C and D the conjugation of agropine-type plasmids (Klapwijk et al. 1978; Petit et al. 1978; Ellis et al. 1982).
Ti plasmid transfer is also regulated by quorum sensing (QS; Piper et al. 1993; Zhang et al. 1993). QS is a widely occurring regulatory process that couples gene expression (in a positive or negative way) with bacterial cell concentration. It relies upon the production and sensing by a bacterial population of diffusible signal(s), the concentration of which indicates that of the microbial cells. Once a threshold concentration of signal is reached in the environment, the presence of the signal is sensed by receptors and translated into activation or repression of the expression of the genes regulated by QS (for recent reviews on QS, see Garg et al. 2014; Grandclément et al. 2016; Papenfort and Bassler 2016).
In the reference Agrobacterium strain C58, the presence of agrocinopines A and B triggers the expression of the acc operon of the Ti plasmid that encodes agrocinopine degradation, and that of the adjacent arc operon by releasing the repression exerted by the master regulator AccR (Beck von Bodman et al. 1992). Agrocinopine A can be cleaved into arabinose-2-P and sucrose by AccF, because only arabinose-2-phosphate (and not agrocinopine A) interacts with AccR (El Sahili et al. 2015). One of the genes of the arc operon is traR. It encodes the regulatory protein TraR that, once bound to the QS signal, dimerizes and activates the transcription of the traAFB, traCDG, and trb operons (Piper et al. 1999). The tra operons encode components of the DNA transfer and replication (DTR) system that recognizes and cleaves plasmid DNA at the origin of transfer (oriT) located between the two tra operons (Farrand et al. 1996; Zechner et al. 2001). The trb operon encodes components of a type IV secretion system (T4SSpTi) that permits the transfer of the plasmid DNA and associated proteins to recipient bacteria (Li et al. 1999). Interestingly, the first gene of the trb operon is traI. The eponym protein TraI is responsible for the synthesis of a diffusible QS signal that belongs to the widely distributed N-acyl homoserine lactone (AHL) class of signals (Hwang et al. 1994). In the presence of agrocinopines but at low cell concentration, the trb operon—hence, the TrbI QS signal synthase—is very weakly expressed and only low amounts of QS signals accumulate in the environment. In the presence of agrocinopines and at high cell concentrations, the QS signal concentration increases and its presence is sensed by TraR that becomes activated and induces the full expression of the T4SSpTi and DTR system, stimulating the transfer of the Ti plasmid (Li et al. 1999; Li and Farrand 2000).
3.6 Cost and Control of Opine-Niche Construction and Exploitation
As indicated above, the key step of opine-niche construction is the transfer of T-DNA to plant cells via a dedicated T4SS (T4SST-DNA) that imposes a fitness cost to agrobacteria (Platt et al. 2012). In a competitive arena, individuals expressing the T4SST-DNA are disadvantaged compared to those impaired for T4SST-DNA or defective for a Ti plasmid. Indeed, in short-term experimental evolution cultures in the presence of acetosyringone (an inducer of T4SST-DNA expression) and in plant tumors, spontaneous mutants arose in the progeny of a virulent Agrobacterium ancestor. These mutants were altered in virulence because of alteration or loss of the Ti plasmid (Bélanger et al. 1995; Fortin et al. 1992, 1993; Llop et al. 2009). Virulent agrobacteria exhibit three potential mechanisms to balance the fitness cost and damage imposed by T-DNA transfer: (i) a tight control of vir gene expression by phenolics, acidic pH, and sugars contributes to optimize the cost/benefit of T4SST-DNA expression, hence the success of T-DNA transfer into plant cells (Nair et al. 2011; He et al. 2009); (ii) Ti plasmid horizontal transfer that may re-introduce the Ti plasmid into those cells which have lost it (Lang et al. 2013); (iii) a fitness gain to agrobacteria—that have kept or acquired a Ti plasmid—because of opine-niche exploitation (Lang et al. 2014). Conditioning the transfer of the Ti plasmid to the tumor environment (opine as ecological proxy) ensures that the Ti plasmid-carrying Agrobacterium individuals will gain a selective advantage in the most compatible ecological niche.
In nature, the Ti plasmid may be transferred to other agrobacteria (other species or clonal lineages) or non-agrobacteria that is free of a Ti plasmid, whereas this transfer could be considered as advantageous for the Ti plasmid per se (selfish gene and reservoir hypotheses), and it could be disadvantageous for the Ti plasmid donor lineage because potential bacterial competitors could acquire the opine-niche exploitation trait. Another important consideration in Ti plasmid transfer is its cost as the process uses a second T4SSpTi. An experimental evolution experiment conducted with an A. tumefaciens C58 derivative expressing QS and T4SSpTi revealed the emergence of mutants defective for QS signal synthesis (mutations in traR) or exhibiting a QS-hijacking behavior or defective for the presence of a Ti plasmid (Tannières et al. 2017). Agrobacterium Ti plasmid donors exhibit several mechanisms to finely control QS, and hence Ti plasmid transfer. QS relates Ti plasmid transfer to a high population level of donors. This major requirement allows a virulent population to become dominant in a plant tumor habitat before activating Ti plasmid transfer, which is costly (growth slowdown) and hazardous (increase of opine-assimilating competitors). Additional mechanisms which are not present in all agrobacteria also contribute to delay QS signaling, therefore leaving time for donors to proliferate before transferring a Ti plasmid. First, the TraM protein encoded by traM on the Ti plasmid interacts with TraR and blocks the formation of an active TraR homodimer at low QS signal concentrations (Khan et al. 2008; Qin et al. 2007). Second, the lactonases BlcC and AiiB open the gamma-butyrolactone ring of AHLs (Haudecoeur et al. 2009). The traM and aiiB genes are encoded by the Ti plasmid and are expressed in the presence of agrocinopines in strain C58. The blcC gene (formerly attM) belongs to the blcABC (formerly the attKLM ) operon located on pAt plasmids. BlcC is activated in the presence of gamma-butyrolactone (GBL), gamma-hydroxybutyric acid (GHB), and succinic semialdehyde (SSA), which are activated and repressed by a high and low gamma-aminobutyric acid (GABA)/proline ratio, respectively (Carlier et al. 2004; Lang et al. 2016), whereas TraM titrates TraR and prevents its early production, intracellular lactonases constrain the level of AHLs in the intra- and extracellular environments, hence their binding to TraR. Both these QS-delaying mechanisms are bypassed when TraR and AHLs are produced at a high level (Khan et al. 2008; Haudecoeur et al. 2009).
3.7 Competition for the Opine Niche by the Plant Microbiota
Although engineered by Agrobacterium as a niche, the tumor can be colonized by other opine-degrading microorganisms, including bacteria such as pseudomonads, Sinorhizobium meliloti, Arthrobacter sp., coryneform isolates (Tremblay et al. 1987; Nautiyal and Dion 1990; Nautiyal et al. 1991; Moore et al. 1997; Faist et al. 2016), or by fungal strains (Cylindrocarpon heteronema and Fusarium solani; Beauchamp et al. 1990). These microorganisms are naturally present in soils of diverse origins, and their growth can be stimulated by opines produced by the tumor and released at the root system of the plant independently of the soil and plant considered (Oger et al. 1997; Mansouri et al. 2002, Mondy et al. 2014; Faist et al. 2016). Interestingly, as indicated earlier, opines are chemoattractants for Agrobacterium (Kim and Farrand 1988). This feature may provide a way for agrobacteria that migrate from the tumor to return to the opine-rich niche of the crown gall. A possibility exists that opines could also attract non-agrobacterial organisms, but to the best of our knowledge, this has not yet been investigated.
3.8 Exploitation of Other Plant Tumor Resources
Besides opines, a wide variety of organic (amino acids, organic acids, oses, polyols, etc.) and mineral compounds, which are potential resources for agrobacteria, accumulate in plant tumors (Deeken et al. 2006; Lang et al. 2016). Unlike opines, these compounds are not specific to tumor tissues and Ti plasmid type. To be considered part of the niche construction process, these compounds should not only accumulate in plant tumors, and their exploitation should also confer a selective advantage to agrobacteria for colonizing this habitat. Numerous traits are potentially consistent with this definition but experimental evidence is missing. A chromosomal locus picA, which may be involved in the degradation of plant polymers and whose expression is induced in the presence of plant tissues, may be such a candidate (Rong et al. 1991).
Numerous Agrobacterium isolates (carrying or lacking a Ti plasmid) harbor larger plasmids known as pAt plasmids (Merlo and Nester 1977; Rosenberg and Huguet 1984; Hynes et al. 1985). pAt plasmids can be very different from one strain to another, whereas they may comprise up to 10% of the agrobacterial genome, only a limited number of pAt functions are known. In A. tumefaciens C58, aside from utilization of GBL, GHB, and SSA (a by-product of GABA) as nutrients (Carlier et al. 2004), the plasmid pAtC58 encodes degradation of the Amadori compound deoxy-fructosyl-glutamine (Vaudequin-Dransart et al. 1995; Baek et al. 2003). Exploitation of these plant compounds could contribute to tumor colonization by virulent (carrying Ti plasmid) and avirulent (free of Ti plasmid) agrobacteria.
The question about the cost associated with At plasmid maintenance has been investigated in Agrobacterium strain C58 by comparing different derivatives carrying two, only one, or none of the plasmids pAtC58 or pTiC58. In culture medium when the T4SST-DNA and T4SSpTi are not expressed, the cost of carrying the pAt plasmid was higher than that of the Ti plasmid (Morton et al. 2014). This may be related to the large size of the pAt plasmid as well as to the constitutive expression of the T4SSpAt that promotes its conjugative transfer (Chen et al. 2002). A fitness gain associated with the pAt plasmid was reported in the rhizosphere of Helianthus annuus (Morton et al. 2014), but this question remains unsolved in plant tumors. Interestingly, in Agrobacterium strain C58 the transfer of the pAt plasmid is co-regulated with that of the Ti plasmid and strongly depends upon the activity of the master regulatory protein AccR encoded by a Ti plasmid gene, the transcription of which is induced in the presence of agrocinopines A and B (Lang et al. 2013). This observation suggests that the tumor habitat stimulates a simultaneous propagation of both the pAt and pTi plasmids, probably meaning that a selective advantage could be conferred by the acquisition of the two plasmids. In some Agrobacterium strains, pTi and pAt plasmids can cointegrate and cooperate for opine degradation (Vaudequin-Dransart et al. 1998). This cooperation has also been observed for some Ri plasmids (Costantino et al. 1980; Petit et al. 1983). In strains devoid of a Ti plasmid, the transfer of the pAt plasmid may also be regulated, by QS, by-products of genes located on this plasmid. In this case, the existence of one or more possible inducers of conjugation has not been demonstrated (Mhedbi-Hajri et al. 2016).
4 Niche Construction and Exploitation by Agrobacterium-Related Genera
All the findings described above paved the path to investigate whether the opine concept can be extended outside the Agrobacterium clade. Experiments performed with closely related Rhizobiaceae revealed that transformed plant roots induced by Rhizobium rhizogenes (formerly Agrobacterium rhizogenes) also contain opines (Petit et al. 1983). These two pathogens are closely related. Indeed, as in Agrobacterium spp., pathogenic strains of R. rhizogenes harbor large plasmids known as Ri plasmids (Moore et al. 1979). A portion of these plasmids, T-DNA, is transferred to the nucleus of the plant cells where it integrates into the genome upon infection (Chilton et al. 1982; Willmitzer et al. 1982; White et al. 1982). R. rhizogenes T-DNA harbors oncogenes that for the most part differ from those of A. tumefaciens and trigger the formation of transformed roots (e.g., Durand-Tardif et al. 1985; Slightom et al. 1986; Cardarelli et al. 1987; Spena et al. 1987). However, the genes involved in opine biosynthesis are often highly related to those of Agrobacterium Ti plasmids, and several of them direct the synthesis of opines, such as cucumopine or mannityl opines (Fig. 2d, e), that are also found in crown gall tumors (Tepfer and Tempé 1981; Jouanin 1984; De Paolis et al. 1985; Petit and Tempé 1985).
A further extension dealt with nitrogen-fixing nodules incited by Sinorhizobium meliloti and Rhizobium leguminosarum strains on leguminous plants. Some of these nodules contain opine-like molecules, identified as scyllo-inosamine (SI) and 3-O-methyl-scyllo-inosamine (3OSI; Murphy et al. 1987; Saint et al. 1993) and collectively termed rhizopines (Fig. 3). However, only a limited number of strains of these species (ca. 11–12% of assayed clones) were able to produce and degrade rhizopines, independent of their geographical origin (Rossbach et al. 1995; Wexler et al. 1995). Genes involved in both the synthesis and degradation of SI and 3OSI have been identified. They are adjacent on the symbiotic plasmid of the bacteria (Murphy et al. 1987). In contrast to the Agrobacterium system, these biosynthetic genes are not transferred to plant cells but are expressed by the bacteria itself in the nodule context only. As with Agrobacterium opines, rhizopines provide a selective advantage for rhizopine utilizers in the plant environment, possibly by providing a selective nutrient to members of the population living around the nodules. This selective advantage has been demonstrated by competition experiments that involved a wild-type S. meliloti strain and a mutant unable to degrade rhizopines (Gordon et al. 1996; Heinrich et al. 1999). For a recent review on rhizopines and more data on genes involved in biosynthesis and degradation, the reader can refer to Savka et al. (2013).
Structural formulas of opine-like molecules found in nodules. The opine-like compound 3-O-methyl-scyllo-inosamine (as well as scyllo-inosamine, not shown) is opine-like molecules detected in nodules incited in alfalfa (Medicago sativa) by some strains of Rhizobium meliloti. Rhizolotine is an opine-like compound found in Lotus spp. nodules incited by Mesorhizobium loti strain NZP2037. This riboside molecule exhibits a tetrahydropyrimidine ring
Two other opine-like molecules have been detected in the nitrogen-fixing nodules induced on Lotus spp. by Mesorhizobium loti. One was identified as the riboside of an alpha-hydroxy-imino acid and named rhizopines (Fig. 3). The second is an unidentified ninhydrin-positive compound (Shaw et al. 1986; Scott et al. 1987). No indication of the competitive advantage given to the rhizolotine-degrading strains in nature is available.
Aside from the above-described interactions, other interactions between bacteria and their hosts involve a trophic link. This is the case, for instance, for rhizobia that induce nodules on mimosa (Acacia dealbata) or Leucaena spp. plants. Plants of both genera produce large amount of mimosine, a toxic amino acid that only rhizobia nodulating these plants can degrade (Soerdajo et al. 1994), providing them with a selective advantage (Soedarjoa et al. 1998). Also the alkaloids calystegins present in the roots and exudates of morning glory (Convolvulus arvensis), hedge bindweed (Calystegia sepium), and belladonna (Atropa belladonna) can be efficiently degraded by Sinorhizobium meliloti strain Rm 41, a strain that is frequently detected in the root system of these plants, though they are not members of the legume clade and not hosts for symbiotic nitrogen fixation (Tepfer et al. 1988). None of these interactions, however, fits the description of the opine concept that remains limited to agrobacteria and—to a certain extent—to some rhizobia. As most rhizobia are symbionts, the opine concept should therefore be reformulated as “opines are chemical mediators of plant-microbe interactions, the synthesis of which is induced by the micro-organism, thus providing an environment favorable to its growth and dissemination of its plant-interacting capacity.”
All the above data prompted scientists to propose that the growth of beneficial microbial populations in the rhizosphere could be engineered and favored by establishing an opine-based, trophic link between the plant to protect and selected microbial population (Savka et al. 2002: Dessaux et al. 2016). Though elegant, and in spite of encouraging preliminary results obtained for some plant growth promoting rhizobacteria (Dessaux et al. 1987; Guyon et al. 1993; Wilson et al. 1995; Savka and Farrand 1997; Oger et al. 1997), this concept has not yet received definitive experimental validation.
5 Unsolved Mysteries in Agrobacterium Ecology
5.1 Where Do Pathogenic Agrobacteria Hide in Nature?
Though some pathogenic Agrobacterium strains can be isolated from uncultivated pasture soil (Schroth et al. 1971), natural soil, and plant rhizospheres, agrobacteria isolates are most often nonpathogenic unless the place of isolation has a history of crown gall contamination (Bouzar et al. 1993; Krimi et al. 2002; Dessaux, unpublished). This feature led scientists to wonder whether pathogenic agrobacteria can be isolated from some nursery soils because plants are contaminated and therefore provide the source of bacteria, or whether the plants are contaminated because virulent agrobacteria are present in these soils. This “chicken or egg” causal dilemma cannot be definitively resolved at this time, but some factual and speculative elements can be proposed. First, it is clear that exchange of contaminated plant material between various locations and countries could be at the origin of crown gall outbursts (Pionnat et al. 1999). Once contaminated, and in spite of seasonal fluctuations, soils can host agrobacterial populations and maintain them for years (Bouzar et al. 1993; Krimi et al. 2002). Second, it cannot be excluded that pathogenic agrobacteria can “hide” in the rhizosphere of non-host plants (i.e., plants that do not develop crown gall symptoms) and, consequently, in places where they will not be searched for. In agreement with this proposal, agrobacteria have been detected at the root system of maize (Gomes et al. 2001) and wheat (Bednárová et al. 1979).
An alternative or complementary hypothesis is that Agrobacterium do not hide, but Ti plasmids do. It could be speculated that Ti plasmids could conjugate in tumors to other, non-agrobacterial isolates where they could replicate. In support of this model, Ti plasmids could conjugate to E. coli under laboratory conditions but they do not replicate in this host (Holsters et al. 1978). They can also be transferred to rhizobia that possess genetic backgrounds in which Ti plasmids can replicate but do not always express their tumorigenic functions (Hooykaas et al. 1977; van Veen et al. 1989; Teyssier-Cuvelle et al. 1999). Though attractive, this later hypothesis is not really supported by probabilistic elements. First, the conjugative transfer frequency of the Ti plasmid in vitro reaches at best 103 per donor (Lang et al. 2013). Second, to generate a pathogenic Agrobacterium, the Ti plasmid would have to conjugate from the replicative bacteria back to an agrobacterial isolate in environments deprived of opines that are precisely the inducers of this conjugative transfer.
Clearly, the question of the reservoirs of Ti plasmids in nature remains an open but critical one. Further investigations are necessary to identify such reservoirs and complement our understanding of the ecology of both agrobacteria and Ti plasmid. Studies that aim to elucidate the genes and functions that contribute to bacterial fitness in tumors, the rhizosphere, bare soil, and possibly surface waters could contribute to reach these objectives.
5.2 Origin of T-DNA, Origin of Opines
Analyses of Ti plasmids revealed that they exhibit a chimeric structure (Otten et al. 1993; Otten and De Ruffray 1994) composed mostly of four key clusters of genes: the T-DNA, the virulence region that encodes the T4SST-DNA involved in T-DNA transfer, the opine catabolic region, and the conjugative transfer regions that includes the T4SSpTi. Interestingly, in A. tumefaciens Ti plasmids, T-DNA, the T4SST-DNA, and the conjugative region(s) are highly related, whereas the opine catabolic regions differ from one plasmid type to another. The homology of several genes that encode the T4SST-DNA and the T4SSpTi (Chen et al. 2002) clearly suggests that both may derive from a common ancestral protein secretion system. Similarly, T-DNA genes responsible for the production of the plant hormone auxin, namely iaaM and iaaH, are orthologues of genes found in members of the Pseudomonas savastanoi species (Yamada et al. 1985). The cytokinin biosynthetic gene, ipt or tmr, is also related to the cytokinin biosynthetic gene ptz of P. savastanoi (Powell and Morris 1986).
As indicated above, A. tumefaciens T-DNAs differ from one another mostly by the nature of the opine anabolic genes. A parsimonious hypothesis therefore implies that T-DNA and the T4SST-DNA have been acquired before the genes involved in opine metabolism in the evolutionary history of the plasmids, possibly as a way to reduce plant defense (Dunoyer et al. 2006; Gohlke and Deeken 2014). Furthermore, opine metabolic genes could have different origins. Some of these opine metabolic genes have evolved by duplication from common ancestor(s). This is the case of the genes involved in the synthesis and degradation of the mannityl opines. The synthesis of mannopine and mannopinic acid proceeds in two steps: (i) the condensation of glucose with glutamine or glutamate, respectively, to Schiff bases and their Amadori rearrangement compounds to form deoxy-fructosyl-glutamine (dfgln) and deoxy-fructosyl-glutamate (dfglu; Fig. 2g); (ii) the reduction of dfgln and dfglu to mannopine or mannopinic acid, respectively (Ellis et al. 1984). Mannopine can be dehydrated to yield the cognate lactone agropine, another mannityl opine (Dessaux et al. 1986a, b). Degradation proceeds almost in the reverse way. Agropine undergoes a lactonolysis to mannopine that is in turn degraded to dfgln and mannose and glutamine. Dfglu is degraded to mannose and glutamate. In this scheme, dfgln appears to play a central role. First, it is also an opine found in the tumors induced by strains of Agrobacterium that harbor a chrysopine-type Ti plasmid; second, and in contrast to most opines, dfgln can be degraded by numerous strains of Agrobacterium irrespective of their virulence (Bouzar et al. 1995; Chilton et al. 1995; Vaudequin-Dransart et al. 1995). Accordingly, the Ti plasmid-free derivative of the reference strain C58 can metabolize dfgln via the product of genes located on the At plasmid that are highly homologous to genes found on the Ti plasmids (Baek et al. 2003). Remarkably, contrary to the situation with other opines, dfgln and dfglu occur widely in nature, i.e., outside Agrobacterium-induced tumors. As with numerous Amadori compounds, dfgln and dfglu form spontaneously in decaying plant material (Anet 1957; Anet and Reynolds 1957). It is tempting to speculate that their common occurrence in nature provides a sufficient selective pressure to account for the emergence and selection of degradative functions, as seen in nonpathogenic strains of Agrobacterium. The duplication of the degradative opine genes and their incorporation into a “proto T-DNA” could have provided Agrobacterium with a way to force living plant cells to produce dfgln and dfglu. A further step in the evolution of the Ti plasmid could be the acquisition of opine anabolic and catabolic functions to allow the conversion of the two Amadori compounds to mannopine and mannopinic acid and later agropine (and vice versa) that are less accessible to competing organisms. Though entirely speculative, this model is nevertheless consistent with the physiological, biochemical, and molecular data available today.
The origins of other opine metabolic functions are even more speculative than those of the dfgln and mannityl opines. Octopine is synthesized in the muscle of marine animals such as octopus and squid during anaerobic muscle contraction (Thoai and Robin 1959) as a way to re-oxidize NADH, regenerate ATP, and reduce the concentrations of both arginine and pyruvate that accumulate under this condition (Grieshaber and Gäde 1976). Because marine agrobacterial isolates have been obtained (Rüger and Höfle 1992), a possibility exists that octopine degradation in these bacteria arose in relation with the presence of octopine in marine animals. In agreement, octopine-degrading bacteria have been isolated from mussels and oysters (Dion 1986). The related structures and sequence homologies of both the catabolic and anabolic genes for octopine and nopaline (Zanker et al. 1992, 1994) also suggest that these two opine systems may have evolved from a common ancestor.
The origin of sugar opines, such as the agrocinopines, is also unclear. Agrocinopine A is composed of sucrose linked to l-arabinose by a phosphodiester bond, whereas in Agrocinopine C, a d-glucose is present instead of the l-arabinose in agrocinopine A. Agrocinopines B and D differ from A and C, respectively, by lacking one sugar from the sucrose moiety (Ellis and Murphy 1981). In Agrobacterium strain C58, agrocinopine A is cleaved into arabinose-2-phosphate that is able to interact with AccR for activating quorum-sensing and Ti plasmid conjugation (El Sahili et al. 2015). Noticeably, arabinose-2-phosphate is uncommon (unique until now) in the living world due to the unusual phosphate linkage on the C2 atom of the pyranose. This exemplifies the capacity of Agrobacterium to innovate by the use of a signal that is discriminable among the diverse sugars in plant hosts.
The occurrence of various opine anabolic and degradative systems may appear puzzling at first glance. However, the occurrence of multiple opine systems could indeed allow the diversification and coexistence of various agrobacterial populations in the same plant environment. These populations can therefore be considered as sympatric, and may eventually evolve novel species in further evolutionary steps (Lassalle et al. 2015). In agreement, whereas octopine or heliopine can be found in tumors incited by numerous Agrobacterium species, a number of opines such as cucumopine or ridéopine have been found only in grapevine tumors induced by members of the A. vitis species (Chilton et al. 2001). Similarly, cucumopine (or mikimopine) are detected only in hairy roots induced by R. rhizogenes (Davioud et al. 1988).
5.3 Is Agrobacterium’s Ability to Transfer DNA to Organisms Belonging to Other Kingdoms Unique?
Agrobacterium spp. and R. rhizogenes, due to the presence of Ti and highly related Ri plasmids, are to the best of our knowledge rare examples of bacteria naturally capable of transferring DNA to members of the eukaryote kingdom (Lacroix and Citovsky 2016). However, Ensifer adhaerens, a related bacterium, has recently been reported to transform plant cells when equipped with an appropriate plant transformation plasmid vector of the pCAMBIA series (Wendt et al. 2012). Aside from Agrobacterium, the only transkingdom DNA transfer that has been reported under laboratory conditions is between the pathogen Bartonella henselae and a human endothelial cell line (Schröder et al. 2011). B. henselae is not a major human pathogen except in immune-compromised patients where it may trigger a disease known as bacillary angiomatosis (Dehio 2005). A mobilisable cryptic plasmid from another Bartonella species was tagged with a fluorescent protein gene expressed only in eukaryotic backgrounds and introduced into a B. henselae strain that was used to infect endothelial cells. Post-infection, a low numbers of cells were fluorescent, indicating a T4SS-mediated transfer frequency of the plasmid of ~2 × 10−4. There is, however, no direct evidence that such a transfer may occur in animals, and no indication that such a transfer may lead to a permanent transformation of the recipient eukaryotic cells.
A recurring question is why no other bacteria have evolved comparable host transformation systems? First, to inquire whether other systems comparable to the Ti and Ri plasmids exist, 21 bp DNA border sequences have been compared to sequences of bacterial genomes or soil microbial metagenomes in data banks. The only hits identified were members of the two former genera (Agrobacterium and Rhizobium; Dessaux, unpublished). Second, the uniqueness of the Agrobacterium spp. and R. rhizogenes transformation machinery could be explained by some of the evolutionary elements presented above which indicate that the occurrence of Ti and Ri plasmids proceeded in several steps, under selective pressures that may have rarely encountered in the living world. In other words, acquisition of Ti and Ri plasmids was quite an exceptional event.
In addition, once Ti and Ri plasmids evolved, it appears that their propagation in other bacteria was restricted by various factors. For instance, Ti plasmids do not replicate in firmicutes and actinobacteria, nor do they in beta- and gamma-proteobacteria such as E. coli or pseudomonads (Holsters et al. 1978; Dessaux, unpublished). Also, cloned Ti plasmid genes such as the opine catabolic genes are generally not expressed in other bacteria, including proteobacteria (Dessaux et al. 1987). Even in the related Rhizobium group where Ti plasmids replicate, tumor-inducing functions may or may not be expressed (Klein and Klein 1953; Hooykaas et al. 1977; van Veen et al. 1989) possibly because chromosomal genes involved in this process (see for instance Douglas et al. 1985; Close et al. 1985; Thomashow et al. 1987) may be missing. These data imply that transfer of the Ti plasmid outside the Agrobacterium genera, the R. rhizogenes species, and some Rhizobium species may be an evolutionary cul-de-sac either because the plasmid does not replicate or because the plasmid functions are not expressed. To a certain extent, and from an anthropomorphic point of view, Agrobacterium drastically protects the invention of the Ti plasmids that allow it to shift from a generalist behavior in the soil and the rhizosphere to a specialist behavior in the tumor where it escapes most microbial competitors and a part of plant defense.
References
Akiyoshi DE, Morris RO, Hinz R et al (1983) Cytokinin/auxin balance in crown gall tumors is regulated by specific loci in the T-DNA. Proc Natl Acad Sci USA 80:407–411
Ampomah OY, Avetisyan A, Hansen E et al (2013) The thuEFGKAB operon of rhizobia and Agrobacterium tumefaciens codes for transport of trehalose, maltitol, and isomers of sucrose and their assimilation through the formation of their 3-keto derivatives. J Bacteriol 195:3797–3807
Anet EFLJ (1957) Chemistry of non-enzymic browning. II. Some crystalline amino acid-deoxy-sugars. Austral J Chem 10:193–197
Anet EFLJ, Reynolds TM (1957) Chemistry of non-enzymic browning. II. Reactions between amino acids, organic acids, and sugars in freeze-dried apricots and peaches. Austral J Chem 10:182–192
Aoki S, Kawaoka A, Sekine M et al (1994) Sequence of the cellular T-DNA in the untransformed genome of Nicotiana glauca that is homologous to ORFs 13 and 14 of the Ri plasmid and analysis of its expression in genetic tumours of N. glauca x N. langsdorffii. Mol Gen Genet 243:706–710
Baek CH, Farrand SK, Lee KE et al (2003) Convergent evolution of Amadori opine catabolic systems in plasmids of Agrobacterium tumefaciens. J Bacteriol 185:513–524
Baude J, Vial L, Villard C et al (2016) Coordinated regulation of species-specific hydroxycinnamic acid degradation and siderophore biosynthesis pathways in Agrobacterium fabrum. Appl Environ Microbiol 82:3515–3524
Beauchamp CJ, Chilton WS, Dion P et al (1990) Fungal catabolism of crown gall opines. Appl Environ Microbiol 56:150–155
Beck von Bodman S, Hayman GT, Farrand SK (1992) Opine catabolism and conjugal transfer of the nopaline Ti plasmid pTiC58 are coordinately regulated by a single repressor. Proc Natl Acad Sci USA 89:643–647
Bednárová M, Stanĕk M, Vancura V et al (1979) Microorganisms in the rhizosphere of wheat colonized by the fungus Gaeumannomyces graminis var. tritici. Folia Microbiol (Praha) 24:253–261
Bélanger C, Canfield ML, Moore LW et al (1995) Genetic analysis of nonpathogenic Agrobacterium tumefaciens mutants arising in crown gall tumors. J Bacteriol 177:3752–3757
Bhattacharya A, Sood P, Citovsky V (2010) The roles of plant phenolics in defence and communication during Agrobacterium and Rhizobium infection. Mol Plant Pathol 11:705–719
Brencic A, Eberhard A, Winans SC (2004) Signal quenching, detoxification and mineralization of vir gene-inducing phenolics by the VirH2 protein of Agrobacterium tumefaciens. Mol Microbiol 51:1103–1115
Bolton GW, Nester EW, Gordon MP (1986) Plant phenolic compounds induce expression of the Agrobacterium tumefaciens loci needed for virulence. Science 232:983–985
Boncompagni E, Osteras M, Poggi MC, le Rudulier D (1999) Occurrence of choline and glycine betaine uptake and metabolism in the family rhizobiaceae and their roles in osmoprotection. Appl Environ Microbiol 65:2072–2077
Bouzar H, Chilton WS, Nesme X, Dessaux Y, Vaudequin V, Petit A, Jones JB, Hodge NC (1995) A new Agrobacterium strain isolated from aerial tumors on Ficus benjamina L. Appl Environ Microbiol 61:65–73
Bouzar H, Moore LW (1987) Isolation of different Agrobacterium biovars from a natural oak savanna and tallgrass prairie. Appl Environ Microbiol 53:717–721
Bouzar H, Ouadah D, Krimi K et al (1993) Correlative association between resident plasmids and the host chromosome in a diverse Agrobacterium soil population. Appl Environ Microbiol 59:1310–1317
Bulgarelli D, Rott M, Schlaeppi K et al (2012) Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 48:91–95
Burr TJ, Katz BH, Bishop AL (1987) Populations of Agrobacterium in vineyard and nonvineyard soils and grape roots in vineyards and nurseries. Plant Dis 71:617–620
Cardarelli M, Mariotti D, Pomponi M et al (1987) Agrobacterium rhizogenes T-DNA genes capable of inducing hairy root phenotype. Mol Gen Genet 209:475–480
Caretto S, Linsalata V, Colella G et al (2015) Carbon fluxes between primary metabolism and phenolic pathway in plant tissues under stress. Int J Mol Sci 16:26378–26394. https://doi.org/10.3390/ijms161125967
Carlier A, Chevrot R, Dessaux Y et al (2004) The assimilation of gamma-butyrolactone in Agrobacterium tumefaciens C58 interferes with the accumulation of the N-acyl-homoserine lactone signal. Mol Plant Microbe Interact 17:951–957
Chen K, Otten L (2017) Natural Agrobacterium transformants: recent results and some theoretical considerations. Front Plant Sci 8:1600. https://doi.org/10.3389/fpls.2017.01600
Chen L, Chen Y, Wood DW et al (2002) A new type IV secretion system promotes conjugal transfer in Agrobacterium tumefaciens. J Bacteriol 184:4838–4845
Chevrot R, Rosen R, Haudecoeur E et al (2006) GABA controls the level of quorum-sensing signal in Agrobacterium tumefaciens. Proc Natl Acad Sci USA 103:7460–7464
Chilton MD, Tepfer DA, Petit A et al (1982) Agrobacterium rhizogenes inserts T-DNA into the genomes of the host plant root cells. Nature 295:432–434
Chilton WS, Chilton MD (1984) Mannityl opine analogs allow isolation of catabolic pathway regulatory mutants. J Bacteriol 158:650–658
Chilton WS, Petit A, Chilton MD, Dessaux Y (2001) Structure and characterization of the crown gall opines heliopine, vitopine and ridéopine. Phytochemistry 58:137–142
Chilton WS, Stomp AM, Beringue V et al (1995) The chrysopine family of amadori-type crown gall opines. Phytochemistry 40:619–628
Christie PJ (2016) The mosaic Type IV secretion systems. EcoSal Plus 7(1). https://doi.org/10.1128/ecosalplus.esp-0020-2015
Close TJ, Tait RC, Kado CI (1985) Regulation of Ti plasmid virulence genes by a chromosomal locus of Agrobacterium tumefaciens. J Bacteriol 164:774–781
Costantino P, Hooykaas PJJ, Dendulk-Ras H et al (1980) Tumor formation and rhizogenicity of Agrobacterium rhizogenes carrying Ti plasmids. Gene 11:79–87
Cushnie TTP, Lamb AJ (2005) Antimicrobial activity of flavonoids. Int J Antimicrob Agents 26:342–356
Davioud E, Petit A, Tate ME et al (1988) Cucumopine—a new T-DNA-encoded opine in hairy root and crown gall. Phytochemistry 27:2429–2433
Deeken R, Engelmann JC, Efetova M et al (2006) An integrated view of gene expression and solute profiles of Arabidopsis tumors: a genome-wide approach. Plant Cell 18:3617–3634
De Greve H, Dhaese P, Seurinck J et al (1982) Nucleotide sequence and transcript map of the Agrobacterium tumefaciens Ti plasmid-encoded octopine synthase gene. J Mol Appl Genet 1:499–511
Dehio C (2005) Bartonella-host-cell interactions and vascular tumour formation. Nat Rev Microbiol 3:621–631
De Lajudie PM, Young JPW (2017) International Committee on Systematics of Prokaryotes Subcommittee for the Taxonomy of Rhizobium and Agrobacterium—minutes of the meeting, Budapest, 25 August 2016. Int J Syst Evol Microbiol. https://doi.org/10.1099/ijsem.0.002144
De Paolis A, Mauro ML, Pomponi M et al (1985) Localization of agropine-synthesizing functions in the TR region of the root-inducing plasmid of Agrobacterium rhizogenes 1855. Plasmid 13:1–7
Dessaux Y, Grandclément C, Faure D (2016) Engineering the rhizosphere. Trends Plant Sci 21:266–278
Dessaux Y, Guyon P, Farrand SK et al (1986a) Agrobacterium Ti and Ri plasmids specify enzymic lactonization of mannopine to agropine. J Gen Microbiol 132:2549–2559
Dessaux Y, Guyon P, Petit A et al (1988) Opine utilization by Agrobacterium spp.: octopine-type Ti plasmids encode two pathways for mannopinic acid degradation. J Bacteriol 170:2939–2946
Dessaux Y, Petit A, Ellis JG et al (1989) Ti plasmid-controlled chromosome transfer in Agrobacterium tumefaciens. J Bacteriol 171:6363–6366
Dessaux Y, Petit A, Farrand SK et al (1998) Opine and opine-like molecules involved in plant-Rhizobiaceaea interactions. In: Spaink HP, Kondorosi A, Hooykaas P (eds) The Rhizobiaceae. Kluwer Academic Publishers, Dordrecht, pp 173–197
Dessaux Y, Petit A, Tempé J et al (1986b) Arginine catabolism in Agrobacterium strains: role of the Ti plasmid. J Bacteriol 166:44–50
Dessaux Y, Tempé J, Farrand SK (1987) Genetic analysis of mannityl opine catabolism in octopine-type Agrobacterium tumefaciens strain 15955. Mol Gen Genet 208:301–308
Dion P (1986) Utilization of octopine by marine bacteria isolated from mollusks. Can J Microbiol 32:959–963
Douglas CJ, Staneloni RJ, Rubin RA et al (1985) Identification and genetic analysis of an Agrobacterium tumefaciens chromosomal virulence region. J Bacteriol 161:850–860
Durand-Tardif M, Broglie R, Slightom J (1985) Structure and expression of Ri T-DNA from Agrobacterium rhizogenes in Nicotiana tabacum. Organ and phenotypic specificity. J Mol Biol 186:557–564
Dunoyer P, Himber C, Voinnet O (2006) Induction, suppression and requirement of RNA silencing pathways in virulent Agrobacterium tumefaciens infections. Nat Genet 38:258–263
Ellis JG, Kerr A, Tempé J et al (1979) Arginine catabolism: a new function of both octopine and nopaline Ti-plasmids of Agrobacterium. Mol Gen Genet 173:263–269
Ellis JG, Kerr A, Petit A et al (1982) Conjugal transfer of nopaline and agropine Ti-plasmids—the role of agrocinopines. Mol Gen Genet 186:269–274
Ellis JG, Murphy PJ (1981) Four new opines from crown gall tumours—their detection and properties. Mol Gen Genet 181:36–43
Ellis JG, Ryder MH, Tate ME (1984) Agrobacterium tumefaciens TR-DNA encodes a pathway for agropine biosynthesis. Mol Gen Genet 195:466–473
El Sahili A, Li SZ, Lang J et al (2015) A pyranose-2-phosphate motif is responsible for both antibiotic import and quorum-sensing regulation in Agrobacterium tumefaciens. PLoS Pathog 11(8):e1005071. https://doi.org/10.1371/journal.ppat.1005071
Engström P, Zambryski P, Van Montagu M et al (1987) Characterization of Agrobacterium tumefaciens virulence proteins induced by the plant factor acetosyringone. J Mol Biol 197:635–645
Farrand SK, Hwang I, Cook DM (1996) The tra region of the nopaline-type Ti plasmid is a chimera with elements related to the transfer systems of RSF1010, RP4, and F. J Bacteriol 178:4233–4247
Flores-Mireles AL, Eberhard A, Winans SC (2012) Agrobacterium tumefaciens can obtain sulphur from an opine that is synthesized by octopine synthase using S-methylmethionine as a substrate. Mol Microbiol 84:845–856
Fortin C, Nester EW, Dion P (1992) Growth inhibition and loss of virulence in cultures of Agrobacterium tumefaciens treated with acetosyringone. J Bacteriol 174:5676–5685
Fortin C, Marquis C, Nester EW et al (1993) Dynamic structure of Agrobacterium tumefaciens Ti plasmids. J Bacteriol 175:4790–4799
Garg N, Manchanda G, Kumar A (2014) Bacterial quorum sensing: circuits and applications. Antonie Van Leeuwenhoek 105:289–305
Gelvin SB (2012) Traversing the Cell: Agrobacterium T-DNA’s journey to the host genome. Front Plant Sci 3:52. https://doi.org/10.3389/fpls.2012.00052
Gelvin SB (2017) Integration of Agrobacterium T-DNA into the plant genome. Annu Rev Genet. https://doi.org/10.1146/annurev-genet-120215-035320
Genetello C, Van Larebeke N, Holsters M et al (1977) Ti plasmids of Agrobacterium as conjugative plasmids. Nature 265:561–563
Gohlke J, Deeken R (2014) Plant responses to Agrobacterium tumefaciens and crown gall development. Front Plant Sci 5:155. https://doi.org/10.3389/fpls.2014.00155
Gomes NCM, Heuer H, Schönfeld J et al (2001) Bacterial diversity of the rhizosphere of maize (Zea mays) grown in tropical soil studied by temperature gradient gel electrophoresis. Plant Soil 232:167–180
Gordon DM, Ryder MH, Heinrich K et al (1996) An experimental test of the rhizopine concept in Rhizobium meliloti. Appl Environ Microbiol 62:3991–3996
Grandclément C, Tannières M, Moréra S et al (2016) Quorum quenching: Role in nature and applied developments. FEMS Microbiol Rev 40:86–116
Grieshaber M, Gäde G (1976) The biological role of octopine in the squid, Loligo vulgaris (Lamarck). J Comp Physiol 108:225–232
Guyon P, Chilton MD, Petit A et al (1980) Agropine in “null-type” crown gall tumors: Evidence for generality of the opine concept. Proc Natl Acad Sci USA 77:2693–2697
Guyon P, Petit A, Tempé J et al (1993) Transformed plants producing opines specifically promote growth of opine-degrading agrobacteria. Mol Plant-Microbe Interact 6:92–98
Haudecoeur E, Tannières M, Cirou A et al (2009) Different regulation and roles of lactonases AiiB and AttM in Agrobacterium tumefaciens C58. Mol Plant Microbe Interact 22:529–537
Hawes MC, Smith LY (1989) Requirement for chemotaxis in pathogenicity of Agrobacterium tumefaciens on roots of soil grown pea plants. J Bacteriol 171:5668–5671
Hawes MC, Pueppke SG (1987) Correlation between binding of Agrobacterium tumefaciens by root cap cells and susceptibility of plants to crown gall. Plant Cell Rep 6:287–290
Hood EE, Chilton WS, Chilton MD et al (1986) T-DNA and opine synthetic loci in tumors incited by Agrobacterium tumefaciens A281 on soybean and alfalfa plants. J Bacteriol 168:1283–1290
He F, Nair GR, Soto CS et al (2009) Molecular basis of ChvE function in sugar binding, sugar utilization, and virulence in Agrobacterium tumefaciens. J Bacteriol 191:5802–5813
Heinrich K, Gordon DM, Ryder MH et al (1999) A rhizopine strain of Sinorhizobium meliloti remains at a competitive nodulation advantage after an extended period in the soil. Soil Biol Biochem 31:1063–1065
Hinsinger P, Bengough AG, VetterleinIain D et al (2009) Rhizosphere: biophysics, biogeochemistry and ecological relevance. Plant Soil 321:117–152
Holsters M, Silva B, Van Vliet F et al (1978) In vivo transfer of the Ti-plasmid of Agrobacterium tumefaciens to Escherichia coli. Mol Gen Genet 163:335–358
Hong SB, Hwang I, Dessaux Y (1997) A T-DNA gene required for agropine biosynthesis by transformed plants is functionally and evolutionarily related to a Ti plasmid gene required for catabolism of agropine by Agrobacterium strains. J Bacteriol 179:4831–4840
Hooykaas PJJ, Klapwijk PM, Nuti MP et al (1977) Transfer of the Agrobacterium tumefaciens Ti-plasmid to avirulent agrobacteria and to Rhizobium ex planta. J Gen Microbiol 98:477–484
Hwang I, Li PL, Zhang L, Piper KR et al (1994) TraI, a LuxI homologue, is responsible for production of conjugation factor, the Ti plasmid N-acylhomoserine lactone autoinducer. Proc Natl Acad Sci USA 91:4639–4643
Hynes MF, Simon R, Pühler A (1985) The development of plasmid-free strains of Agrobacterium tumefaciens by using incompatibility with a Rhizobium meliloti plasmid to eliminate pAtC58. Plasmid 13:99–105
Iacobellis NS, Devay JE (1986) Long-term storage of plant-pathogenic bacteria in sterile distilled water. Appl Environ Microbiol 52:388–389
Idnurm A, Bailey AM, Cairns TC et al (2017) A silver bullet in a golden age of functional genomics: the impact of Agrobacterium-mediated transformation of fungi. Fungal Biol Biotechnol 4:6. https://doi.org/10.1186/s40694-017-0035-0
Inceoglu Ö, Al-Soud WA, Salles JF et al (2011) Comparative analysis of bacterial communities in a potato field as determined by pyrosequencing. PLoS One 63(2):460–470
Jones DL, Nguyen C, Finlay RD (2009) Carbon flow in the rhizosphere: carbon trading at the soil–root interface. Plant Soil 321:5–33
Joos H, Inzé D, Caplan A (1983) Genetic analysis of T-DNA transcripts in nopaline crown galls. Cell 32:1057–1067
Jouanin L (1984) Restriction map of an agropine-type Ri plasmid and its homologies with Ti plasmids. Plasmid 12:91–102
Kanemoto RH, Powell AT, Akiyoshi DE et al (1989) Nucleotide sequence and analysis of the plant-inducible locus pinF from Agrobacterium tumefaciens. J Bacteriol 171:2506–2512
Kefeli VI, Kalevitch MV, Borsari B (2003) Phenolic cycle in plants and environment. J Cell Mol Biol 2:13–18
Kerr A, Manigault P, Tempé J (1977) Transfer of virulence in vivo and in vitro in Agrobacterium. Nature 265:560–561
Khan SR, Gaines J, Roop RM et al (2008) Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl Environ Microbiol 74:5053–5062
Khanaka H, Catteau M, Tailliez R (1981) Antibiotic sensitivity in Rhizobium and Agrobacterium. Zentralbl Bakt Mikrobiol Hygiene I Abt Orig C2:282–288
Kim H, Farrand SK (1997) Characterization of the acc operon from the nopaline-type Ti plasmid pTiC58, which encodes utilization of agrocinopines A and B and susceptibility to agrocin 84. J Bacteriol 179:7559–7572
Kim H, Farrand SK (1988) Opine catabolic loci from Agrobacterium plasmids confer chemotaxis to their cognate substrates. Mol Plant Microbe Interact 11:131–143
Kim KS, Chilton WS, Farrand SK (1996) A Ti plasmid-encoded enzyme required for degradation of mannopine is functionally homologous to the T-region-encoded enzyme required for synthesis of this opine in crown gall tumors. J Bacteriol 178:3285–3292
Kim KS, Farrand SK (1996) Ti plasmid-encoded genes responsible for catabolism of the crown gall opine mannopine by Agrobacterium tumefaciens are homologs of the T-region genes responsible for synthesis of this opine by the plant tumor. J Bacteriol 178:3275–3284
Klapwijk PM, Oudshoorn M, Schilperoort RA (1977) Inducible permease involved in the uptake of octopine, lysopine and octopinic acid by Agrobacterium tumefaciens strains carrying virulence-associated plasmids. J Gen Microbiol 102:1–11
Klapwijk PM, Scheulderman T, Schilperoort RA (1978) Coordinated regulation of octopine degradation and conjugative transfer of Ti plasmids in Agrobacterium tumefaciens: evidence for a common regulatory gene and separate operons. J Bacteriol 136:775–785
Klein DT, Klein RM (1953) Transmittance of tumor-inducing ability to avirulent crown-gall and related bacteria. J Bacteriol 66:220–228
Koncz C, De Greve H, André D et al (1983) The opine synthase genes carried by Ti plasmids contain all signals necessary for expression in plants. EMBO J 2:1597–1603
Krimi Z, Petit A, Mougel C et al (2002) Seasonal fluctuations and long-term persistence of pathogenic populations of Agrobacterium spp. in soils. Appl Environ Microbiol 68:3358–3365
Kuzmanović N, Puławska J, Prokić A et al (2015) Agrobacterium arsenijevicii sp. nov., isolated from crown gall tumors on raspberry and cherry plum. Syst Appl Microbiol 38:373–378
Kyndt T, Quispe D, Zhai H et al (2015) The genome of cultivated sweet potato contains Agrobacterium T-DNAs with expressed genes: an example of a naturally transgenic food crop. Proc Natl Acad Sci USA 112:5844–5849
Lacroix B, Citovsky V (2013) The roles of bacterial and host plant factors in Agrobacterium-mediated genetic transformation. Int J Dev Biol 57:467–481
Lacroix B, Citovsky V (2016) Transfer of DNA from bacteria to eukaryotes. MBio 7(4). pii: e00863-16
Lang J, Gonzalez-Mula A, Taconnat L et al (2016) The plant GABA signaling downregulates horizontal transfer of the Agrobacterium tumefaciens virulence plasmid. New Phytol 210:974–983
Lang J, Planamente S, Mondy S et al (2013) Concerted transfer of the virulence Ti plasmid and companion At plasmid in the Agrobacterium tumefaciens-induced plant tumour. Mol Microbiol 90:1178–1189
Lang J, Vigouroux A, Planamente S et al (2014) Agrobacterium uses a unique ligand-binding mode for trapping opines and acquiring a competitive advantage in the niche construction on plant host. PLoS Pathog 10(10):e1004444
Lassalle F, Muller D, Nesme X (2015) Ecological speciation in bacteria: reverse ecology approaches reveal the adaptive part of bacterial cladogenesis. Res Microbiol 166:729–741
Lattanzio V, Kroon PA, Quideau S et al (2008) Plant phenolics—secondary metabolites with diverse functions. In: Daayf F, Lattanzio V (eds) Recent advances in polyphenol research. Wiley, Oxford, pp 1–35
Li PL, Hwang I, Miyagi H et al (1999) Essential components of the Ti plasmid trb system, a type IV macromolecular transporter. J Bacteriol 181:5033–5041
Li PL, Farrand SK (2000) The replicator of the nopaline-type Ti plasmid pTiC58 is a member of the repABC family and is influenced by the TraR-dependent quorum-sensing regulatory system. J Bacteriol 182:179–188
Llop P, Murillo J, Lastra B et al (2009) Recovery of nonpathogenic mutant bacteria from tumors caused by several Agrobacterium tumefaciens strains: a frequent event? Appl Environ Microbiol 75:6504–6514
Lundberg DS, Lebeis SL, Paredes SH et al (2012) Defining the core Arabidopsis thaliana root microbiome. Nature 488:86–90
Ma LS, Hachani A, Lin JS et al (2014) Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe 16:94–104
Mansouri H, Petit A, Oger P et al (2002) Engineered rhizosphere: the trophic bias generated by opine-producing plants is independent of the opine type, the soil origin, and the plant species. Appl Environ Microbiol 68:2562–2566
Marasco R, Lago CT, De Felice M (1995) Utilization of cellobiose and other beta-d-glucosides in Agrobacterium tumefaciens. Res Microbiol 146:485–492
Marty L, Vigouroux A, Aumont-Nicaise M et al (2016) Structural basis for high specificity of Amadori compound and mannopine opine binding in bacterial pathogens. J Biol Chem 291:22638–22649
Matveeva TV, Bogomaz DI, Pavlova OA et al (2012) Horizontal gene transfer from genus Agrobacterium to the plant Linaria in nature. Mol Plant Microbe Interact 25:1542–1551
Merlo DJ, Nester EW (1977) Plasmids in avirulent strains of Agrobacterium. J Bacteriol 129:76–80
Mhedbi-Hajri N, Yahiaoui N, Mondy S et al (2016) Transcriptome analysis revealed that a quorum sensing system regulates the transfer of the pAt megaplasmid in Agrobacterium tumefaciens. BMC Genomics 17:661
Mondy S, Lenglet A, Beury-Cirou A et al (2014) An increasing opine carbon bias in artificial exudation systems and genetically modified plant rhizospheres leads to an increasing reshaping of bacterial populations. Mol Ecol 23:4846–4861
Montoya AL, Chilton MD, Gordon MP et al (1977) Octopine and nopaline metabolism in Agrobacterium tumefaciens and crown gall tumor cells: role of plasmid genes. J Bacteriol 129:101–107
Moore LW, Chilton WS, Canfield ML (1997) Diversity of opines and opine-catabolizing bacteria isolated from naturally occurring crown gall tumors. Appl Environ Microbiol 63:201–207
Moore L, Warren G, Strobel G (1979) Involvement of a plasmid in the hairy root disease of plants caused by Agrobacterium rhizogenes. Plasmid 2:617–626
Morton ER, Platt TG, Fuqua C et al (2014) Non-additive costs and interactions alter the competitive dynamics of co-occurring ecologically distinct plasmids. Proc Biol Sci 281(1779):20132173
Mousavi SA, Österman J, Wahlberg N et al (2014) Phylogeny of the Rhizobium-Allorhizobium-Agrobacterium clade supports the delineation of Neorhizobium gen. nov. Syst Appl Microbiol 37:208–215
Murphy PJ, Heycke N, Banfalvi Z et al (1987) Genes for the catabolism and synthesis of an opine-like compound in Rhizobium meliloti are closely linked and on the Sym plasmid. Proc Natl Acad Sci USA 84:493–497
Nair GR, Lai X, Wise AA et al (2011) The integrity of the periplasmic domain of the VirA sensor kinase is critical for optimal coordination of the virulence signal response in Agrobacterium tumefaciens. J Bacteriol 193:1436–1448
Nautiyal CS, Dion P (1990) Characterization of the opine-utilizing microflora associated with samples of soil and plants. Appl Environ Microbiol 56:2576–2579
Nautiyal CS, Dion P, Chilton WS (1991) Mannopine and mannopinic acid as substrates for Arthrobacter sp. strain MBA209 and Pseudomonas putida NA513. J Bacteriol 173:2833–2841
Nester EW (2015) Agrobacterium: nature’s genetic engineer. Front Plant Sci 5:730
Nobile S, Deshusses J (1986) Transport of gamma-butyrobetaine in an Agrobacterium species isolated from soil. J Bacteriol 168:780–784
Nüsslein K, Tiedje JM (1998) Characterization of the dominant and rare members of a young Hawaiian soil bacterial community with small-subunit ribosomal DNA amplified from DNA fractionated on the basis of its guanine and cytosine composition. Appl Environ Microbiol 64:1283–1289
Ooms G, Hooykaas PJ, Moolenaar G et al (1981) Grown gall plant tumors of abnormal morphology, induced by Agrobacterium tumefaciens carrying mutated octopine Ti plasmids; analysis of T-DNA functions. Gene 14:33–50
Ong SA, Peterson T, Neilands JB (1979) Agrobactin, a siderophore from Agrobacterium tumefaciens. J Biol Chem 254:1860–1865
Oger P, Petit A, Dessaux Y (1997) Genetically engineered plants producing opines alter their biological environment. Nat Biotechnol 15:369–372
Otten L, De Ruffray P (1994) Agrobacterium vitis nopaline Ti plasmid pTiAB4: relationship to other Ti plasmids and T-DNA structure. Mol Gen Genet 245:493–505
Otten L, Gérard JC, De Ruffray P (1993) The Ti plasmid from the wide host range Agrobacterium vitis strain Tm4: map and homology with other Ti plasmids. Plasmid 29:154–159
Parke D, Ornston LN, Nester EW (1987) Chemotaxis to plant phenolic inducers of virulence genes is constitutively expressed in the absence of the Ti plasmid in Agrobacterium tumefaciens. J Bacteriol 169:5336–5338
Palanichelvam K, Oger P, Clough SJ et al (2000) A second T-region of the soybean-supervirulent chrysopine-type Ti plasmid pTiChry5, and construction of a fully disarmed vir helper plasmid. Mol Plant Microbe Interact 13:1081–1091
Palumbo JD, Kado CI, Phillips DA (1998) An isoflavonoid-inducible efflux pump in Agrobacterium tumefaciens is involved in competitive colonization of roots. J Bacteriol 180:3107–3113
Papenfort K, Bassler BL (2016) Quorum sensing signal-response systems in Gram-negative bacteria. Nat Rev Microbiol 14:576–588
Penyalver R, Oger P, López MM et al (2001) Iron-binding compounds from Agrobacterium spp.: biological control strain Agrobacterium rhizogenes K84 produces a hydroxamate siderophore. Appl Environ Microbiol 67:654–664
Petit A, David C, Dahl G (1983) Further extension of the opine concept: plasmids in Agrobacterium rhizogenes cooperate for opine degradation. Mol Gen Genet 190:204–214
Petit A, Tempé J (1985) The function of T-DNA in nature. In: Van Vloten-Doting L, Groot GSP, Hall TC (eds) Molecular form and function of the plant genome. Plenum Press, New York/London, pp 625–636
Petit A, Tempé J, Kerr A et al (1978) Substrate induction of conjugative activity of Agrobacterium tumefaciens Ti plasmids. Nature 271:570–572
Pionnat S, Keller H, Héricher D et al (1999) Ti plasmids from Agrobacterium characterize rootstock clones that initiated a spread of crown gall disease in Mediterranean countries. Appl Environ Microbiol 65:4197–4206
Piper KR, Beck von Bodman S, Farrand SK (1993) Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature 362:448–450
Piper KR, Beck Von Bodman S, Hwang I (1999) Hierarchical gene regulatory systems arising from fortuitous gene associations: controlling quorum sensing by the opine regulon in Agrobacterium. Mol Microbiol 32:1077–1089
Pitzschke A, Hirt H (2010) New insights into an old story: Agrobacterium-induced tumour formation in plants by plant transformation. EMBO J 29:1021–1032
Platt TG, Bever JD, Fuqua C (2012) A cooperative virulence plasmid imposes a high fitness cost under conditions that induce pathogenesis. Proc Biol Sci 279:1691–1699
Powell GK, Morris RO (1986) Nucleotide sequence and expression of a Pseudomonas savastanoi cytokinin biosynthetic gene: homology with Agrobacterium tumefaciens tmr and tzs loci. Nucleic Acids Res 14:2555–2565
Qin Y, Su S, Farrand SK (2007) Molecular basis of transcriptional antiactivation. TraM disrupts the TraR-DNA complex through stepwise interactions. J Biol Chem 282:19979–19991
Ream LW, Gordon MP, Nester EW (1983) Multiple mutations in the T region of the Agrobacterium tumefaciens tumor-inducing plasmid. Proc Natl Acad Sci USA 80:1660–1664
Riley PS, Weaver RE (1977) Comparison of thirty-seven strains of Vd-3 bacteria with Agrobacterium radiobacter: morphological and physiological observations. J Clin Microbiol 5:172–177
Rondon MR, Ballering KS, Thomas MG (2004) Identification and analysis of a siderophore biosynthetic gene cluster from Agrobacterium tumefaciens C58. Microbiology 150:3857–3866
Rong LJ, Karcher SJ, Gelvin SB (1991) Genetic and molecular analyses of picA, a plant-inducible locus on the Agrobacterium tumefaciens chromosome. J Bacteriol 173:5110–5120
Rosenberg S, Huguet T (1984) The pAtC58 plasmid of Agrobacterium tumefaciens is not essential for tumor induction. Mol Gen Genet 196:533–536
Rossbach S, Rasul G, Schneider M et al (1995) Structural and functional conservation of the rhizopine catabolism (moc) locus is limited to selected Rhizobium meliloti strains and unrelated to their geographical origin. Mol Plant Microbe Interact 8:549–559
Rüger HJ, Höfle MG (1992) Marine star-shaped-aggregate-forming bacteria: Agrobacterium atlanticum sp. nov.; Agrobacterium meteori sp. nov.; Agrobacterium ferrugineum sp. nov., nom. rev.; Agrobacterium gelatinovorum sp. nov., nom. rev.; and Agrobacterium stellulatum sp. nov., nom. rev. Int J Syst Bacteriol 42:133–143
Ryu CM (2015) Against friend and foe: type 6 effectors in plant-associated bacteria. J Microbiol 53:201–208
Saint CP, Wexler M, Murphy PJ et al (1993) Characterization of genes for synthesis and catabolism of a new rhizopine induced in nodules by Rhizobium meliloti Rm220-3: extension of the rhizopine concept. J Bacteriol 175:5205–5215
Sánchez-Cañizares C, Jorrín B et al (2017) Understanding the holobiont: the interdependence of plants and their microbiome. Curr Opin Microbiol 38:188–196
Savka MA, Black RC, Binns AN et al (1996) Translocation and exudation of tumor metabolites in crown galled plants. Mol Plant Microbe Interact 9:310–313
Savka MA, Dessaux Y, McSpadden-Gardener BB et al (2013) The “biased rhizosphere” concept and advances in the omics era to study bacterial competitiveness and persistence in the phytosphere. In: de Bruijn FJ (ed) Molecular microbial ecology of the rhizosphere, vol 2. Wiley, Hoboken, pp 1147–1161
Savka MA, Dessaux Y, Oger P et al (2002) Engineering bacterial competitiveness and persistence in the phytosphere. Mol Plant Microbe Interact 15:866–874
Savka MA, Farrand SK (1992) Mannityl opine accumulation and exudation by transgenic tobacco. Plant Physiol 98:784–789
Savka MA, Farrand SK (1997) Modification of rhizobacterial populations by engineering bacterium utilization of a novel plant-produced resource. Nat Biotechnol 15:363–368
Schell J, Van Montagu M, De Beuckeleer M et al (1979) Interactions and DNA transfer between Agrobacterium tumefaciens, the Ti-plasmid and the plant host. Proc R Soc Lond B Biol Sci 204:251–266
Schröder G, Schuelein R, Quebatte M, Dehio C (2011) Conjugative DNA transfer into human cells by the VirB/VirD4 type IV secretion system of the bacterial pathogen Bartonella henselae. Proc Natl Acad Sci USA 108:14643–14648
Schroth MN, Weinhold AR, McCain AH (1971) Biology and control of Agrobacterium tumefaciens. Hilgardia 40:537–552
Scott DB, Wilson R, Shaw GJ (1987) Biosynthesis and degradation of nodule-specific Rhizobium loti compounds in Lotus nodules. J Bacteriol 169:278–282
Shaw CH (1991) Swimming against the tide: chemotaxis in Agrobacterium. Bioessays 13:25–29
Shaw GJ, Wilson RD, Lane GA (1986) Structure of rhizolotine, a novel opine-like metabolite from Lotus tenuis nodules. J Chem Soc Chem Commun 2:180–181
Slightom JL, Durand-Tardif M (1986) Nucleotide sequence analysis of TL-DNA of Agrobacterium rhizogenes agropine type plasmid. Identification of open reading frames. J Biol Chem 261:108–121
Smith LT, Smith GM, Madkour MA (1990) Osmoregulation in Agrobacterium tumefaciens: accumulation of a novel disaccharide is controlled by osmotic strength and glycine betaine. J Bacteriol 172:6849–6855
Soedarjo M, Hemscheidt TK, Borthakur D (1994) Mimosine, a toxin present in leguminous trees (Leucaena spp.), induces a mimosine-degrading enzyme activity in some Rhizobium strains. Appl Environ Microbiol 60:4268–4272
Soedarjo M, Borthakur D (1998) Mimosine, a toxin produced by the tree-legume Leucaena provides a nodulation competition advantage to mimosine-degrading Rhizobium strains. Soil Biol Biochem 30:1605–1613
Spena A, Schmülling T, Koncz C et al (1987) Independent and synergistic activity of rolA, B and C loci in stimulating abnormal growth in plants. EMBO J 6:3891–3899
Subramoni S, Nathoo N, Klimov E (2014) Agrobacterium tumefaciens responses to plant-derived signaling molecules. Front Plant Sci 5:322
Sukanya NK, Vaidyanathan CS (1964) Aminotransferases of Agrobacterium tumefaciens: transamination between tryptophan and phenylpyruvate. Biochem J 92:594–598
Süle S (1978) Biological control of crown gall by a peat cultured antagonist. Phytopath Z 91:273–275
Suzuki K, Yamashita I, Tanaka N (2002) Tobacco plants were transformed by Agrobacterium rhizogenes infection during their evolution. Plant J 32:775–787
Tannières M, Lang J, Barnier C et al (2017) Quorum-quenching limits quorum-sensing exploitation by signal-negative invaders. Sci Rep 7:40126
Teixeira LC, Peixoto RS, Cury JC et al (2010) Bacterial diversity in rhizosphere soil from Antarctic vascular plants of Admiralty Bay, maritime Antarctica. ISME J 4(2010):989–1001
Tempé J, Petit A (1983) La piste des opines. In: Pühler A (ed) Molecular genetics of the bacteria-plant interaction. Springer, Berlin-Heidelberg, pp 14–32
Tennigkeit J, Matzura H (1991) Nucleotide sequence analysis of a chloramphenicol-resistance determinant from Agrobacterium tumefaciens and identification of its gene product. Gene 8:113–116
Tepfer D, Goldmann A, Pamboukdjian N et al (1988) A plasmid of Rhizobium meliloti 41 encodes catabolism of two compounds from root exudate of Calystegium sepium. J Bacteriol 170:1153–1161
Tepfer DA, Tempé J (1981) Production d’agropine par des racines formées sous l’action d’Agrobacterium rhizogenes souche A4. C R Hebd Séances Acad Sci Ser III 292:153–156
Teyssier-Cuvelle S, Mougel C, Nesme X (1999) Direct conjugal transfers of Ti plasmid to soil microflora. Mol Ecol 8:1273–1284
Thoai NV, Robin Y (1959) Métabolisme des dérivés guanidylés: VIII. Biosynthèse de l’octopine et répartition de l’enzyme l’opérant chez les invertebrés. Biochimica Biophysica Acta 35:446–453
Thomashow MF, Karlinsey JE, Marks JR et al (1987) Identification of a new virulence locus in Agrobacterium tumefaciens that affects polysaccharide composition and plant cell attachment. J Bacteriol 169:3209–3216
Tremblay G, Gagliardo R, Chilton WS et al (1987) Diversity among opine-utilizing bacteria: identification of coryneform isolates. Appl Environ Microbiol 53:1519–1524
Quispe-Huamanquispe DG, Gheysen G, Kreuze JF (2017) Horizontal gene transfer contributes to plant evolution: the case of Agrobacterium T-DNAs. Front Plant Sci 8:2015
Vain P (2007) Thirty years of plant transformation technology development. Plant Biotechnol J 5:221–229
Van Larebeke N, Engler G, Holsters M et al (1974) Large plasmid in Agrobacterium tumefaciens essential for crown gall-inducing ability. Nature 252:169–170
van Veen RJM, den Dulk-Ras H, Schilperoort RA et al (1989) Ti plasmid containing Rhizobium meliloti are non-tumorigenic on plants, despite proper virulence gene induction and T-strand formation. Arch Microbiol 153:85–89
Vaudequin-Dransart V, Petit A, Poncet C et al (1995) Novel Ti plasmids in Agrobacterium strains isolated from fig tree and chrysanthemum tumors and their opinelike molecules. Mol Plant Microbe Interact 8:311–321
Vaudequin-Dransart V, Petit A, Chilton WS et al (1998) The cryptic plasmid of Agrobacterium tumefaciens cointegrates with the Ti plasmid and cooperates for opine degradation. Mol Plant Microbe Interact 11:583–591
Vigouroux A, El Sahili A, Lang J et al (2017) Structural basis for high specificity of octopine binding to exploit the constructed octopine-niche on plant host. Sci Rep 7:18033
Watson B, Currier TC, Gordon MP, Chilton MD, Nester EW (1975) Plasmid required for virulence of Agrobacterium tumefaciens. J Bacteriol 123:255–264
Wendt T, Doohan F, Mullins E (2012) Production of Phytophthora infestans-resistant potato (Solanum tuberosum) utilising Ensifer adhaerens OV14. Transgenic Res 21:567–578
Wexler M, Gordon D, Murphy PJ (1995) The distribution of inositol rhizopine genes in Rhizobium populations. Soil Biol Biochem 27:531–537
White FF, Garfinkel DJ, Huffman GA et al (1983) Sequences homologous to Agrobacterium rhizogenes T-DNA in the genomes of uninfected plants. Nature 301:348–350
White FF, Ghidossi G, Gordon MP et al (1982) Tumor induction by Agrobacterium rhizogenes involves the transfer of plasmid DNA to the plant genome. Proc Natl Acad Sci USA 79:3193–3197
Wichelecki DJ, Vetting MW, Chou L et al (2015) ATP-binding cassette (ABC) transport system solute-binding protein-guided identification of novel d-altritol and galactitol catabolic pathways in Agrobacterium tumefaciens C58. J Biol Chem 290:28963–28976
Willmitzer L, Sanchez-Serramo J, Duschelder E et al (1982) DNA from Agrobacterium rhizogenes is transferred to and expressed in axemic hairy root plant tissue. Mol Gen Genet 186:16–22
Wilson M, Savka MA, Hwang I et al (1995) Altered epiphytic colonization of mannityl opine-producing transgenic tobacco plants by a mannityl opine-catabolizing strain of Pseudomonas syringae. Appl Environ Microbiol 61:2151–2158
Wu HY, Chung PC, Shih HW et al (2008) Secretome analysis uncovers an Hcp-family protein secreted via a type VI secretion system in Agrobacterium tumefaciens. J Bacteriol 190:2841–2850
Yamada T, Palm CJ, Brooks B et al (1985) Nucleotide sequences of the Pseudomonas savastanoi indoleacetic acid genes show homology with Agrobacterium tumefaciens T-DNA. Proc Natl Acad Sci USA 82:6522–6526
Zanker H, Lurz G, Langridge U et al (1994) Octopine and nopaline oxidases from Ti plasmids of Agrobacterium tumefaciens: molecular analysis, relationship, and functional characterization. J Bacteriol 176:4511–4517
Zanker H, von Lintig J, Schröder J (1992) Opine transport genes in the octopine (occ) and nopaline (noc) catabolic regions in Ti plasmids of Agrobacterium tumefaciens. J Bacteriol 174:841–849
Zechner EL, de la Cruz F, Eisenbrandt R et al (2001) Conjugative-DNA transfer processes. In: Thomas CM (ed) The horizontal gene pool: bacterial plasmids and gene spread. Harwood Academic Publishers, Amsterdam, p 87
Zhang L, Murphy PJ, Kerr A et al (1993) Agrobacterium conjugation and gene regulation by N-acyl-l-homoserine lactones. Nature 362:446–448
Zhao J, Binns AN (2014) GxySBA ABC transporter of Agrobacterium tumefaciens and its role in sugar utilization and vir gene expression. J Bacteriol 196:3150–3159
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG, part of Springer Nature
About this chapter
Cite this chapter
Dessaux, Y., Faure, D. (2018). Niche Construction and Exploitation by Agrobacterium: How to Survive and Face Competition in Soil and Plant Habitats. In: Gelvin, S. (eds) Agrobacterium Biology. Current Topics in Microbiology and Immunology, vol 418. Springer, Cham. https://doi.org/10.1007/82_2018_83
Download citation
DOI: https://doi.org/10.1007/82_2018_83
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-03256-2
Online ISBN: 978-3-030-03257-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)