Abstract
There is a complex interplay between the regulation of flagellar motility and the expression of virulence factors in many bacterial pathogens. Here, we review the literature on the direct and indirect roles of flagellar motility in mediating the tripartite interaction between entomopathogenic bacteria (Photorhabdus and Xenorhabdus), their nematode hosts, and their insect targets. First, we describe the swimming and swarming motility of insect pathogenic bacteria and its impact on insect colonization. Then, we describe the coupling between the expression of flagellar and virulence genes and the dynamic of expression of the flagellar regulon during invertebrate infection. We show that the flagellar type 3 secretion system (T3SS) is also an export apparatus for virulence proteins in X. nematophila. Finally, we demonstrate that phenotypic variation, a common property of the bacterial symbionts of nematodes, also alters flagellar motility in Photorhabdus and Xenorhabdus. Finally, the so-called phenotypic heterogeneity phenomenon in the flagellar gene expression network will be also discussed. As the main molecular studies were performed in X. nematophila, future perspectives for the study of the interplay between flagellum and invertebrate interactions in Photorhabdus will be discussed.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Bacterial Motility and Virulence in Pathogenic Bacteria
Many bacteria are motile through the action of large complex protein assemblages called flagella. Flagellum-mediated motility often plays an essential role in colonization by facilitating bacterial motility in mediating different bacterial interactions with plants, vertebrates, and invertebrates. In addition to motility, flagellum per se can participate in adhesion, invasion, or biofilm formation (Josenhans and Suerbaum 2002). Flagellin or depolymerised flagellum is recognized by the innate immune system in organisms as diverse as flies, plants, and mammals. Specific domains of flagellin trigger innate immune response through TLR5 and FLS2 receptors in mammals and plants, respectively (Josenhans and Suerbaum 2002). Flagellar transcriptional regulators can also contribute to virulence by regulating the expression of virulence factors and, as discussed thereafter, the flagellum is able to function as an export apparatus that mediates extracellular secretion of non-flagellar effector proteins.
A complex interplay between flagella and virulence has been demonstrated in entomopathogenic bacteria such as Bacillus thuringiensis (Ghelardi et al. 2002), Xenorhabdus nematophila (Givaudan and Lanois 2000), Photorhabdus luminescens (Easom and Clarke 2008), and P. temperata (Hurst et al. 2015) (see Table 1).
This chapter is focused on describing the interplay between bacterial flagellar-driven motility (see Sect. 2) and the tripartite interaction between bacteria (Photorhabdus and its sister species, Xenorhabdus), the nematode hosts, and the insect targets. The more detailed scientific studies on this topic have been realized in X. nematophila. They depicted the regulation of flagellar gene expression (in vitro and during invertebrate infection, see Sect. 3 and Sect. 5) and the flagellar T3SS (type 3 secretion system) as an export apparatus for virulence proteins (Sect. 4). Phenotypic variation is a common property of the bacterial symbionts of nematodes and here we show that this phenomenon also alters flagellar motility in both Photorhabdus and Xenorhabdus bacteria. In addition, the so-called phenotypic noise phenomenon in the flagellar gene expression network will be discussed (see Sect. 6). Future perspectives for the study of the interplay between flagellum and invertebrate interactions in Photorhabdus will be proposed.
2 Flagella Affect Virulence by Facilitating Motility
Flagella-driven swimming motility is a self-propulsion movement through water-filled channels within the agar (0.2–0.4% agar). Flagella can also allow one particular type of surface motility, the so-called swarming motility. Swarming has been studied extensively in Proteus mirabilis, and swarmer cells are usually hyper-flagellated and move in a cell groups called rafts, and bacteria maintain close contact with other swarmer cells (Allison et al. 1992). It has been demonstrated that P. luminescens, P. temperata, and Xenorhabdus spp. display swimming and swarming (Derzelle et al. 2004a; Givaudan et al. 1995; Michaels and Tisa 2011). The macroscopic swarming pattern of X. nematophila and P. temperata colonies displays concentric zonation cells or ‘‘fried egg’’ appearance when grown on low agar concentrations (about 0.8–1%). Xenorhabdus and Photorhabdus use peritrichous flagella for both swimming and swarming motility. As expected, mutations in genes encoding flagellar structural proteins such as flgE (flagellar hook protein) and FlgK (flagellar hook-associated protein 1) in P. temperata (Hurst et al. 2015; Michaels and Tisa 2011), flgG (component of the distal rod) and motAB (flagellar motor proteins) in P. luminescens (Easom and Clarke 2008) and fliC (flagellin) in X. nematophila (Herbert and Goodrich-Blair 2007) were nonmotile for both swimming and swarming motility.
The assessment of the role of flagellar-driven motility per se could be achieved by the construction of mutants altering merely bacterial locomotion. To study the impact of flagellar-driven motility in the Photorhabdus–nematode–insect tripartite association, Easom and Clarke (2008) constructed deletion mutants of flgG (blocking flagella production) and motAB (blocking flagella rotation) in P. luminescens TT01. Virulence, nematode growth, and development were not impaired in non-motile mutants suggesting that flagellum and motility are not required for pathogenicity or mutualism. However, authors showed that both the ΔflgG and the ΔmotAB mutants of P. luminescens are out-competed by the wild-type strain illustrating that motile bacteria have a slight competitive fitness advantage during colonization of the insect larvae (Easom and Clarke 2008). Moreover, non-motile fliC mutant of X. nematophila (which lacks the flagellin subunit) has been shown to display wild-type virulence in insects (Herbert and Goodrich-Blair 2007). Thus, flagellar-driven motility per se (swarming or swimming) is not required for the virulence of EPN symbionts following injection into insect larvae but bacterial motility may be involved in more subtle advantage during bacterial colonization.
3 Flagellar Regulators Affect Virulence by Regulating Non-flagellar Virulence Factors
For a long time, it has been recognized that the flagellar regulatory proteins were specific transcription factors only for flagellar component genes, having a restricted role in swimming motility. However, in the early 2000s, flagellar transcriptional regulators were shown to control the expression of non-flagellar genes in Escherichia coli and Xenorhabdus (Givaudan and Lanois 2000; Pruss et al. 2003). Actually, more than 50 genes are involved in the biogenesis and function of a flagellum in E. coli or Salmonella Typhimurium. These genes are transcriptionally regulated as a cascade and are coordinated with the flagellar hierarchy. At the top of the hierarchy is the class I operon, flhDC, whose products are required for the expression of all other flagellar genes (see Fig. 1). The E. coli FlhD and FlhC proteins act as an activator for class II operons including most of the structural genes for the flagellar hook–basal body complexes plus the alternative sigma factor fliA. The product of the fliA gene, σ28, directs the transcription of class III genes that encode the filament protein, hook-associated proteins, motor proteins, and various chemotaxis proteins. The central channel is believed to work as a passage not only for flagellar component proteins, but also for flagellar regulatory protein FlgM, an anti-sigma factor. Accumulation of FlgM in the cell by preventing its export blocks the transcription. Two other genes within the flagellar regulon, fliT and fliZ, have been shown to regulate class II gene transcription (Kalir et al. 2001).
Model of flagellar regulation cascade in X. nematophila. In this regulatory hierarchy model, arrows and crossbars indicate positive and negative regulations, respectively, that are either direct or indirect. Direct binding has been demonstrated for FliZ at flhDC and hemolysin (xaxAB and xhlBA) promoters (Lanois et al. 2008). All other regulatory relationships are inferred from genetic or bioinformatic analyses. For simplicity, some known regulatory connections are not shown. In this model, the flagellar master operon flhDC is positively regulated by Lrp and LrhA (red boxes and arrows) (Richards et al. 2008) and negatively regulated by OmpR (blue boxes and arrows) (Park and Forst 2006). Class II–III flagellar genes (strictly FlhD-dependent) and exoprotein genes (lipase, protease, and hemolysins) are coregulated through fliAZ operon products (Givaudan and Lanois 2000; Park and Forst 2006). FliZ has been shown to directly control the flhDC operon through a positive feedback (Lanois et al. 2008). In addition, FliZ directly or indirectly upregulates the expression of genes encoding insecticidal toxins and antimicrobial compounds and downregulates the expression of two genes encoding the transcriptional regulators RpoS and NilR, and a structural gene, feoABC (Jubelin et al. 2013)
First in E. coli, it has been demonstrated that FlhD, the flagellar master regulator, is involved in processes other than flagellar expression that occurs when cells enter the stationary phase (Pruss and Matsumura 1996). In addition, by using flhD mutants and phenotypic characterization, Givskov et al. (1995) and Young et al. (1999) have shown that the flhDC operon controls phospholipase expression in Serratia liquefaciens and Yersinia enterocolitica, respectively. We previously showed that FlhDC positively regulates swarming behavior, hemolysin, and lipase production in the insect pathogenic bacteria, X. nematophila (Givaudan and Lanois 2000). In addition, we demonstrated that FlhDC regulatory proteins of Xenorhabdus are involved in host interactions being essential for full virulence after injection into insects. In X. nematophila, the transcription of flhDC is also negatively influenced by the two-component regulator EnvZ/OmpR (Park and Forst 2006) and is positively regulated by LrhA (Richards et al. 2008) and the global regulator Lrp (Cowles et al. 2007) (see Fig. 1). Consequently, mutations in the genes encoding these regulators have also been shown to affect swimming and swarming motilities, lipase and protease production, and hemolysis.
In this manner, the main regulatory pathway controlling these coregulated phenotypes was identified, and the precise hierarchy in the flagellar cascade was documented. First, FliA, the sigma 28 factor, co-ordinates the expression of two non-flagellar genes, xlpA and xrtA, encoding, respectively, a lipase and a protease and the flagellar motility (Park and Forst 2006). Next, another flagellar regulator, FliZ, is encoded by the second gene in the fliAZ operon. Surprisingly, the motility displayed by the Xenorhabdus fliZ mutant was strongly altered relative to the wild-type strain, whereas the fliZ mutants of E. coli and S. Typhimurium are fully motile. However, unlike the polar fliAZ mutation, fliZ deletion did not fully abolish the swimming capacity of Xenorhabdus (Jubelin et al. 2013; Lanois et al. 2008). A detailed molecular study was undertaken to identify the gene network controlled by the X. nematophila flagellar regulator FliZ. A global RNA-Seq analysis demonstrated that FliZ controls 278 coding sequences (Jubelin et al. 2013). FliZ has a positive impact on the expression of all the genes belonging to the flagellar cascade. This effect is due to the FliZ positive feedback loop on flhDC expression and the levels of FlhDC are critical for efficient motility in Xenorhabdus (Jubelin et al. 2013; Lanois et al. 2008). FliZ up- or downregulated the expression of genes encoding many non-flagellar proteins potentially involved in key steps of the Xenorhabdus life cycle. FliZ was found to upregulate hemolysin gene expression by binding directly to the xaxAB and xhlAB promoter regions (Lanois et al. 2008). The two directly FliZ-dependent genes, termed xaxAB and xhlBA, encode hemolysins from two different families. XaxAB is the prototype of a new extensive family of hemolysins with apoptotic and pore-forming activities in mammalian and invertebrate cells (Vigneux et al. 2007). The second FliZ-dependent hemolysin, XhlA, belongs to a family characterized by a two-partner secretion system (TPSS). This cell surface-associated hemolysin has been shown to be required for the full virulence of X. nematophila in insect larvae (Cowles and Goodrich-Blair 2005). Moreover, through positive feedback on fliAZ expression, FliZ also modulates the expression of the FliA-dependent gene prtA (also called xrtA), which encodes a protease (Park and Forst 2006). Another FliZ-dependent gene is xptA, a gene encoding a high molecular weight toxin complex (Tc) protein with insecticidal effects found in Xenorhabdus and Photorhabdus (Waterfield et al. 2001). Therefore, it is likely that downregulation of the expression of genes encoding the hemolysins and Tc toxins observed in the fliZ mutant explains the delayed virulence pattern observed with the fliZ mutant (Jubelin et al. 2013). In conclusion, FliZ has been shown to mediate the coordinate regulation of flagellum synthesis and virulence in the insect pathogen X. nematophila.
In P. luminescens, no mutants in flagellar regulators have been described. However, mutations in global transcriptional regulators such as the two-component system, PhoPQ (Derzelle et al. 2004b), and AstR affect the ability of Photorhabdus cells to swim and swarm on agar surfaces. Further, a transcriptional analysis in P. luminescens indicated that the negative regulation by AstR occurs at the flhDC transcription level and explains the early onset of swarming observed in the astR mutant. However, this astR mutation has no apparent impact on virulence in insects (Derzelle et al. 2004a).
4 The Flagella Apparatus as Virulence Factor Secretion System
Sequence similarities exist between components of bacterial type III secretion systems (T3SS) and those of the flagellar assembly machinery (Blocker et al. 2003). T3SSs are essential determinants of the interaction of many Gram-negative bacteria with animal or plant cells and facilitate the translocation of bacterial proteins into eukaryotic host cells. Similarly, the flagellum has also been shown to function as an export apparatus that mediates extracellular secretion of non-flagellar virulence-associated effector proteins. In contrast to P. luminescens (Brugirard-Ricaud et al. 2005), it is noteworthy that X. nematophila does not encode a dedicated (i.e., non-flagellar) type III secretion system (Brugirard-Ricaud et al. 2004). As described above, xlpA (lipase) and xrtA (protease) are both regulated by FliA-dependent promoters in X. nematophila in a similar way to class III flagellar genes. Therefore, the question remained on the secretory pathways responsible for enzyme export. One way to solve this question is the use of mutants in export apparatus components, such as FlhA and FlhB. Indeed loss of lipase, but not protease, activity was reported in the flhA strain of X. nematophila suggesting that XlpA is secreted via the flagellar export apparatus while XrtA is secreted by a dedicated ABC transport system (Park and Forst 2006). Moreover, the FliZ-dependent XaxAB hemolysin activity was not abolished in the flhA strain indicates that this hemolysin is secreted by an unknown pathway other than the flagellar export system. It was also recently proposed that the type III export apparatus of the flagellar system transports virulence-associated phospholipase A in Y. enterocolitica (Young et al. 1999) and Serratia liquefaciens (Givskov et al. 1995). Finally, the expression and secretion of lipase XlpA in Xenorhabdus is dependent on a functional flagellar system.
The bacteriocin xenocin complex is also exported through the flagellar type III system in X. nematophila. Indeed, the xenocin operon of X. nematophila consists of xciA and ximB genes encoding a xenocin and immunity protein able to kill competing microbes in the insect. The expression of xenocin operon is not dependent on flagellar regulators but is dependent on SOS induction. Using an flhA strain, authors suggested the involvement of the flagellar type III secretory system in xenocin export (Singh et al. 2013). These data demonstrate a surprising uncoupling between expression and secretion of this non-flagellar protein.
5 In Situ Flagellar Gene Expression During Invertebrate Infection
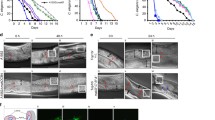
The X. nematophila life cycle has been divided into three phases: (1) insect infection, (2) bacterial and nematode reproduction in the insect cadaver, and (3) nematode transmission of X. nematophila to insect hosts (Richards and Goodrich-Blair 2009). Gene inactivation studies have identified regulatory proteins involved in adaptation to the shifting between these different hosts. Based on such data, Richard and Goodrich-Blair (2009) proposed a regulatory model that highlights the temporal changes in symbiotic and virulence factor expression that may allow X. nematophila to cope with the shifts in host environments inherent to the successful colonization of the two invertebrate hosts. However, few studies have investigated gene expression during transitions between X. nematophila life stages. Jubelin et al. (2011) investigated the dynamics of the FliA-dependent flagellin gene fliC and FliZ-dependent hemolysin genes (xaxAB and xhlBA) during insect infection and nematode association by carrying out real-time expression analysis using an unstable GFP monitoring system. Regardless of the route of infection (bacterial injection or infestation by EPN complexes), expression of flagellar genes and flagellar regulated-hemolysin genes was high only in dead insects (see Fig. 2A) and these genes were not expressed in infective juvenile stages of nematodes (see Fig. 2B). It has been proposed that activation of the flagellar cascade in dead insects may allow bacteria to move through cadaver tissues toward nutrient-rich niches by chemotaxis, facilitating their growth. In addition, other activities (lipases, proteases, and hemolysins) controlled by the flagellar regulon may also contribute to the degradation of insect tissues, releasing nutrients required for nematode feeding. Also, the expression of FliZ-dependent hemolysin genes coincided with the increase in iron availability detected at the time of insect death, suggesting that iron availability is a signal governing the adaptation of X. nematophila to changes in host environments. This study demonstrated for the first time that the flagellar regulon in X. nematophila is transiently expressed for the period of the shift between the insect infection and nematode reproduction phases. Interestingly, this paper also revealed that the expression of the flagellar and hemolysin genes in Xenorhabdus was heterogeneous at the individual cell level (see Sect. 6).
Expression of hemolysin and flagellin genes within infected insects and infective juvenile (IJ) nematodes. A Temporal and spatial expression of hemolysin and flagellin genes during the late stage of insect infection. For each experiment, at time zero, we injected 104 bacteria (F1 strains carrying PD31–gfp[AAV], PxaxAB–gfp[AAV] and PfliC–gfp[AAV] constructs) into the hemocoel of the insect larvae. Whole larvae were observed with a fluorescence macroscope, with an exposure time set to 600 ms for photography (bars, 2.5 mm). B The flagellin and hemolysin genes are not expressed in X. nematophila bacteria associated with IJ nematodes. Axenic nematodes of S. carpocapsae were associated with F1 strains carrying PD31–gfp[AAV], PxaxAB–gfp[AAV], and PfliC–gfp[AAV] constructs. Larvae of S. littoralis were infested with the various bacterium–nematode complexes and IJs were observed by fluorescence microscopy shortly after their emergence from insect cadavers. Arrows indicate the bacterium-containing vesicle of the IJs. Bars represent 30 mm. Adapted from Jubelin et al. (2011)
6 Phenotypic Variation and Bimodal Expression of Flagellar and Virulence Genes
All Xenorhabdus species and both P. luminescens and P. temperata have been reported to undergo “phenotypic variation” characterized by a switching between two cell types known as “primary” and “secondary” forms or variants. Although the phenotypic differences between primary and secondary forms can vary depending on strain and species (Hurlbert et al. 1989; Smigielski et al. 1994), typically the primary, but not secondary, cells are more highly pigmented, agglutinate red blood cells, produce fimbriae, hemolysins, proteases, antimicrobials, and crystalline inclusion bodies, and are impaired in bioluminescence for Photorhabdus (Gerritsen et al. 1995; Givaudan et al. 1995; Hurlbert et al. 1989; Smigielski et al. 1994; Volgyi et al. 2000). The swimming motility of variants in P. temperata was also investigated (Michaels and Tisa 2011) but both the primary and the secondary variants were able to swim in liquid or semisolid media under appropriate conditions. However, variation in the oxygen level greatly influenced the behavior of the secondary form and not motility of primary form. Also, it is demonstrated that the LysR-type transcriptional regulator HexA from P. temperata is able to repress several primary-specific phenotypes, i.e., exoenzyme production, pigmentation, and antibiotic activity but no effect was observed on motility (Joyce and Clarke 2003). In P. luminescens, a study showed that mutation in astRS genes, encoding a two-component signal transduction system, induces an earlier transition to the secondary phenotype than the wild type (Derzelle et al. 2004a). This mutation also affected motility in P. luminescens.
In Xenorhabdus, (Givaudan et al. 1995) revealed that most primary variants exhibit swimming and swarming behavior, whereas secondary variants in X. nematophila and X. bovienii are non-motile. Exhaustive molecular studies in X. nematophila showed that the secondary form was unable to synthesize flagellar filaments and that the flagellin-encoding gene, fliC, and the hook-associated protein 2 gene, fliD, were switched off at the transcriptional level in the variants (Givaudan et al. 1996). These results suggested that the expression of a gene earlier in the transcriptional hierarchy of the flagellar regulon was impaired. However, it showed that flhDC gene structure and expression of the master regulator FlhDC are not altered in variants (Givaudan and Lanois 2000), suggesting that this locus is not responsible for flagellar variation phenomenon in X. nematophila.
Another phenomenon observed in the flagellar regulon of X. nematophila is the so-called “phenotypic heterogeneity”. This phenomenon describes usually non-genetic variations that are observed between individual cells in an isogenic population (Veening et al. 2008). The terms “bistability” or “bimodality” describe the situation in which the population has bifurcated into coexisting cell types one representing the “OFF” state, and the other representing the “ON” state. Using single-cell analysis, (Jubelin et al. 2013) showed a bimodal expression of FliZ-dependent genes (class II–III flagellar genes and hemolytic genes) during exponential growth of the bacterial population. Actually, this bimodality generated a mixed population of subsets of cells in which FliZ-dependent genes are expressed at high- or low-level cells either expressing (‘‘ON state’’) or not expressing (‘‘OFF state’’) FliZ-dependent genes (see Fig. 3). It is known that in the presence of positive regulatory feedback, a graded expression can be converted into a binary response, in which cells express a certain gene at high or low levels (Veening et al. 2008). In addition, studies of a bacterial population exposed to a graded series of FliZ concentrations showed that FliZ controls the rate of transition between the ‘‘OFF’’ and ‘‘ON’’ states in individuals (see Fig. 3). FliZ thus plays a key role in cell fate decisions, by transiently creating individuals with different potentials for motility and host interaction genes encoding non-flagellar proteins potentially involved in key steps of the Xenorhabdus life cycle (see Fig. 1). It is likely that the heterogeneity already observed in the course of insect infection (see Sect. 5) is due to this bimodality phenomenon.
Model depicting the regulatory network underlying the regulation of flagella and the coregulation of insect virulence. The diagram shows a mixed bacterial population coexisting during the exponential growth phase of X. nematophila. On the left, a motile bacterium is shown. When the stochastic expression of the circuit generates large amounts of FliZ, this molecule exerts positive feedback on flhDC expression, upregulating the flagellar cascade and FliZ-dependent hemolysin genes. Consequently, the noisy expression of class II flagellar genes is reduced and cells fully express classes II and III flagellar genes, resulting in the motility phenotype. On the right, a non-motile bacterium is shown. Authors suggest that the lower level of FlhD-FliZ output delays and desynchronizes class II genes expression, probably impairing completion of the basal body–hook structure. The FlgM protein, an anti-sigma-28 factor, binds the FliA sigma-28 factor directly in E. coli, preventing class III promoter transcription until after hook–basal body completion (Kutsukake 1994). Thus, the accumulation of FlgM in cells probably blocks the transcription of class III genes, including that encoding flagellin, resulting in a non-motile state. The threshold of FliZ controls the rate of transition between the ‘‘OFF’’ and ‘‘ON’’ states in individuals in Xenorhabdus. adapted from Jubelin et al. (2013)
The relationship between the FliZ-mediated bimodality and the phenotypic variation in X. nematophila is still therefore an open question. In a preliminary experiment, we investigated the form status (primary or secondary forms) in ON and OFF populations after cell sorting, but no variant forms were identified (our unpublished data) even in the OFF population in which transient expression patterns were similar to that observed in the secondary form.
7 Conclusions and Future Work on P. luminescens
Flagellar motility plays an important role in the life cycle of the entopathogenic bacteria. However for Photorhabdus and Xenorhabdus, it is clear that the bacterial locomotion per se has little impact on the interaction with nematodes or insects. However, the coupling of motility with other bacterial virulence factors appears to be mediated in part by master regulatory proteins in motility systems (FlhDC, FliA, and FliZ) of X. nematophila. It is therefore clear that virulence and motility are coordinately regulated to best support the lifestyle of the pathogenic bacteria in the course of insect infection. Further research will improve our understanding of how virulence and motility are coordinately regulated in Photorhabdus. One additional aspect which has to be investigated further is the role of flagella in innate immunity of insects. While flagellin triggers humoral immunity in fly Samakovlis et al. (1992), no flagellin TLR-like ligands have yet been identified in other insects. Finally, the switch-off of the expression of the flagellar regulon during the colonization of nematode infective juvenile larva suggests that the flagellar regulon is not involved in mutualism in Xenorhabdus. However, this question remains open for the complex association between Photorhabdus and Heterorhabditis.
References
Allison C, Lai HC, Hughes C (1992) Co-ordinate expression of virulence genes during swarm-cell differentiation and population migration of Proteus mirabilis. Mol Microbiol 6:1583–1591
Blocker A, Komoriya K, Aizawa S (2003) Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc Natl Acad Sci USA 100:3027–3030. doi:10.1073/pnas.0535335100
Brugirard-Ricaud K, Givaudan A, Parkhill J, Boemare N, Kunst F, Zumbihl R, Duchaud E (2004) Variation in the effectors of the type III secretion system among Photorhabdus species as revealed by genomic analysis. J Bacteriol 186:4376–4381. doi:10.1128/JB.186.13.4376-4381.2004
Brugirard-Ricaud K, Duchaud E, Givaudan A, Girard PA, Kunst F, Boemare N, Brehelin M, Zumbihl R (2005) Site-specific antiphagocytic function of the Photorhabdus luminescens type III secretion system during insect colonization. Cell Microbiol 7:363–371. doi:10.1111/j.1462-5822.2004.00466.x
Cowles KN, Goodrich-Blair H (2005) Expression and activity of a Xenorhabdus nematophila haemolysin required for full virulence towards Manduca sexta insects. Cell Microbiol 7:209–219
Cowles KN, Cowles CE, Richards GR, Martens EC, Goodrich-Blair H (2007) The global regulator Lrp contributes to mutualism, pathogenesis and phenotypic variation in the bacterium Xenorhabdus nematophila. Cell Microbiol 9:1311–1323
Derzelle S, Ngo S, Turlin E, Duchaud E, Namane A, Kunst F, Danchin A, Bertin P, Charles JF (2004a) AstR-AstS, a new two-component signal transduction system, mediates swarming, adaptation to stationary phase and phenotypic variation in Photorhabdus luminescens. Microbiology 150:897–910. doi:10.1099/mic.0.26563-0
Derzelle S, Turlin E, Duchaud E, Pages S, Kunst F, Givaudan A, Danchin A (2004b) The PhoP-PhoQ two-component regulatory system of Photorhabdus luminescens is essential for virulence in insects. J Bacteriol 186:1270–1279
Easom CA, Clarke DJ (2008) Motility is required for the competitive fitness of entomopathogenic Photorhabdus luminescens during insect infection. BMC Microbiol 8:168. doi:10.1186/1471-2180-8-168
Gerritsen L, van der Wolf JM, van Vuurde J, Ehlers R, Krasomil-Osterfel KC, Smits PH (1995) Polyclonal Antisera to distinguish strains and form variants of Photorhabdus (Xenorhabdus) luminescens. Appl Environ Microbiol 61:284–289
Ghelardi E, Celandroni F, Salvetti S, Beecher DJ, Gominet M, Lereclus D, Wong AC, Senesi S (2002) Requirement of flhA for swarming differentiation, flagellin export, and secretion of virulence-associated proteins in Bacillus thuringiensis. J Bacteriol 184:6424–6433
Givaudan A, Lanois A (2000) flhDC, the flagellar master operon of Xenorhabdus nematophilus: requirement for motility, lipolysis, extracellular hemolysis, and full virulence in insects. J Bacteriol 182:107–115
Givaudan A, Baghdiguian S, Lanois A, Boemare N (1995) Swarming and swimming changes concomitant with phase variation in Xenorhabdus nematophilus. Appl Environ Microbiol 61:1408–1413
Givaudan A, Lanois A, Boemare N (1996) Cloning and nucleotide sequence of a flagellin encoding genetic locus from Xenorhabdus nematophilus: phase variation leads to differential transcription of two flagellar genes (fliCD). Gene 183:243–253
Givskov M, Eberl L, Christiansen G, Benedik MJ, Molin S (1995) Induction of phospholipase- and flagellar synthesis in Serratia liquefaciens is controlled by expression of the flagellar master operon flhD. Mol Microbiol 15:445–454
Herbert EE, Goodrich-Blair H (2007) Friend and foe: the two faces of Xenorhabdus nematophila. Nat Rev Microbiol 5:634–646
Hurlbert RE, Xu J, Small CL (1989) Colonial and cellular polymorphism in Xenorhabdus luminescens. Appl Environ Microbiol 55:1136–1143
Hurst S, Rowedder H, Michaels B, Bullock H, Jackobeck R, Abebe-Akele F, Durakovic U, Gately J, Janicki E, Tisa LS (2015) Elucidation of the Photorhabdus temperata Genome and generation of a transposon mutant library to identify motility mutants altered in Pathogenesis. J Bacteriol 197:2201–2216. doi:10.1128/JB.00197-15
Josenhans C, Suerbaum S (2002) The role of motility as a virulence factor in bacteria. Int J Med Microbiol 291:605–614
Joyce SA, Clarke DJ (2003) A hexA homologue from Photorhabdus regulates pathogenicity, symbiosis and phenotypic variation. Mol Microbiol 47:1445–1457
Jubelin G, Pages S, Lanois A, Boyer MH, Gaudriault S, Ferdy JB, Givaudan A (2011) Studies of the dynamic expression of the Xenorhabdus FliAZ regulon reveal atypical iron-dependent regulation of the flagellin and haemolysin genes during insect infection. Environ Microbiol 13:1271–1284. doi:10.1111/j.1462-2920.2011.02427.x
Jubelin G, Lanois A, Severac D, Rialle S, Longin C, Gaudriault S, Givaudan A (2013) FliZ is a global regulatory protein affecting the expression of flagellar and virulence genes in individual Xenorhabdus nematophila bacterial cells. PLoS Genet 9:e1003915. doi:10.1371/journal.pgen.1003915
Kalir S, McClure J, Pabbaraju K, Southward C, Ronen M, Leibler S, Surette MG, Alon U (2001) Ordering genes in a flagella pathway by analysis of expression kinetics from living bacteria. Science 292:2080–2083. doi:10.1126/science.1058758
Kutsukake K (1994) Excretion of the anti-sigma factor through a flagellar substructure couples flagellar gene expression with flagellar assembly in Salmonella typhimurium. Mol Gen Genet 243:605–612
Lanois A, Jubelin G, Givaudan A (2008) FliZ, a flagellar regulator, is at the crossroads between motility, haemolysin expression and virulence in the insect pathogenic bacterium Xenorhabdus. Mol Microbiol 68:516–533
Michaels B, Tisa LS (2011) Swarming motility by Photorhabdus temperata is influenced by environmental conditions and uses the same flagella as that used in swimming motility. Can J Microbiol 57:196–203. doi:10.1139/W10-119
Park D, Forst S (2006) Co-regulation of motility, exoenzyme and antibiotic production by the EnvZ-OmpR-FlhDC-FliA pathway in Xenorhabdus nematophila. Mol Microbiol 61:1397–1412
Pruss BM, Matsumura P (1996) A regulator of the flagellar regulon of Escherichia coli, flhD, also affects cell division. J Bacteriol 178:668–674
Pruss BM, Campbell JW, Van Dyk TK, Zhu C, Kogan Y, Matsumura P (2003) FlhD/FlhC is a regulator of anaerobic respiration and the Entner-Doudoroff pathway through induction of the methyl-accepting chemotaxis protein Aer. J Bacteriol 185:534–543
Richards GR, Goodrich-Blair H (2009) Masters of conquest and pillage: Xenorhabdus nematophila global regulators control transitions from virulence to nutrient acquisition. Cell Microbiol 11:1025–1033. doi:10.1111/j.1462-5822.2009.01322.x
Richards GR, Herbert EE, Park Y, Goodrich-Blair H (2008) Xenorhabdus nematophila lrhA is necessary for motility, lipase activity, toxin expression, and virulence in Manduca sexta insects. J Bacteriol 190:4870–4879. doi:10.1128/JB.00358-08
Samakovlis C, Asling B, Boman HG, Gateff E, Hultmark D (1992) In vitro induction of cecropin genes–an immune response in a Drosophila blood cell line. Biochem Biophys Res Commun 188:1169–1175
Singh P, Park D, Forst S, Banerjee N (2013) Xenocin export by the flagellar type III pathway in Xenorhabdus nematophila. J Bacteriol 195:1400–1410. doi:10.1128/JB.01532-12
Smigielski AJ, Akhurst RJ, Boemare NE (1994) Phase Variation in Xenorhabdus nematophilus and Photorhabdus luminescens: Differences in Respiratory Activity and Membrane Energization. Appl Environ Microbiol 60:120–125
Veening JW, Smits WK, Kuipers OP (2008) Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol 62:193–210. doi:10.1146/annurev.micro.62.081307.163002
Vigneux F, Zumbihl R, Jubelin G, Ribeiro C, Poncet J, Baghdiguian S, Givaudan A, Brehelin M (2007) The xaxAB genes encoding a new apoptotic toxin from the insect pathogen Xenorhabdus nematophila are present in plant and human pathogens. J Biol Chem 282:9571–9580
Volgyi A, Fodor A, Forst S (2000) Inactivation of a novel gene produces a phenotypic variant cell and affects the symbiotic behavior of Xenorhabdus nematophilus. Appl Environ Microbiol 66:1622–1628
Waterfield NR, Bowen DJ, Fetherston JD, Perry RD, ffrench-Constant RH (2001) The tc genes of Photorhabdus: a growing family. Trends Microbiol 9:185–191
Young GM, Schmiel DH, Miller VL (1999) A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc Natl Acad Sci U S A 96:6456–6461
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing AG
About this chapter
Cite this chapter
Givaudan, A., Lanois, A. (2016). Flagellar Regulation and Virulence in the Entomopathogenic Bacteria—Xenorhabdus nematophila and Photorhabdus luminescens . In: ffrench-Constant, R. (eds) The Molecular Biology of Photorhabdus Bacteria . Current Topics in Microbiology and Immunology, vol 402. Springer, Cham. https://doi.org/10.1007/82_2016_53
Download citation
DOI: https://doi.org/10.1007/82_2016_53
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-52714-7
Online ISBN: 978-3-319-52715-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)