Abstract
Cholera is still a major public health problem. The underlying bacterial pathogen Vibrio cholerae (V. cholerae) is evolving and some of its mutations have set the stage for outbreaks. After V. cholerae acquired the mobile elements VSP I & II, the El Tor pandemic began and spread across the tropics. The replacement of the O1 serotype encoding genes with the O139 encoding genes triggered an outbreak that swept across the Indian subcontinent. The sxt element generated a third selective sweep and most recently a fourth sweep was associated with the exchange of the El Tor ctx allele for a classical ctx allele in the El Tor background. In Kenya, variants of this fourth selective sweep have differentiated and become endemic residing in and emerging from environmental reservoirs. On a local level, studies in Bangladesh have revealed that outbreaks may arise from a nonrandom subset of the genetic lineages in the environment and as the population of the pathogen expands, many novel mutations may be found increasing the amount of genetic variation, a phenomenon known as a founder flush. In Haiti, after the initial invasion and expansion of V. cholerae in 2010, a second outbreak occurred in the winter of 2011–2012 driven by natural selection of specific mutations.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Background
Cholera outbreaks have historically represented major public health events, with the potential for large numbers of cases and deaths. The largest outbreaks have moved across multiple continents (meeting the definition of pandemics), with case counts in the millions. Pandemics are traditionally referred to on the basis of the year in which they commenced: the first started in 1817, the second in 1829, with subsequent ones starting in 1852, 1861, 1881, 1899, and the seventh and most recent (still ongoing) in 1961. More localized outbreaks are known by the location and the year: Orissa 1999, Dhaka 2006, Zimbabwe 2008, Haiti 2010, and Kenya 2010. Left undefined by the term outbreak is the source, the mode of transmission, and the genetic diversity of the causative bacterium, V. cholerae.
On the broadest scale there are two modes of transmission, emergence of an endemic strain from environmental reservoirs and/or introduction of strain from a visitor. Multiple port cities, such as New York, Hamburg, St. Petersburg, Alexandria, and Gran Canaria, can trace introductions to specific ships arriving from ports with ongoing outbreaks. More recently, the 2010 outbreak in Haiti has been traced to the UN peacekeeping troops from Nepal. In each of these cases, there is a single source and the tracing of the outbreak involves finding the common origin.
V. cholerae is a very diverse species, including virulent and avirulent strains. The disease cholera is caused by strains that produce cholera toxin, and which, traditionally, have been in a limited number of serotypes, including serotypes O1 and O139. The avirulent strains do not produce cholera toxin, and usually fall into one of the over 200 other V. cholerae serotypes recognized to date. There are no data about the strains in the first five pandemics because the discovery that cholera was caused by a bacterium (by Pacini 1855), its isolation (by Koch 1884), and its preservation did not occur until the twentieth century and after the start of the sixth pandemic. Numerous isolates are available from the seventh pandemic. All of the isolates in the sixth and seventh pandemics appear to be derived from a single ancestor: that is, they are clonally related. They differ from each other by nucleotide variants, some of which are de novo mutations, and others the result of recombination events (Garg, Aydanian et al. 2003; Salim, Lan et al. 2005).
2 Genetic Variation and Selective Sweeps
In addition to the point mutations, there are numerous mobile elements that have been incorporated into the genome of some of the sublineages (Chun, Grim et al. 2009). Among the mobile elements are pathogenic islands (described below) and the serotype: although often thought as a phylogenetic marker and by definition not subject to lateral gene transfer, the first evidence that it might be mobile came from the discovery of the O139 morph of the O1–O139 lineage. It has been shown by three groups that the O1 encoding genes were replaced by O139 encoding genes (Bik, Bunschoten et al. 1995; Comstock, Johnson et al. 1996; Mooi and Bik 1997). Subsequently, the O1 encoding genes were shown to transfer between SNP-defined lineages, and furthermore the “jump start” sequence (Hobbs and Reeves 1994) was identified as the junction point and shown to be similar to a DNA uptake sequence leading to the suggestion that serotype genes are mechanistically prone to being mobile (Gonzalez-Fraga, Pichel et al. 2008). After the unprecedented epidemic caused by serotype O139 strains, considerable attention has been paid to non O1–O139 serotypes in the O1–O139 lineage: this includes two serotypes that have caused significant outbreaks of O37 in Czechoslovakia in 1965 and in Sudan in 1968. Other serotypes that have been reported in the O1–O139 lineage include O10, O26, O27, O53, O65, O75, and O141 (Rudra, Mahajan et al. 1996; Dalsgaard, Serichantalergs et al. 2001; Li, Shimada et al. 2002; Octavia, Salim et al. 2013). While these have been associated with disease usually found in patients and occasionally in small outbreaks, they have not caused a major outbreak.
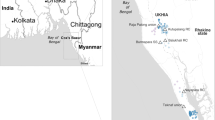
When V. cholerae, or any bacteria, acquires a novel genetic element(s) that increases its fitness, a selective sweep will occur. The first selective sweep to be identified was the emergence of O139. Isolates with the serotype O139 did not cross-react serologically with O1 V. cholerae and so older persons who were immune to O1 fell ill to O139. The sweep appears to have started near Madras (now known as Chennai) and spread across the Indian subcontinent over the next 2 years. Tracking the epidemic was easy because of its unique serotype and its propensity to strike older individuals (Nair et al. 1994). As shown in Fig. 1, the new serotype spread from Madras south to Madurai and north to Kolkata and then west to Lucknow and eventually to Delhi. Each of these cities represents introduction of the new O139 form of V. cholerae to a new geographic region.
Map of India detailing the spread of O139 V. cholerae. The dates indicate the first observation of O139 in that city and the arrows represent the likely directions of the spread. From Nair et al. 1994
Among the O1 strains, the O1 classical strains associated with the sixth pandemic (collected during the first quarter of the twentieth century) are the ancestral isolates (Salim, Lan et al. 2005). The seventh pandemic has been characterized by four selective sweeps. The first sweep chronologically coincided with the emergence of the El Tor strains in 1961. The ancestor of the O1 El Tor strains acquired two major pathogenic islands: Vibrio Seventh Pandemic (VSP) I & II that have 11 and 7 genes, respectively (Dziejman, Balon et al. 2002). Additional genes were also acquired, however, the selective advantage conferred by VSP I & II and the other genes is not known. The second selective sweep began when the novel element sxt was acquired around 1981. The sxt element is an integrating conjugative element and in addition to the core region of about two dozen genes that encode its ability to transfer and to be regulated, there are several antibiotic resistance genes. These latter genes are considered to confer the selective advantage on the genomes containing them (Waldor, Tschape et al. 1996). This second selective sweep is referred to as wave two (see Fig. 2) that spread around the world. The third selective sweep was the O139 sweep mentioned above. The fourth and the most recent selective sweep was initiated after an sxt-positive El Tor strain acquired the classical allele of the cholera toxin gene. The classical cholera toxin allele isolates selectively replaced the El Tor ctx allele carrying isolates (Raychoudhuri, Patra et al. 2009) and may be associated with more severe disease (Nair, Faruque et al. 2002). This fourth wave expanded in Asia and across the world including Kenya and Haiti (Mutreja, Kim et al. 2011). During each selective sweep, the common ancestor differentiates with the acquisition of many novel variant nucleotides that produce a radiation of different but closely related genotypes. While the nucleotide changes may be fit neatly into a “phylogenetic” tree, the bacteria are also differentiating by the gain and loss of mobile elements (Chun, Grim et al. 2009). It should be noted that while the selected variants in these expansions become the most frequent type, there is no reason to expect they entirely outcompete the previous forms. Despite claims of the extinction of earlier types, evidence of their existence continues to appear. Classical strains (sixth pandemic) have been identified in the last 20 years (Boyd, Heilpern et al. 2000; Alam, Islam et al. 2012) and typical El Tor strains in the last five (Rashed et al. 2013), though they were competitively swept aside by the most recent variant 10 or more years ago. Although there is no way to predict where or when the next acquisition of a novel element or novel mutations will occur, it is a safe prediction that such an event will occur and it will be selectively spread wherever there is cholera.
Phylogenetic tree based on variable nucleotides. Points at which selective sweeps begin are indicated by the arrows. The right side of the figure indicates the presence of various mobile elements. From Mutreja et al. 2011
3 V. cholerae is Endemic in Kenya
In Africa, an open question is whether or not V. cholerae is endemic. Although all three waves of O1 have invaded the continent (Mutreja, Kim et al. 2011), one working hypothesis is that V. cholerae has not become an endemic part of the biological landscape. Around Lake Victoria, V. cholerae is considered to move from place to place, and when the cases in any locale subside V. cholerae is thought to go extinct, with the next outbreak occurring when the roulette wheel carry V. cholerae returns to that locale (Nkoko, Giraudoux et al. 2011). Until recently, there has been little genetic research on whether this is correct. One genetic-based study examined the Kenyan 2010 outbreak which began in the Lake region of Kenya, and based on the epidemiology of the initial cases, onset was attributed to introduction and subsequent spread (Mohamed, Oundo et al. 2012). This conclusion was consistent with pulse field gel electrophoretic data that show that all the isolates were identical or differed at a single band. However, analysis of pulse field genotypes is not as discriminatory as multilocus variable tandem repeat analysis (MLVA).
MLVA for V. cholerae relies on five or six loci that contain tandem repeats of six or seven nucleotides that are repeated from 4 to 31 times. The length of the repeats is measured and used as the allele number. The alleles at each locus in order produce a five-digit genotype. Genotypes that differ from another at a single locus are related and considered part of a clonal complex, i.e., derived from the same ancestor. Genotypes that differ from all members of a clonal complex at two or more loci are considered unrelated. The application of MLVA to a series of Kenyan isolates revealed that there were five clonal complexes (Mohamed, Oundo et al. 2012). The genotypes in a single clonal complex are related to each other, but not to genotypes in other clonal complexes. The presence of five distinct clonal complexes rejects unequivocally the hypothesis that this outbreak is the result of introduction and spread of a clone. More recent data using nucleotide variants from whole genome sequencing also showed that there are multiple genetic lineages within the 2010 outbreak (Kiiru, Mutreja et al. 2013). In Kenya, as in other countries, when a selective sweep occurs, the isolates may diverge. In this case there are two large well-separated groups by SNP analysis, from an earlier introduction (estimated to be about 1990), confirming that some of these strains have resided in Kenya for at least a decade. Thus both MLVA and whole genome sequencing data lead to the same conclusion that, in Kenya, the only African country with data, the question is settled, V. cholerae is endemic (Fig. 3).
Months in which various clonal complexes appeared in a Highland region including Nairobi, b coastal region, c Semi-arid Northern region, and d Lower Eastern region. Distinct clonal complexes occur simultaneously in different regions, thus the isolates could not have been transported between regions. From Mohamed et al. 2012
4 Molecular Epidemiology in Local Outbreaks
Tracking V. cholerae when it is endemic presents a more difficult problem because V. cholerae can alter its physiological state and the question of transmission includes whether novel isolates are introduced into the region or native isolates are emerging from the environment into the human population. Throughout south Asia, especially around the Bay of Bengal, V. cholerae is endemic. V. cholerae survive in ponds, streams, and estuaries (Huq, Colwell et al. 1990; Huq, Parveen et al. 1993). The disease-causing strains are rare in the environment being heavily outnumbered by avirulent strains of V. cholerae. Although unusual in the environment, the O1–O139 s can survive long periods in the environment either attached to copepods or in an altered physiological state: viable but nonculturable state (VBNC) (Huq, Grim et al. 2006), or possibly a newly described “persister” state (Jubair, Morris et al. 2012). The VBNC state may be associated with biofilms in which V. cholera can survive months in sterilized water. V. cholerae reside in the environment and their numbers fluctuate with the seasons. When they reach sufficient numbers and densities to provide an infectious dose, they emerge from their environmental refuge and wreak havoc among the human population (Huq, Colwell et al. 1990). At this stage there are many potential sources, the affected humans and the environmental refuge. The relative proportion that these two sources contribute to the outbreak has been disputed. The extreme position is that all cases result from the bacteria emerging from the environment. Complicating the issue, is the recent observation that after traveling through either a mouse or a human intestine, V. cholerae enters a second altered physiological state referred to as “hyperinfective.” When it is hyperinfective, the infectious dose is decreased to 104, and after ~18 h in water, V. cholerae reverts to normal infectiveness (~106) (Merrell et al. 2002).
Mathematical modeling of incidence data from outbreaks has been used to estimate the contribution of “fast” and “slow” transmission. The initial slow transmission is from the environment to humans, while the accelerated fast transmission occurs when V. cholerae is hyperinfective. In the outbreak in Zimbabwe in 2008–2009, a total of 98,585 cases and 4,285 deaths were reported from multiple provinces with data being collected on a weekly basis. A simple model with both fast and slow transmission revealed that 41–95 % of the transmission, depending on province, was attributed to the fast process (Mukandavire, Liao et al. 2011). A second use of the same model was applied the data from the 2010–2011 outbreak in Haiti with a similar result, fast transmission was an essential amplifier in the outbreak and it varied by region of the country (Mukandavire, Smith et al. 2013). While being promising, these estimates have been based on official symptomatic case reports without more detailed underlying epidemiologic data. Furthermore, it may not be possible to estimate the contribution of two transmission mechanisms from incidence data alone when the timescale of the slow is similar to that of fast transmission (Eisenberg, Robertson et al. 2013).
Dissecting the alternative routes of transmission in seasonal outbreaks in endemic requires the ability to distinguish between isolates occurring in a limited geographic area. Most of the work distinguishing among isolates within a geographic region has been based on MLVA. Extensive variation has been found in and between every location that has been examined (Stine, Alam et al. 2008). While it is not surprising to find different genetically distinguishable isolates in a geographically restricted area because O1 Inaba and Ogawa (the two common biotypes of O1) and O139 isolates are occasionally observed in a single sampling location, it was completely unexpected to find multiple genetic lineages of V. cholerae within a single individual (Kendall, Chowdhury et al. 2010). Interestingly, the three loci on the first large chromosome seem to vary at a slower rate than those on the second or small chromosome (Kendall, Chowdhury et al. 2010). Even if the analysis is limited to the slower evolving loci, multiple genetic lineages were found in a single stool. Whether this increases the difficultly of tracking, V. cholerae as it spreads because tracking needs to account for all the lineages, or narrows the sources because of having greater genetic differentiation between samples, remains to be seen.
5 Genetic Variation and Founder Flush Events
Notably missing from the above discussion is any consideration of the number of bacteria. The numbers are enormous. A single case represents the growth of an infectious dose (~106) to 1014 (100 trillion) bacteria that are excreted. Multiplying a single case ten million times and the number reaches 1021, a sextillion. The sixth pandemic caused an estimated 1.5 million deaths in India (800,000), Russia (500,000), and the Philippines (200,000) and it devastated other numerous countries.
Each case, outbreak, and pandemic represents an exponential expansion of the population of V. cholerae and then an exponential decrease. Yet the implications of these fluctuations on the genetics of V. cholerae have seldom been discussed. Multilocus sequence typing of O139 isolates from Kolkata demonstrated that although one allele was the most frequently found allele (77–99 % depending on the locus), alternative alleles were observed and the same alternative allele could be found in multiple years implying a persistence of the minor alleles. The authors (Garg, Aydanian et al. 2003) suggested the founder flush phenomenon might be why novel alleles were seen. Founder flush, initially described by H.D. and E.B. Ford (1930) and recounted by Wallace (1981), posits that as a population expands, variants that otherwise would not survive, do survive and expand in number along with the rest of the population.
The founder flush phenomenon may also apply in short seasonal outbreaks. In October and November of 2010, 138 isolates from Chhatak, Bangladesh were genotyped using MLVA (Rashed et al. 2014). Twenty-six genotypes were found in a single clonal complex. A “founder” genotype defined as the genotype with the most single locus variants related to it was identified. It was one of several genotypes observed on the first day of sampling clinic patients. Of the 25 derived genotypes, 23 were observed temporally after the genotype closer to the founder than from which they were derived. This observation is consistent with successive mutations occurring during the expansion. Although these polymorphic alleles occur within coding sequences (thus increasing the size of the protein by two amino acids for each additional repeat) in four of the five loci, whether or not these have a selective value is not known. In Haiti in 2010, the founder genotype radiated into eight additional MLVA genotypes (Ali, Chen et al. 2011). In Dhaka from 2004–2006, the three major clonal complexes all included additional genotypes as the years progressed (Kendall, Chowdhury et al. 2010). Thus, it is clear that during an expansion, novel MLVA alleles can be detected (Fig. 4).
Clonal complex found in Chhatak 2010. The dates of the first observation of each isolate are indicated. Genotypes seen only in isolates from clinic patients have a yellow background, while those in environmental isolates only have a green background and when the genotype was isolated from both patient and environmental sources it had a blue background. From Rashed et al. 2014
Apropos to the discussion of the role of the environment in the transmission of the V. cholerae, the outbreaks in Chhatak 2010 and Mathbaria 2011 revealed that the genotypes in the patients were a nonrandom subset of the genotypes in the environment (Rashed et al. 2014). In both outbreaks, multiple genotypes were observed in the environment whether the analysis included the second chromosome loci or not and a single genotype or if the second chromosome loci were included a single clonal complex or genetic lineage was found in the patients. This is very strong support for an accelerated mode of transmission. However, whether the accelerated mode incorporates the hyperinfective state or involves massive numerical increases of a genetic lineage from the earliest cases cannot be distinguished from these analyses.
V. cholerae readily accepts novel DNA elements. It has been shown to recombine in the presence of chitin (Blokesch and Schoolnik 2007). It may have a mechanism for the uptake of specific DNA sequences. Serotype encoding genes (Gonzalez-Fraga, Pichel et al. 2008) and housekeeping genes have been shown to recombine (Garg, Aydanian et al. 2003; Salim, Lan et al. 2005; Octavia, Salim et al. 2013). The incorporation of these novel elements is an ongoing process and numerous novel combinations have been detected recently with the advent of genomic sequencing. However, the key for these variants is whether or not they are favored by natural selection. If not, they will eventually disappear, but if so they will be swept into the population causing disease as new variant expands in frequency.
One example of natural selection for SNPs comes from V. cholerae in Haiti. Consistent with a founder flush event, as the pathogen population expands, the number of polymorphic sites expands from approximately 0 in 2010 to 195 SNPs in 2012 (Fig. 5) (Salemi et al. 2014). For these SNPs, the presumptive role of selection can be ascertained. The number of nonsynonymous substitutions (those that change the amino acid) continues to increase and at a rate exceeding that of synonymous substitutions. The excess of nonsynonymous to synonymous substitutions (a 2:1 ratio, in this case) is a hallmark of positive selection. Although the precise selective force is not known, it may be the pathogen adapting to expand the number of individuals that it successfully infects. In Fig. 5b, the bars labeled I, II, and III mark periods of increased incidence of cases, a pattern consistent of the selective sweeps described above and the radiation or expansion of genetic variation often found during a flush.
Genetic and population variation in Haiti 2010–2012. Panel a graphs the effective population size with its 95 % confidence limits over time with an overlay of the phylogram reflecting the genetic variation. Panel b graphs the proportion of SNPs that are synonymous (purple) versus those that nonsynonymous (green). In each case, the white line is the mean and the colored area represents the 95 % confidence limits. From Salemi et al. 2014
In summary, V. cholerae is evolving. The circulating clones are expected to vary over time. Genetic changes, both mutations and acquisitions of new genetic sequences, provide the substrate for natural selection to shape the population. Unfortunately for us, a major selected path is to increase the spread of V. cholerae, so in the foreseeable future, there will be an increasing number of cases of cholera.
References
Alam M, Islam MT et al (2012) Vibrio cholerae classical biotype strains reveal distinct signatures in Mexico. J Clin Microbiol 50(7):2212–2216
Ali A, Chen Y et al (2011) Recent clonal origin of cholera in Haiti. Emerg Infect Dis 17(4):699–701
Bik EM, Bunschoten AE et al (1995) Genesis of the novel epidemic Vibrio cholerae O139 strain: evidence for horizontal transfer of genes involved in polysaccharide synthesis. EMBO J 14(2):209–216
Blokesch M, Schoolnik GK (2007) Serogroup conversion of Vibrio cholerae in aquatic reservoirs. PLoS Pathog 3(6):e81
Boyd EF, Heilpern AJ et al (2000) Molecular analyses of a putative CTXphi precursor and evidence for independent acquisition of distinct CTX(phi)s by toxigenic Vibrio cholerae. J Bacteriol 182(19):5530–5538
Chun J, Grim CJ et al (2009) Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci U S A 106(36):15442–15447
Comstock LE, Johnson JA et al (1996) Cloning and sequence of a region encoding a surface polysaccharide of Vibrio cholerae O139 and characterization of the insertion site in the chromosome of Vibrio cholerae O1. Mol Microbiol 19(4):815–826
Dalsgaard A, Serichantalergs O et al (2001) Clinical and environmental isolates of Vibrio cholerae serogroup O141 carry the CTX phage and the genes encoding the toxin-coregulated pili. J Clin Microbiol 39(11):4086–4092
Dziejman M, Balon E et al (2002) Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc Natl Acad Sci U S A 99(3):1556–1561
Eisenberg MC, Robertson SL et al (2013) Identifiability and estimation of multiple transmission pathways in cholera and waterborne disease. J Theor Biol 324C:84–102
Garg P, Aydanian A et al (2003) Molecular epidemiology of O139 Vibrio cholerae: mutation, lateral gene transfer, and founder flush. Emerg Infect Dis 9(7):810–814
Gonzalez-Fraga S, Pichel M et al (2008) Lateral gene transfer of O1 serogroup encoding genes of Vibrio cholerae. FEMS Microbiol Lett 286(1):32–38
Hobbs M, Reeves PR (1994) The JUMP start sequence: a 39 bp element common to several polysaccharide gene clusters. Mol Microbiol 12(5):855–856
Huq A, Colwell RR et al (1990) Detection of Vibrio cholerae O1 in the aquatic environment by fluorescent-monoclonal antibody and culture methods. Appl Environ Microbiol 56(8):2370–2373
Huq A, Grim C et al (2006) Detection, isolation, and identification of Vibrio cholerae from the environment. Curr Protoc Microbiol Chapter 6(Unit6A):5
Huq A, Parveen S et al (1993) Comparison of Vibrio cholerae serotype 01 strains isolated from patients and the aquatic environment. J Trop Med Hyg 96(2):86–92
Jubair M, Morris JG Jr et al (2012) Survival of Vibrio cholerae in nutrient-poor environments is associated with a novel “persister” phenotype. PLoS ONE 7(9):e45187
Kendall EA, Chowdhury F et al (2010) Relatedness of Vibrio cholerae O1/O139 isolates from patients and their household contacts, determined by multilocus variable-number tandem-repeat analysis. J Bacteriol 192(17):4367–4376
Kiiru J, Mutreja A et al (2013) A Study on the geophylogeny of clinical and environmental Vibrio cholerae in Kenya. PLoS ONE 8(9):e74829
Koch R (1884) Funfter Bericht Leiters der Deutschen wissenschaftlichen Commission zur Erforschung der Cholera, Geheimen Regierungsraths Dr Koch. Dtsch Med Wochenschr 10:221–222
Li M, Shimada T et al (2002) Evidence for the emergence of non-O1 and non-O139 Vibrio cholerae strains with pathogenic potential by exchange of O-antigen biosynthesis regions. Infect Immun 70(5):2441–2453
Merrell DS, Butler SM, Qadri F, Dolganov NA, Alam A, Cohen MB, Calderwood SB, Schoolnik GK, Camilli A (2002) Host-induced epidemic spread of the cholera bacterium. Nature 417(6889):642–645
Mohamed AA, Oundo J et al (2012) Molecular epidemiology of geographically dispersed Vibrio cholerae, Kenya, Jan 2009–May 2010. Emerg Infect Dis 18(6):925–931
Mooi FR, Bik EM (1997) The evolution of epidemic Vibrio cholerae strains. Trends Microbiol 5(4):161–165
Mukandavire Z, Liao S et al (2011) Estimating the reproductive numbers for the 2008–2009 cholera outbreaks in Zimbabwe. Proc Natl Acad Sci U S A 108(21):8767–8772
Mukandavire Z, Smith DL et al (2013) Cholera in Haiti: reproductive numbers and vaccination coverage estimates. Sci Rep 3:997
Mutreja A, Kim DW et al (2011) Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477(7365):462–465
Nair GB, Faruque SM et al (2002) New variants of Vibrio cholerae O1 biotype El Tor with attributes of the classical biotype from hospitalized patients with acute diarrhea in Bangladesh. J Clin Microbiol 40(9):3296–3299
Nair GB, Ramamurthy T, Bhattacharya SK, Mukhopadhyay AK, Garg S, Bhattacharya MK, Takeda T, Shimada T, Takeda Y, Deb BC (1994) Spread of Vibrio cholerae O139 Bengal in India. J Infect Dis 169(5):1029–1034
Nkoko DB, Giraudoux P et al (2011) Dynamics of cholera outbreaks in great lakes region of Africa, 1978–2008. Emerg Infect Dis 17(11):2026–2034
Octavia S, Salim A et al (2013) Population structure and evolution of non-O1/non-O139 Vibrio cholerae by multilocus sequence typing. PLoS ONE 8(6):e65342
Pacini (1855) http://en.wikipedia.org/wiki/Filippo_Pacini
Rashed SM, Iqbal A, Mannan SB, Islam T, Rashid MU, Johura FT, Watanabe H, Hasan NA, Huq A, Stine OC, Sack RB, Colwell RR, Alam M (2013) Vibrio cholerae O1 El Tor and O139 Bengal strains carrying ctxB(ET). Emerg Infect Dis 19(10):1713–1715
Rashed, SM, Azman AS, Alam M, Li S, Sack DA, Morris JG, Longini I, Siddique AK, Iqbal A, Huq A, Colwell RR, Sack RB, Stine OC (2014) Genetic analysis of Vibrio cholerae in the human population of two rural communities in Bangladesh during outbreaks. Emerg Infect Dis 20(1)
Raychoudhuri A, Patra T et al (2009) Classical ctxB in Vibrio cholerae O1, Kolkata, India. Emerg Infect Dis 15(1):131–132
Rudra S, Mahajan R et al (1996) Cluster of cases of clinical cholera due to Vibrio cholerae 010 in east Delhi. Indian J Med Res 103:71–73
Salemi M, Ali A, Azarian T, Johnson JA, Mohr D, Prosperi M, Veras NM, Jubair M, Strickland S, Allem MT, Weppelmann TA, Morris JG (2014) Phylodynamics of Vibrio cholerae O1 in haiti demonstrates evidence of sequential population bottlenecks driven by positive selection. Proc Nat Acad Sci USA
Salim A, Lan R et al (2005) Vibrio cholerae pathogenic clones. Emerg Infect Dis 11(11):1758–1760
Stine OC, Alam M et al (2008) Seasonal cholera from multiple small outbreaks, rural Bangladesh. Emerg Infect Dis 14(5):831–833
Waldor MK, Tschape H et al (1996) A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J Bacteriol 178(14):4157–4165
Wallace B (1981) Basic population genetics. Columbia University Press, New York
Acknowledgments
OCS & JGM are supported in part by RO-1A1039129 and RO1AI097405 from NIAID.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Stine, O.C., Morris, J.G. (2013). Circulation and Transmission of Clones of Vibrio cholerae During Cholera Outbreaks. In: Nair, G., Takeda, Y. (eds) Cholera Outbreaks. Current Topics in Microbiology and Immunology, vol 379. Springer, Berlin, Heidelberg. https://doi.org/10.1007/82_2013_360
Download citation
DOI: https://doi.org/10.1007/82_2013_360
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-55403-2
Online ISBN: 978-3-642-55404-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)