Abstract
Bipolar disorder (BP) is a highly heritable disease, with heritability estimated between 60 and 85% by twin studies. The underlying genetic architecture was poorly understood for years since the available technology was limited to the candidate gene approach that did not allow to explore the contribution of multiple loci throughout the genome. BP is a complex disorder, which pathogenesis is influenced by a number of genetic variants, each with small effect size, and environmental exposures. Genome-wide association studies (GWAS) provided meaningful insights into the genetics of BP, including replicated genetic variants, and allowed the development of novel multi-marker methods for gene/pathway analysis and for estimating the genetic overlap between BP and other traits. However, the existing GWAS had also relevant limitations. Notably insufficient statistical power and lack of consideration of rare variants, which may be responsible for the relatively low heritability explained (~20% in the largest GWAS) compared to twin studies. The availability of data from large biobanks and automated phenotyping from electronic health records or digital phenotyping represent key steps for providing samples with adequate power for genetic analysis. Next-generation sequencing is becoming more and more feasible in terms of costs, leading to the rapid growth in the number of samples with whole-genome or whole-exome sequence data. These recent and unprecedented resources are of key importance for a more comprehensive understanding of the specific genetic factors involved in BP and their mechanistic action in determining disease onset and prognosis.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Bipolar disorder
- Gene
- Genetics
- Genome-wide association study
- Heritability
- Polygenic risk score
- Polymorphism
1 Introduction: Why Genetics Matters in the Susceptibility to Bipolar Disorder?
Each human being is different from another one in many different ways, and it is such a common experience that we probably do not spend much time thinking about the underlying reasons. However, that diversity has fascinating biological bases explaining its reason for being there: nature has favored flexibility and change in order to make life adaptable to the environment. The way we respond to environmental stimuli is dependent from our genes, or, in other words, we all start our lives with a certain genetic makeup that defines our potential in terms of personality, intelligence, body shape, disease risk, and so on, and then the environment acts on this potential and leads to different outcomes in each one of us, at least for most of the traits. The percentage of contribution coming from genes is indeed variable, depending on the condition (trait or phenotype) that we consider. Some traits have complete penetrance, meaning that 100% of the subjects carrying a certain genetic variant or polymorphism exhibit the trait, independently from environmental exposures. For example, a specific polymorphism in the fibroblast growth factor receptor 3 (FGFR3) gene always results in achondroplasia, a disorder characterized by dwarfism. However, most of the traits have incomplete penetrance and variable expressivity, meaning that not all the subjects carrying certain genetic variant(s) manifest the trait, and in those who manifest it, there are different degrees of expressivity. These phenomena are caused not only by interactions with the environment but also by interactions with modifier genes (Lobo 2008). Traits showing incomplete penetrance and variable expressivity are indeed typically influenced by a number of genetic variants, and they are defined as complex or polygenic. Examples of this type of trait are intelligence, height, weight, and psychiatric disorders (Plomin and Deary 2015). Intelligence, height, and weight are normally distributed and can be quite easily interpreted as a result of the adaptation to the requests of different environments, but it is more difficult to interpret psychiatric disorders under an evolutionary perspective. However, psychiatric disorders can also be seen in a continuum spectrum, where the pathological manifestation is at one extreme of the distribution, and there are different degrees of subthreshold manifestations. Under this perspective, depressive and hypomanic subthreshold symptoms may confer an advantage in situations that require learning from negative experiences and creativity/high productivity, respectively. This mechanism maintains more or less constant the frequency of genetic variants conferring risk for potentially lethal diseases, such as sickle cell anemia and cystic fibrosis, wherein heterozygous individuals (one copy of the risk allele) may benefit from increased resistance to malaria and tuberculosis, respectively. However, homozygous individuals (two copies of the risk allele) develop a life-threatening disease. In the case of psychiatric disorders, of course, the genetic factors implicated are more complex; as discussed above they are not monogenic diseases, but the underlying theory explaining the maintenance of the risk variants in the population is comparable (Keller 2018). If there is not an individual genetic variant involved in the risk of bipolar disorder (BP), then how many variants or genes are involved? There is no definitive answer to this question, and different methodological approaches were applied, depending on the working hypothesis, as discussed in Sect. 3. However, as introductory information, it is useful to know that two individuals differ for five million genetic variants on average, the greatest number of which are represented by single nucleotide polymorphisms (SNPs, i.e. substitutions of a single DNA base pair). Copy number variations (CNVs, i.e. insertions or deletions of a certain number of base pairs) are less numerous than SNPs, but they are responsible for a higher percentage of inter-individual variability in terms of number of base pairs (The 1000 Genomes Project Consortium, 2015). SNPs are responsible for the lowest part of the variability between two individuals in terms of number of DNA base pairs, but they are more frequent in the general population (i.e., the same SNP is often found in more than 1% of the population). At the same time, the other types of genetic variants are rarer and somehow more unique of a relatively restricted group of individuals. For this reason, the largest part of existing studies was focused on investigating the role of SNPs in the susceptibility to BP. It is indeed much easier to design genetic studies looking at variants which are known to be represented in a population with a certain frequency instead of genotyping known rare variants which may not be seen at all in the studied sample or search for unknown rare variants by DNA sequencing. Whole-genome or whole-exome sequencing is becoming more and more feasible in the last years in terms of costs, but it is still marginally represented in the existing literature. The starting point for developing all the available methodological approaches to the study of the genetic basis of BP was however the same: the evidence of a high genetic component of this disorder found by family, adoption, and twin studies, as discussed in the next section.
2 Bipolar Disorder Is Heritable: Twin, Adoption, and Family Studies
The observation that some disorders recur within the same families for generations goes back in ancient times, starting from the Old Testament passages which say that God “punishes the children and their children for the sins of the fathers to the third and fourth generation” (Exodus 34:7). BP is one of those diseases which recurs among generations of the same family since it shows a high heritability as demonstrated by a number of studies published since the 1960s. Heritability estimates the degree of variation in a trait that is due to genetic variation between individuals, and it can vary between zero (no genetic component at all) and one (disorders with complete penetrance, for which the presence of a genotype is necessary and sufficient to manifest the trait). Traits with a heritability of one are often autosomal dominant, such as achondroplasia that was taken as an example in the “Introduction” section, meaning that the inheritance of one copy of the risk variant from one of the parents is associated with the manifestation of the trait. Thus, these traits are often seen in many generations within the same family. BP is not a monogenic trait, and it has incomplete penetrance. However, it has quite high heritability that was estimated to be between 60 and 85% by twin studies (Smoller and Finn 2003). Twin studies estimate the heritability of a trait by comparing the concordance of the trait between dizygotic twins (DZ, who share on average 50% of their genetic variation) and monozygotic twins (MZ, who share 100% of their genetic variation), providing a natural condition optimal for performing genetic studies. Assuming that shared environmental influences on MZ twins are not different from environmental influences on DZ twins, significantly higher concordance rates in MZ twins reflect the effect of genes. Other types of studies which contributed to estimating the genetic contribution to BP were adoption studies and family studies. Adoption studies compare the rate of a disorder in biological family members to those in adoptive family members in order to distinguish the genetic component from the environmental one. These studies are logistically difficult to conduct, and their availability is limited. However, they confirmed that there is a significantly higher rate of affective illness in the biological parents (31%) than in the adoptive parents (12%) of bipolar probands (Smoller and Finn 2003). Finally, family studies investigate if a disorder aggregates in families by comparing the prevalence of the disorder among first-degree relatives of affected probands (cases) to the prevalence in the population or among relatives of unaffected probands (controls). A relevant limitation of these studies consists in the fact that they cannot distinguish if a condition aggregating in families is caused by genetic or environmental factors or a combination of the two; however, they are still useful to estimate if there is an increased risk in relatives of cases compared to controls and to quantify this risk. According to the results of family studies, the recurrence risk of bipolar disorder for first-degree relatives of bipolar probands is 8.7%. In comparison, the risk for unipolar depression is 14.1%, indicating that positive family history for BP increases the risk of both bipolar and unipolar disorders, while interestingly family history of unipolar depression does not seem to increase the risk of BP (Smoller and Finn 2003). The estimated absolute risks correspond to a recurrence risk ratio for BP and major depressive disorder (MDD) in relatives of bipolar probands of around 4 and 2, respectively. Several studies have also demonstrated that clinical features might be associated with greater familiarity with BP. The most replicated are represented by early onset of the disease, presence of psychotic symptoms, and good lithium response. Early-onset BP may represent a more severe subtype with stronger genetic loading, as demonstrated by a greater familial risk of mood disorders in relatives of probands with early-onset BP, and it could also represent a distinct subtype of BP, characterized by a greater neurodevelopmental component compared to non-early-onset BP. Patients with early-onset BP and psychotic symptoms have more frequently cognitive impairment and neurological signs, reduced frontal gray matter at the time of their first psychotic episode, and greater brain changes than healthy controls, in a pattern similar to early-onset schizophrenia cases (Arango et al. 2014). The prepubertal onset of psychopathology was associated with a poorer response to lithium, and lithium responsiveness was reported to aggregate in families, supporting the hypothesis of a genetic component (Smoller and Finn 2003).
3 How Many Genes Modulate the Risk of Bipolar Disorder? Linkage Studies, Candidate Gene Studies, and Genome-Wide Association Studies
Starting from the evidence emerged from twin studies, the first experimental approach used to identify the specific genetic regions responsible for the heritability of BP was represented by linkage studies. These studies use information from family members who are affected and unaffected with the disorder and examine which genetic regions are coinherited with the disease within the family. More than 40 linkage scans for BP, including three meta-analyses, have been published, implicating many areas of the genome but with little consistency between studies. The strongest evidence was found for linkage on chromosomes 6q for BP type I and 8q for BP type I and type II; however, the genes responsible for these linkage signals have not been identified (Barnett and Smoller 2009). The main reason for these inconsistent and inconclusive findings is that BP has a polygenic architecture, while linkage studies work well when the genetic risk is conferred by a relatively small number of genes, such as for cystic fibrosis. Candidate gene association studies suffered from the same limitation. In this case, specific alleles in genes with a hypothesized link with the pathogenesis of BP are investigated in terms of possible different distribution between cases affected with BP and healthy controls. The most part of the investigated candidate genes plays a central role in the activity of the dopaminergic, serotoninergic, and glutamatergic pathways or the modulation of circadian rhythms. Examples of genes which have been associated with the risk of BP in independent samples or meta-analyses include disrupted in schizophrenia 1 (DISC1), D-amino acid oxidase activator (DAOA, or G72), the dopamine transporter (SLC6A3) and the serotonin transporter (SLC6A4), brain-derived neurotrophic factor (BDNF), the NMDA glutamate receptor subunit 2B (GRIN2B), the kainate class ionotropic glutamate receptor gene GRIK4, and neuregulin 1 (NRG1) (Barnett and Smoller 2009). However, none of these has been established as a BP susceptibility gene. Candidate gene association studies showed several relevant limitations: (1) earlier studies reported more often positive and larger effect associations than later studies, a phenomenon which is also referred to as “winner’s curse”; (2) most of the studies were performed on underpowered sample sizes and without applying appropriate multiple testing correction; and (3) many genes are involved in the risk of BP, part of which has likely no known connection with BP and cannot be investigated using the candidate gene method. Given these limitations, genome-wide association studies (GWAS) became the most common approach to the study of the genetics of complex disorders such as BP in the last 10 years. GWAS are based on a technology called microarray, which can genotype hundreds of thousands or millions of genetic variants cost-effectively, by multiplexed and parallel processing. GWAS include common genetic variants in a population, typically those variants having a frequency above 1%. As noted in the “Introduction” section, these common variants are usually represented by SNPs, which can be studied in an easier way than rare variants (in terms of costs, time, and sample size needed). GWAS have been successful for identifying susceptibility alleles for a broad range of common complex disorders including diabetes, cardiovascular disease, prostate and breast cancer, and many others. The early GWAS of BP did not identify any loci achieving genome-wide significance, that is, a p threshold <5 × 10−8, which takes into account multiple testing correction. The top variants emerging from these early GWAS included DGKH, coding for diacylglycerol kinase eta, an enzyme involved in modulating the effects of mood stabilizers, and CACNA1C, coding for the alpha-1C subunit of the L-type voltage-gated calcium channel, which was then confirmed by later studies (Barnett and Smoller 2009). The main issue of GWAS is represented by the need for large samples to have adequate power to detect small effect sizes at the genome-wide significance threshold. Recent studies indeed showed that most of the genome-wide significant loci have an effect size measured as odds ratio (OR) ≤ 1.1 (Stahl et al. 2019). In order to have an adequate power (i.e., ≥80%) for detecting this magnitude of effect size for a common variant with the frequency of 10% in a population, a sample of about one million subjects was estimated to be needed (Visscher et al. 2017), while currently, the largest GWAS of BP included 20,352 cases and 31,358 controls (Stahl et al. 2019). This study identified 30 independent loci associated with BP at the genome-wide significance threshold that is expected to be a much lower number compared to those that would be identified in a sample size providing adequate power. The significant loci contain genes encoding ion channels and transporters (e.g., CACNA1C, SCN2A (sodium voltage-gated channel alpha subunit 2), SLC4A1 (solute carrier family 4 member 1)), neurotransmitter receptors (e.g., GRIN2A), and synaptic components (e.g., ANK3 (ankyrin 3) and RIMS1 (regulating synaptic membrane exocytosis 1)) (Stahl et al. 2019). These processes are important in neuronal hyperexcitability, which is a pathogenetic mechanism implicated in BP using pluripotent stem cell-derived neurons from patients, and by the fact that the reduction of neuronal hyperexcitability is one of the mechanisms of action of mood stabilizers such as lithium (Mertens et al. 2015). Other significant loci which were reported in earlier GWAS were within the TRANK1 (tetratricopeptide repeat and ankyrin repeat containing 1), NCAN (neurocan), and TENM4 (teneurin transmembrane protein 4) genes. In the same study, the integration of the GWAS results with information on SNPs modulating gene expression implicated GLT8D1 (glycosyltransferase 8 domain containing 1), which is involved in proliferation and differentiation of neural stem cells. Pathway analyses reveal genetic evidence for insulin secretion and endocannabinoid signaling. Top genes in these pathways included calcium and potassium channel subunits, MAP kinase, and GABA-A receptor subunit genes. The SNP-based heritability estimated by this study was 17–23% depending from the considered population prevalence of BP (between 0.5 and 2%), and heritability estimates were higher for BP type I than BP type II (25% vs 11%) (Stahl et al. 2019). These heritability estimates are much lower than the heritability estimated by twin studies, because they are based on common genetic variants only. An overview of the significant findings of GWAS is provided in Table 1.
4 Genetic Overlap Between Bipolar Disorder and Other Brain Disorders: Disorder-Specific or General Genetic Influences?
New methods recently developed to study the genetics of complex traits estimate the genetic correlation between different traits and the predictive ability of the genetics of one trait on another one. One of the most used approaches is linkage disequilibrium score regression (LDSR) because it can be performed using GWAS summary statistics (no need for raw genotype data). LDSR was initially developed to differentiate between GWAS p-values inflation caused by polygenicity and by confounding effects, such as population stratification. Other applications were developed; in particular, the method can be used to estimate genetic correlations between different traits. For providing these estimations, LDSR uses the test statistics of the SNPs from a GWAS (summary statistics) and the degree of linkage disequilibrium (LD) between SNPs. LD refers to the nonrandom association of alleles at two or more loci in a population, or, in other words, to the fact that two or more alleles are found together more often than expected by chance, and this phenomenon is influenced by different factors (e.g., rate of genetic recombination, mutation rate). The genetic variants associated with BP are expected to show a high degree of LD among each other (LD score), because the trait is polygenic, and for each associated SNPs, a number of nearby SNPs is expected to show association signals as well, inflating the GWAS statistics. In contrast, inflation caused by other factors would not be associated with LD score. Thus, the regression of the SNP test statistics from a GWAS against the LD score provides an estimation of the polygenicity of a trait and was used to confirm the polygenic architecture of psychiatric disorders (Bulik-Sullivan et al. 2015a). The same approach can be adapted to estimate the genetic correlation between two traits (Bulik-Sullivan et al. 2015b), and it has been largely applied in the field of neuropsychiatry.
Using LDSR, BP was demonstrated to be genetically correlated with other psychiatric disorders, particularly with schizophrenia (SCZ) and secondly with MDD and obsessive-compulsive disorder, while less strongly with anorexia nervosa and autism spectrum disorders (Stahl et al. 2019; Brainstorm Consortium et al. 2018). BP type I was estimated to be more strongly genetically correlated with SCZ, while BP type II was more strongly genetically correlated with MDD. The genetic correlation between BP and SCZ was estimated to be 0.70, supporting a widely shared genetic predisposition. In a large GWAS of SCZ and BP, 114 independent genetic loci were found associated with both disorders, and genetic pathways implicated in the common genetic susceptibility were modulators of neuron projection development, synaptic plasticity, and neurogenesis. On the other hand, response to potassium ion was reported to be the only pathway associated with SCZ only and not BP (Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium 2018).
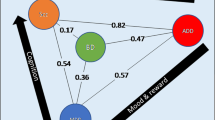
Moving beyond psychiatric disorders, positive genetic correlations were reported for BP and education years, as well as college attendance, but not with either adult or childhood IQ, suggesting that the role of BP genetics in educational attainment may be independent of general intelligence (Stahl et al. 2019). This finding is consistent with evidence obtained in clinical samples, suggesting that IQ in BP may be affected by the onset and progression of the disease, while in the pre-onset phase, prodromal symptoms like elevated energy may contribute to good educational attainment (Vreeker et al. 2016). BP does not seem to be genetically correlated with a number of neurological diseases, such as Alzheimer’s disease, Parkinson’s disease, multiple sclerosis, and ischemic stroke, despite a nonsignificant trend of positive genetic correlation was reported with the last (Brainstorm Consortium et al. 2018). An overview of the genetic correlations of BP with other disorders is reported in Fig. 1.
Barplot representing the genetic correlations between bipolar disorder and other psychiatric and nonpsychiatric traits. Genetic correlations were estimated using linkage disequilibrium score regression (LDSR), and references to the corresponding literature are reported in Sect. 4. AN anorexia nervosa, ASD autism spectrum disorder, college_attend college attendance, dep_symptoms depressive symptoms, edu_years years of education, MDD major depressive disorder, OCD obsessive-compulsive disorder, SCZ schizophrenia, subj_wellbeing subjective well-being
Another approach used to estimate the genetic overlap between two traits is represented by polygenic risk scores (PRS). This method, contrary to LDSR, requires raw genotypes, though some recent methods can be applied to summary statistics. PRS are used to estimate if the genetics of one trait (according to the GWAS results obtained in a base sample) is able to predict the same trait or a different trait in an independent sample (target sample) and the strength of prediction. PRS are calculated in the target sample as the sum of the risk alleles associated with the trait weighted by their effect size calculated in the base sample. This approach contributed to a better understanding of the genetic overlap between BP and transdiagnostic clinical dimensions. Among 24 clinical dimensions or characteristics, a positive correlation was reported between BP PRS and manic symptoms in SCZ, between SCZ PRS and psychosis in BP cases, and between BP+SCZ PRS and psychotic features in BP and negative symptoms in SCZ (Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium 2018). Overall, these findings support the hypothesis that the main clinical dimension associated with the shared genetic liability between BP and SCZ is represented by psychosis. Consistently, across a number of disorders, including BP, SCZ PRS was shown to predict the risk of psychotic symptoms, despite the phenotypic variance explained was very small (4.4%) (Calafato et al. 2018).
5 The Role of Rare Genetic Variants
Large (>100 kb), rare copy number variants (CNVs) have been shown to confer risk for SCZ and other neurodevelopmental disorders, while the available evidence suggests that this type of variant is less strongly involved in the risk of BP. The strongest evidence for association with BP was obtained for duplications at 16p11.2, which were found in 0.13% of BP cases compared with 0.03% of healthy controls. Duplications at ATF7IP2, upstream of GRIN2A, a gene associated with BP by GWAS, and at the gene CGNL1 (cingulin-like 1) were also reported as risk factors. The burden of very large (>500 kb) CNVs in BP was shown to be not different from controls, contrary to what was observed in SCZ (Green et al. 2016).
Whole-exome sequencing in families including affected individuals led to the identification of rare variants segregating with BP in the G protein-coupled receptor (GPCR) family genes, including the corticotropin-releasing hormone receptor 2 (CRHR2) gene and the metabotropic glutamate receptor 1 (GRM1). GPCR are involved in the transduction of cellular signals, in response to stimuli such as neurotransmitters and neuropeptides. Receptors that activate G proteins include serotonin, glutamatergic, and dopaminergic receptors (e.g., HTR1B, GRM1, GRM4, and DRD5). Through the activation of G proteins, these receptors modulate two major signaling pathways, cAMP and phosphatidylinositol signaling pathways, both of which are associated with the pathophysiology of BP and the mechanism of action of drugs commonly prescribed in BP (Cruceanu et al. 2018).
Rare damaging variants were hypothesized to play a role in severe BP cases comorbid with other neuropsychiatric disorders, particularly those having a neurodevelopmental component, such as attention deficit hyperactivity disorder, seizure disorders, and learning disabilities. In such cases, rare variants damaging the protein structure or function were identified in affected individuals and not in controls, particularly in genes coding for proteins with GTPase-activating function which are among the targets of lithium (Rao et al. 2017). CNVs in the neurexin 1 (NRXN1) gene have been associated with BP comorbid with intellectual disability and have replicated associations with cognitive ability, autism, and SCZ. Neurexins are crucial for the regulation of neurotransmission and the formation of synaptic contacts. Thus, they are hypothesized to play a role in neurodevelopment (Viñas-Jornet et al. 2014, 2018).
6 Gene × Environment Studies
Apart from genetic risk factors, a number of environmental exposures including urbanicity, stressful life events, early life stress, and substance abuse may underlie the development of BP. Environmental risk factors are hypothesized to interact with the individual’s genetic predisposition to BP, and they may partly explain the incomplete penetrance of the disorder. Gene x environment (G × E) studies aim to understand how different genotypes affect the risk of developing a disorder depending on the exposure to environmental factors. Most of the G × E studies were focused on variants in candidate genes rather than multiple variants across the genome (GWAS), and BP was under-investigated compared to other psychiatric disorders, leading to inconclusive results so far. Most studies considered the impact of childhood trauma in interaction with the rs6265 SNP of the brain-derived neurotrophic factor (BDNF) gene or SNPs in genes regulating the monoaminergic neurotransmitter system (the 5-HTTLPR variant of the SLC6A4 gene and the rs4680 SNP in the catechol-O-methyltransferase (COMT) gene). However, none of these findings was convincingly replicated, and the effect of potential confounders was implicated as a possible reason (Misiak et al. 2018).
More extensive G × E research has been conducted on the risk of developing a psychotic disorder, including but not limited to a psychotic affective disorder. A strong candidate gene which has modulating effect on the risk of psychosis in cannabis users and has been replicated is serine/threonine kinase 1 (AKT1), a gene encoding a serine/threonine kinase involved in the transduction of signal following cannabinoid receptor activation. Another group of studies used individual’s family history and environmental exposures to derive a proxy of G × E interaction. Individuals with a family history of psychotic illness appear to be particularly sensitive to the effects of multiple environmental risk factors, including cannabis use and urban upbringing. However, positive family history can also be affected by environmental factors and not only genetics; furthermore, genetic variants that increase sensitivity to the environment may be distinct from genetic variants that directly increase the risk of illness. The demonstration corroborated this last hypothesis that a PRS of overall sensitivity to the environment (both positive and negative) predicted both the effects of parenting on emotional problems and response to psychological treatment among children with anxiety disorders. However, it did not directly predict psychopathology (Zwicker et al. 2018). Further research is needed to understand how genetic variants modulate psychopathology in interaction with the environment, particularly in BP, which has been under-investigated so far.
7 Nongenetic Mechanisms Contributing to the Regulation of Gene Expression: Epigenetics
Gene expression is modulated not only by genetic variants affecting gene transcription or transduction in proteins but also through fluid mechanisms that can respond to internal or external stimuli, in a way to adapt gene expression profiles to the environment, namely, epigenetics. Epigenetics may partly explain the mechanism by which environmental factors modulate the risk of BP. Examples of epigenetic mechanisms of gene expression regulation are DNA methylation and histone modification. DNA methylation consists in the addition of a methyl (CH3) group to the carbon-5 of cytosines in the DNA sequence, generating a modified nucleotide called 5-methylcytosine (5mC). DNA methylation occurs throughout the entire genome at cytosines of CpG dinucleotides, which are frequently enriched around promoter regions, in so-called CpG islands. These regions are responsible for DNA methylation-dependent control of gene promoter activity, and higher methylation is typically associated with gene repression. Mechanistically, it represses the activation of a promoter by either sterically inhibiting the binding of transcription factors or by actively recruiting repressor proteins such as histone deacetylases (HDAC), which will ultimately lead to the formation of heterochromatin (a highly condensed form of DNA which is not accessible by transcription factors). DNA methylation patterns are heritable, possibly explaining part of the heritability of BP. However, they are also modulated by environmental exposure through the activity of a specific group of enzymes called DNA methyltransferases (DNMTs) and a number of translocation (TET) family of proteins, responsible for DNA methylation and demethylation, respectively.
The published studies were heterogeneous in terms of the applied methodology to evaluate DNA methylation (type of assay), genes which were tested (global genome methylation, methylation in selected candidate genes, or genome-wide methylation), and type of tissue used (blood in most studies, but in some cases postmortem brain tissues). DNA methylation is tissue-specific, but high concordance between blood and brain was reported, suggesting the utility of peripheral blood as a proxy of brain tissues. Studies examining global genome methylation in BP compared to healthy controls did not find consistent findings. Moreover, the interpretation of alternations in global methylation levels would not be easy under the pathogenetic point of view. The most investigated candidate genes were in line with those reported by studies looking at genetic variation, namely, BDNF; monoaminergic genes, particularly SCL6A4; the serotonin receptors 1A, 2A, and 3A (HTR1A, HTR2A, and HTR3A); and the membrane-bound catechol-O-methyltransferase (COMT). In addition, methylation in the dystrobrevin-binding protein 1 (DTNBP1) gene was implicated in BP, which product is involved in the modulation of glutamatergic neurotransmission by influencing exocytotic glutamate release. Methylation candidate gene association studies had sparse and often not replicated findings, mainly because they do not capture the polygenicity of the disease, as discussed in Sect. 3. Genome-wide methylation studies did not provide encouraging findings either since they did not identify replicated findings. They used different arrays and different preprocessing and analysis methods and considered different tissues, making them poorly comparable. However, an interesting finding was that differentially methylated regions between BP and controls depend from the brain region considered. For example, in the frontal cortex (particularly in Brodmann’s area 9) and anterior cingulate, only a few differentially methylated regions overlapped with promoters, whereas a greater proportion occurred in introns and intergenic regions (Fries et al. 2016). These noncoding regions may contribute to gene expression regulation since they were found to overlap with long intergenic noncoding RNAs (lincRNAs) and microRNAs. The function of these noncoding RNAs is only partially known, but they are hypothesized to regulate gene expression by interacting with transcription factors and by regulating posttranscriptional processing, such as mRNA transport, translation, and degradation (Ransohoff et al. 2018).
The role of epigenetics in the pathogenesis of BP remains poorly understood so far, in terms of genetic regions/genes showing abnormal methylation, differences between tissues, and causal mechanisms responsible for the different methylation patterns seen in BP compared to controls. A possible mechanism that may partially be responsible for alterations in methylation in BP is represented by overexpression of DNMT1 found by several independent studies in Brodmann’s area 9, which was associated with the downregulation of specific GABAergic and glutamatergic genes (Fries et al. 2016). Under this perspective, DNMT1 may be the target of future new treatments.
8 Current and Future Lines of Research
From the previous sections of this chapter, it is quite evident that a strong genetic basis of BP has been established, but the specific genetic variants or regions, as well as the mechanistic process through which they lead to the disease, are still largely unknown. However, a number of new and promising research resources have emerged in the last years.
The recent improvements in genotyping/sequencing technologies but also in the available storing and computational resources allowed the creation and growth of large biobanks in a number of countries, such as the UK Biobank in the UK and All of Us in the USA (UK Biobank 2019) (National Institute of Health (NIH) 2019). These initiatives collected a wide range of health-related measures and biomarkers in hundreds of thousand subjects from the general population, in order to study the individual (genetic and nongenetic) factors associated with the development of diseases, but also with well-being, lifestyles, and other traits of interest such as aging. Thus, not only disease risk factors can be identified, but it is possible also to study those variables modulating disease onset and prognosis and, in this way, develop preventive strategies. These unprecedented resources have been available in the last few years and are in an expansion phase, in terms of number of subjects but also phenotypic information, genetic data, and other biomarkers (e.g., brain imaging). For example, whole-exome sequencing is expected to be completed in all 500,000 subjects included in UK Biobank at the end of 2020, providing the complete DNA sequence of the coding regions of the genome in a sample of unprecedented size and richness of phenotypic characterization. Digital phenotyping has been increasingly used to collect phenotype data from personal digital devices, for example, for remotely monitoring psychiatric symptoms over time and collecting activity data (Insel 2018). Another very powerful resource for extracting phenotypic information is represented by electronic health records (EHR), which typically include both structured information (e.g., diagnostic codes) and narrative notes from which the information of interest can be automatically extracted using natural language processing. Automated definitions of BP using three rule-based classifiers based on information extracted from EHR were demonstrated to have genetic correlations of 0.66, 0.74, and 1 (the most stringent definition) with a traditional interview-based diagnosis of BP, confirming that EHR can be used for automated definition of diagnosis (Chen et al. 2018).
Data from different biobanks are more and more often included in collaborative research efforts, thanks to international consortia, such as the Psychiatric Genomics Consortium (PGC), that made possible the largest GWAS of BP so far (Stahl et al. 2019), and it is actively committed to the recruitment of more samples and the development of new analysis methods. The last point has been object of particular interest since the analysis approaches used in psychiatric genetics had suffered from important limitations for many years. A poor understanding of the genetic architecture of BP and other psychiatric disorders, alongside with limited technological resources, had restricted the existing research to the study of candidate genes and individual variant effects for years. The drop in the costs of genotyping and next-generation sequencing has played a fundamental role in the shift from individual genes and individual variants to the study of the combined effect of a number of variants across the genome. Methods such as LDSR and PRS are examples of analysis approaches reflecting the polygenicity of complex traits, being able to incorporate the effects of all the SNPs with evidence of association with a trait. Future improvements of polygenic analysis methods are going to (a) better estimate the effects of individual variants contributing to a trait, (b) take into account interaction effects, and (c) include also rare genetic variants, which analysis has been challenging.
Another recent interest in psychiatric genetics has been the identification of causality between traits. Genetics can indeed contribute to understanding the causal relationship between traits that are commonly seen together, but where there is no clear knowledge indicating which one is causing the other or if they have independent causes. Mendelian randomization (MR) allows to test this kind of hypothesis on observational data, using genetic variants randomly allocated at conception as instrumental variables for a modifiable exposure (or trait) of interest. MR is analogous to an RCT where instead of the allocation of participants to different treatment groups, individuals are randomized by nature to carry or not carry genetic variants that may modify the risk of exposure. MR uses genetic variants associated with exposure (hypothetical causal trait) as instruments to infer causality on an outcome. For a genetic variant to be a valid instrument, three assumptions must be satisfied: (1) the genetic variant is strongly associated with the exposure of interest (at the genome-wide level); (2) the genetic variant is not associated with any confounder of the exposure-outcome association; and (3) the genetic variant is only associated with the outcome through the exposure (Pagoni et al. 2019). Interestingly, MR was used to assess the causal relationship between BP and a number of cardiometabolic traits, being increased rates of cardiovascular diseases a major concern in patients with BP. Using BP as the exposure, no causal relationship with cardiometabolic traits was identified, suggesting that cardiometabolic abnormalities in these patients may be mostly secondary (e.g., to lifestyles and medications) and not related to the underlying pathophysiology of BP (So et al. 2019). These results further corroborate the importance of preventing and monitoring environmental risk factors of cardiovascular disease in BP.
9 Conclusion
Notable progress has been achieved toward a better understanding of the genetics of BP in the last years, thanks to technological advances and development of new analysis methods. The genetic pathways mediating the disease, as well as the genetic overlap of BP with other disorders or traits, have been partially elucidated. However, the genetic contribution to BP and its interactions with the environment are still not completely understood, and the available scientific findings have no produced clinical applications so far.
The use of PRS to predict the risk of psychiatric disorders is currently one of the most promising future applications, and it has attracted the interest of public and private enterprises. The PRS for a trait can be converted into a standardized score that follows a normal distribution, with higher PRS corresponding to higher risk, in a way that could be used to determine an individual’s risk of the corresponding trait based on his/her position on this distribution. Individuals falling above a certain threshold could be informed of their risk and benefit from preventive strategies. It is unclear how extreme a score would have to be to achieve clinical relevance. However, it was speculated that a PRS in the top 1–5% of the population would warrant feedback (Lewis and Vassos 2017) (Fig. 2). PRS could be used as a screening tool to identify individuals at risk and allow an evidence-based allocation of health-care resources (e.g., clinical monitoring, education about the environmental risk factors associated with the disease). PRSs are currently able to explain between 1 and 15% of the variation between cases and controls (Visscher et al. 2017); however in an individual at the top end of the risk distribution (top 1–5%), the risk could be significantly increased compared to the general population. Apart from the need for further data for assessing the clinical relevance of PRS in psychiatry, other issues should be addressed in order to develop clinical applications of psychiatric genetics. The most relevant of them are represented by ethical concerns, education of physicians, and availability of tools for standard interpretation of genetic results and standardized corresponding clinical interventions. Potential ethical concerns include misinterpretation of findings due to insufficient education and information, stigma and discrimination, commercial use of PRS having unclear clinical relevance as direct-to-consumer product, and PRS use for embryo selection (eugenics). Another issue that GWAS have only recently started to address is the inclusion of people of non-European ancestry, in order to improve our knowledge about the genetic variants that may be specifically observed only in some ethnic groups and relevant to disease risk (Sullivan et al. 2018).
Example of bipolar disorder polygenic risk score (PRS) distribution. Subjects in the highest 1–5% of the PRS distribution are those who might warrant feedback and might benefit from preventive interventions. This plot is meant to serve as a conceptual example and does not reflect the distribution of real data
Hopefully, the valuable resources available to researchers within biobanks and consortia will contribute in solving the still unanswered questions and to the translation of genetic findings in clinical applications with demonstrated beneficial effects to the individual and the society. Appropriate regulations about the consented use of genetic information should be developed in order to avoid discrimination and nonethical uses.
References
Arango C, Fraguas D, Parellada M (2014) Differential neurodevelopmental trajectories in patients with early-onset bipolar and schizophrenia disorders. Schizophr Bull 40(Suppl 2):S138–S146
Barnett JH, Smoller JW (2009) The genetics of bipolar disorder. Neuroscience 164(1):331–343
Baum AE, Akula N, Cabanero M, Cardona I, Corona W, Klemens B et al (2008a) A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry 13(2):197–207
Baum AE, Hamshere M, Green E, Cichon S, Rietschel M, Noethen MM et al (2008b) Meta-analysis of two genome-wide association studies of bipolar disorder reveals important points of agreement. Mol Psychiatry 13(5):466–467
Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium (2018) Genomic dissection of bipolar disorder and schizophrenia, including 28 subphenotypes. Cell 173(7):1705–1715.e16
Brainstorm Consortium, Anttila V, Bulik-Sullivan B, Finucane HK, Walters RK, Bras J et al (2018) Analysis of shared heritability in common disorders of the brain. Science 360(6395):eaap8757
Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Patterson N et al (2015a) LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47(3):291–295
Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh P-R et al (2015b) An atlas of genetic correlations across human diseases and traits. Nat Genet 47(11):1236–1241
Calafato MS, Thygesen JH, Ranlund S, Zartaloudi E, Cahn W, Crespo-Facorro B et al (2018) Use of schizophrenia and bipolar disorder polygenic risk scores to identify psychotic disorders. Br J Psychiatry 213(3):535–541
Charney AW, Ruderfer DM, Stahl EA, Moran JL, Chambert K, Belliveau RA et al (2017) Evidence for genetic heterogeneity between clinical subtypes of bipolar disorder. Transl Psychiatry 7(1):e993
Chen DT, Jiang X, Akula N, Shugart YY, Wendland JR, Steele CJM et al (2013) Genome-wide association study meta-analysis of European and Asian-ancestry samples identifies three novel loci associated with bipolar disorder. Mol Psychiatry 18(2):195–205
Chen C-Y, Lee PH, Castro VM, Minnier J, Charney AW, Stahl EA et al (2018) Genetic validation of bipolar disorder identified by automated phenotyping using electronic health records. Transl Psychiatry 8(1):86
Cichon S, Mühleisen TW, Degenhardt FA, Mattheisen M, Miró X, Strohmaier J et al (2011) Genome-wide association study identifies genetic variation in neurocan as a susceptibility factor for bipolar disorder. Am J Hum Genet 88(3):372–381
Cruceanu C, Schmouth J-F, Torres-Platas SG, Lopez JP, Ambalavanan A, Darcq E et al (2018) Rare susceptibility variants for bipolar disorder suggest a role for G protein-coupled receptors. Mol Psychiatry 23(10):2050–2056
Ferreira MAR, O’Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L et al (2008) Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet 40(9):1056–1058
Fries GR, Li Q, McAlpin B, Rein T, Walss-Bass C, Soares JC et al (2016) The role of DNA methylation in the pathophysiology and treatment of bipolar disorder. Neurosci Biobehav Rev 68:474–488
Green EK, Hamshere M, Forty L, Gordon-Smith K, Fraser C, Russell E et al (2013) Replication of bipolar disorder susceptibility alleles and identification of two novel genome-wide significant associations in a new bipolar disorder case-control sample. Mol Psychiatry 18(12):1302–1307
Green EK, Rees E, Walters JTR, Smith K-G, Forty L, Grozeva D et al (2016) Copy number variation in bipolar disorder. Mol Psychiatry 21(1):89–93
Hou L, Bergen SE, Akula N, Song J, Hultman CM, Landén M et al (2016) Genome-wide association study of 40,000 individuals identifies two novel loci associated with bipolar disorder. Hum Mol Genet 25(15):3383–3394
Ikeda M, Takahashi A, Kamatani Y, Okahisa Y, Kunugi H, Mori N et al (2018) A genome-wide association study identifies two novel susceptibility loci and trans population polygenicity associated with bipolar disorder. Mol Psychiatry 23(3):639–647
Insel TR (2018) Digital phenotyping: a global tool for psychiatry. World Psychiatry 17(3):276–277
Keller MC (2018) Evolutionary perspectives on genetic and environmental risk factors for psychiatric disorders. Annu Rev Clin Psychol 14:471–493
Lewis CM, Vassos E (2017) Prospects for using risk scores in polygenic medicine. Genome Med 9(1):96
Lobo I (2008) Three individuals carry the same disease-causing mutation; two suffer from the disease but exhibit different symptoms, while the third is completely unaffected. Why? Nat Educ 1(1):64
Mertens J, Wang Q-W, Kim Y, Yu DX, Pham S, Yang B et al (2015) Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature 527(7576):95–99
Misiak B, Stramecki F, Gawęda Ł, Prochwicz K, Sąsiadek MM, Moustafa AA et al (2018) Interactions between variation in candidate genes and environmental factors in the etiology of schizophrenia and bipolar disorder: a systematic review. Mol Neurobiol 55(6):5075–5100
Mühleisen TW, Leber M, Schulze TG, Strohmaier J, Degenhardt F, Treutlein J et al (2014) Genome-wide association study reveals two new risk loci for bipolar disorder. Nat Commun 5:3339
National Institute of Health (NIH) (2019) All of us. https://www.joinallofus.org/en/biobank. Accessed 21 Nov 2019
Pagoni P, Dimou NL, Murphy N, Stergiakouli E (2019) Using Mendelian randomisation to assess causality in observational studies. Evid Based Ment Health 22(2):67–71
Plomin R, Deary IJ (2015) Genetics and intelligence differences: five special findings. Mol Psychiatry 20(1):98–108
Psychiatric GWAS Consortium Bipolar Disorder Working Group (2011) Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet 43(10):977–983
Ransohoff JD, Wei Y, Khavari PA (2018) The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol 19(3):143–157
Rao AR, Yourshaw M, Christensen B, Nelson SF, Kerner B (2017) Rare deleterious mutations are associated with disease in bipolar disorder families. Mol Psychiatry 22(7):1009–1014
Scott LJ, Muglia P, Kong XQ, Guan W, Flickinger M, Upmanyu R et al (2009) Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proc Natl Acad Sci U S A 106(18):7501–7506
Sklar P, Smoller JW, Fan J, MAR F, Perlis RH, Chambert K et al (2008) Whole-genome association study of bipolar disorder. Mol Psychiatry 13(6):558–569
Smith EN, Bloss CS, Badner JA, Barrett T, Belmonte PL, Berrettini W et al (2009) Genome-wide association study of bipolar disorder in European American and African American individuals. Mol Psychiatry 14(8):755–763
Smoller JW, Finn CT (2003) Family, twin, and adoption studies of bipolar disorder. Am J Med Genet C Semin Med Genet 123C(1):48–58
So H-C, Chau K-L, Ao F-K, Mo C-H, Sham P-C (2019) Exploring shared genetic bases and causal relationships of schizophrenia and bipolar disorder with 28 cardiovascular and metabolic traits. Psychol Med 49(8):1286–1298
Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V et al (2019) Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet 51(5):793–803
Sullivan PF, Agrawal A, Bulik CM, Andreassen OA, Børglum AD, Breen G et al (2018) Psychiatric genomics: an update and an agenda. Am J Psychiatry 175(1):15–27
The 1000 Genomes Project Consortium (2015) A global reference for human genetic variation. Nature 526(7571): 68–74
UK Biobank (2019) UK Biobank: improving the health of future generations. https://www.ukbiobank.ac.uk. Accessed 21 Nov 2019
Viñas-Jornet M, Esteba-Castillo S, Gabau E, Ribas-Vidal N, Baena N, San J et al (2014) A common cognitive, psychiatric, and dysmorphic phenotype in carriers of NRXN1 deletion. Mol Genet Genomic Med 2(6):512–521
Viñas-Jornet M, Esteba-Castillo S, Baena N, Ribas-Vidal N, Ruiz A, Torrents-Rodas D et al (2018) High incidence of copy number variants in adults with intellectual disability and co-morbid psychiatric disorders. Behav Genet 48(4):323–336
Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA et al (2017) 10 years of GWAS discovery: biology, function, and translation. Am J Hum Genet 101(1):5–22
Vreeker A, Boks MPM, Abramovic L, Verkooijen S, van Bergen AH, Hillegers MHJ et al (2016) High educational performance is a distinctive feature of bipolar disorder: a study on cognition in bipolar disorder, schizophrenia patients, relatives and controls. Psychol Med 46(4):807–818
Wellcome Trust Case Control Consortium (2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447(7145):661–678
Zwicker A, Denovan-Wright EM, Uher R (2018) Gene-environment interplay in the etiology of psychosis. Psychol Med 48(12):1925–1936
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Fabbri, C. (2020). The Role of Genetics in Bipolar Disorder. In: Young, A.H., Juruena, M.F. (eds) Bipolar Disorder: From Neuroscience to Treatment. Current Topics in Behavioral Neurosciences, vol 48. Springer, Cham. https://doi.org/10.1007/7854_2020_153
Download citation
DOI: https://doi.org/10.1007/7854_2020_153
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-72142-8
Online ISBN: 978-3-030-72143-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)