Abstract
In addition to his pioneering work utilizing quantitative fluorescence techniques to study biological macromolecules and assemblies, Gregorio Weber contributed substantially to our understanding of the thermodynamic relationships between different ligand-binding equilibria and subunit association equilibria in proteins. These ideas have proven to be very valuable in clarifying the basis for allosteric modification of enzyme behavior when allosteric ligands act predominantly to modify the affinity of an enzyme’s substrate. Dr. Weber’s influence, both scientific and personal, on the author’s efforts to understand the allosteric behavior of prokaryotic and eukaryotic phosphofructokinase (PFK) in particular is described. These observations not only serve to illuminate the regulatory properties of this important enzyme, but they also, in turn, serve to illustrate the validity of the principles that Weber originally described. Not surprisingly fluorescence techniques have played important roles in elucidating many of these insights.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Allosteric regulation
- Coupling free energy
- Entropy domination
- Fluorescence polarization
- Phosphofructokinase
1 Early Years
On paper, at least that which contains my CV, my connection with Gregorio Weber seems tenuous as my only formal relationship with him stems from the single year I spent in his lab as an undergraduate researcher my senior year from 1972 to 1973. However, the impact of my association with “the Professor” could not have been more profound or formative for my eventual career. This dichotomy is made all the more ironic when I reflect on my utter intimidation at the time that caused me to adopt a primarily observational role in the lab while meekly pursuing my own experiments. Never before had I been exposed to so many homemade precision scientific instruments and, more importantly, so many individuals engaged in such high-level scientific discourse. And I must confess, it took me a while to get used to the accent…
My project was to synthesize a fluorescent probe, chrysenebutyric acid, that could be used in a manner similar to pyrenebutyric that Weber and Knopp [1, 2] had recently put forth as a long-lifetime probe suitable for studying very large proteins. Chrysenebutyric acid was expected to have a shorter, but still relatively long, lifetime that could be used for studying somewhat smaller proteins.
Eventually I succeeded with the synthesis and therefore needed to venture into the realm of fluorescence characterization using the aforementioned homebuilt instruments. Fortunately (for me anyway) Dave Jameson, then a second year graduate student in the lab, took me under his wing at that point, and through his efforts I was able to utilize most of the various spectrometers, spectrofluorometers, and fluorometers in the lab. I remember in particular collecting the fluorescence polarization data, that were eventually to appear as Figure 11 in my undergraduate thesis, with a sample of chrysenebutyric acid in propylene glycol at –55°C on the T-format polarization instrument designed and built by Weber and Bablouzian some years before [3]. What made this experiment particularly memorable was that Dave and I collected the data in the wee hours of a Saturday night/Sunday morning – thus defining “nerd-like” behavior long before the term was in common use. The most lasting benefit of this exercise for me, though, was the concomitant discussions with Dave, and subsequently Dr. Weber, about the meaning and significance of fluorescence polarization.
Fast forward to the latter part of my graduate career that I spent, largely at Dr. Weber’s recommendation, at the University of Wisconsin in the laboratory of Dr. Henry Lardy. Unbeknownst to me, a postdoc had begun lobbying for the lab to acquire a research-grade spectrofluorometer better than the old Aminco-Bowman that was then ubiquitous in non-fluorescence-centric research labs such as ours. Dr. Lardy had delegated the task of new instrument acquisition to a research scientist, Fritz Stratman, in the lab (it was a big lab), and one day Fritz came up to me and asked if I knew anything about the company “SLM” that was headquartered in Urbana and the instrument they had just put on the market – a state-of-the-art photon-counting spectrofluorometer. After quickly connecting a couple of dots in my mind, I replied: “Not specifically, but if the ‘S’ stands for Spenser and the ‘M’ for Mitchell, I’m sure it’s a good instrument.” Dick Spenser and George Mitchell had been postdocs in Weber’s lab when I was there, and Dick and Dave had been working on applying the then new technology of photon-counting detection to a scanning spectrofluorometer, the end result of which had just been described [4]. (Dick Spenser had previously developed the first 2-frequency phase/modulation fluorometer as a graduate student [5]. I had used prototypes of each of these instruments to characterize the chrysenebutyric acid I synthesized, along with the polarization instrument built by Weber and Bablouzian.) My surmise proved to be correct (the ‘L’ stood for Dave Laker – the talented machinist in the Departmental machine shop who actually fabricated much of the hardware for Weber’s instruments whom I had not met as an undergraduate), and we soon acquired one of the early SLM Model 8000 photon-counting spectrofluorometers.
Over the next few months my project in Lardy’s lab was increasingly focused on understanding the tendency of liver phosphofructokinase (PFK) to self-associate to extremely large aggregates. Liver PFK was (and is) thought to be an important component to the regulation of blood sugar homeostasis in mammals [6, 7]. I was beginning to suspect that the self-association behavior might play a significant role in PFK’s physiological regulation. I recalled somewhat vaguely the technique of fluorescence polarization that I had been introduced to in Weber’s lab and thought it might be useful in following this self-association reaction. Also, the postdoc who had pushed to acquire our SLM instrument had moved on so the instrument, along with its nice set of Glan-Thompson polarizers, was sitting there “begging” for me to take advantage. So I made a “cold” call to Dave Jameson (still in Weber’s lab) roughly 4-years after our late-night experiments to inquire (1) whether labeling PFK with a commercially available reactive pyrene probe would be straightforward; and (2) whether embarking on such an effort would likely be a scientific excursion of no more than 6 months since I was already feeling pressures to bring my Ph.D. research to a culmination. Dave immediately invited me to come down to Urbana for a couple of weeks during which time he was convinced that we would not only label the enzyme but also make the majority of the critical measurements I was interested in. Suffice it to say, my trip down to Urbana initiated a 2-year investigation, involving several trips to Urbana, which completely changed the nature of my Ph.D. dissertation.
During that first visit my old feelings of intimidation came back, exacerbated by my uncertainty as to whether Dr. Weber would even remember this former undergraduate who had made a point of being as inconspicuous as possible. My concern was not alleviated when, in the first conversation Dave and I had with Weber, which Dave has recently recounted in his book [8], Dr. Weber indicated that “of course” the commercially available pyrene probe we were using would have a short lifetime, rather than the long lifetime I had expected, because there was a carbonyl conjugated to the pyrene ring system. That night, the three of us went out to eat and while we were in the car, I thanked Dr. Weber for allowing me to try to get this project started by working once again in his lab, and that I had been worried that he might not remember me or my previous time in the lab. Dr. Weber once again chose the perfect words to address my concerns both spoken and unspoken at the time – he said: “Yes Greg, once you are in the lab you are always a member of the family.” I immediately felt at ease, and over the next 20 years Dr. Weber became one of my two closest scientific mentors (Dr. Lardy, of course, being the other).
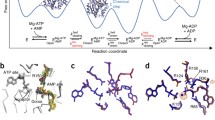
It was during this re-association with Dr. Weber and his lab toward the end of my graduate training in Wisconsin that I became aware of the papers that Dr. Weber had written in the early to mid-1970s regarding ligand binding to proteins and the thermodynamic constraints that must occur when multiple ligands bind to the same protein [9–11]. Indeed, Weber always looked upon fluorescence techniques as tools that he implemented to gain greater insight into biological macromolecules and assemblies. In particular he focused much of his early attention on proteins and protein-ligand interactions. (Once when I was still an undergraduate in his lab, Weber entered the lab from his nearby office to excitedly announce to all who quickly assembled that he thought he “finally understood BSA.” Only later did I connect the “dots” that he was working on the ligand-binding papers [10, 11] at the time (Fig. 1).)
A simple linkage argument, as framed by Weber, was soon to take center stage as the foundational principle upon which the central thesis of my Ph.D. research rested. My overall objective had been to understand the molecular basis by which the hormone glucagon effected an inhibition of the glycolytic enzyme PFK in rat liver, concomitant with glucagon’s well-known stimulation of hepatic gluconeogenesis. However, when I isolated the enzyme, it was already completely inhibited when assessed with physiological concentrations of substrates and allosteric regulators [12]. My problem, therefore, morphed into trying to understand how PFK was active in the first place before I felt it was reasonable to address the glucagon-dependent inhibition mechanism.
The various allosteric activators and inhibitors of PFK share a common feature – they act by modifying the apparent binding affinity the enzyme displays for its substrate, fructose 6-phosphate (Fru-6-P). The activation I was seeking, therefore, was one that substantially improved the apparent binding affinity of Fru-6-P, particularly in the face of an ever-present, and surprisingly constant concentration of, MgATP – both the co-substrate for the enzyme and more importantly under physiological concentrations, an important allosteric inhibitor. At the average intracellular concentration of MgATP (~3 mM), the apparent affinity for Fru-6-P is in the mM range while the physiological concentration of Fru-6-P is roughly only 10 μM [12].
It soon became apparent when working with the enzyme isolated from rat livers that it had a strong tendency to self-associate to very large species that remained active. Analogous species had been observed previously to be formed by the much more studied PFK from rabbit muscle [13–15], but rat liver PFK tended to form these high molecular weight forms at much lower protein concentrations [16]. The smallest active form of mammalian PFK is a homotetramer comprised of subunits with molecular weights of approximately 80,000 (i.e., tetramer Mr equal to 320,000). Yet the high molecular weight self-association represented an association of these tetramers – my early estimates based on ultracentrifugation behavior were that species comprised of about 12 tetramers (molecular weight equal to 3.8 million) were being formed. It was to study this self-association that sent me back to Weber’s lab with the intent to label PFK with pyrene to allow the monitoring of the extent of association using fluorescence polarization.
Once this assay was in place (using a more appropriate derivative of pyrene), it quickly became apparent that the high molecular weight self-association was dramatically influenced by ligand binding [16, 17]. The ligands with the most pronounced effects were MgATP and Fru-6-P. When observed at an enzyme concentration comparable to that which exists in the cell, MgATP favored the formation of the tetrameric form of the enzyme while Fru-6-P promoted the self-association of the tetramers to the high molecular weight form(s). Weber, of course, had also considered the coupling of ligand binding to self-association, what he termed “micro- and macro-associations,” respectively, in his ligand-binding papers of the early 1970s (Fig. 2) [10, 11]. This coupling dictates a simple prediction: If a ligand promotes oligomerization, then the oligomer itself must have a higher affinity for that ligand. Since Fru-6-P promoted the formation of the high molecular weight species, I concluded that those species must exhibit a higher affinity for Fru-6-P than the tetramer.
Free energy diagram from [10] with which Weber illustrates the relationship between cooperative ligand binding and protomer–protomer interactions
We observed these effects at a physiological enzyme concentration that was 2 orders of magnitude higher than the enzyme concentration that was normally employed when studying enzyme kinetics. We concluded, as had those working with the rabbit muscle enzyme, that PFK was largely, if not completely, tetrameric at that dilution. Therefore, the self-association likely to occur at the physiological PFK concentration, particularly given the other features of the cellular milieu, would promote the binding of Fru-6-P beyond what was evident during in vitro kinetic assessment. Lardy and I proposed that the self-association of the tetramers represented the activation mechanism I had been looking for [17]. Weber’s ligand-binding analyses, and the utility of fluorescence polarization measurements, were the key to this proposal.
As luck (and the insights of other researchers) would have it, about a year after my thesis defense, several labs nearly simultaneously discovered the metabolite, fructose 2,6-bisphosphate, and found that it was a profound activator of PFK [18–22]. Moreover its synthesis and degradation proved to be manipulated by glucagon in a manner that was consistent with the proposed stimulation of gluconeogenesis through the inhibition of PFK [23, 24]. This more dramatic discovery overshadowed our proposal, although subsequently I examined the effect of Fru-2,6-BP on the self-association properties of PFK and concluded that they may still play a role in fully rationalizing the apparent activity of PFK in vivo even in the presence of the activator Fru-2,6-BP [25].
2 Ligand Binding and Allosteric Regulation of Enzymes
Weber’s ligand-binding papers had a far more profound impact on my ensuing career as an independent investigator. After some reflection, I felt that the Professor’s papers on ligand binding provided a better basis for analyzing allosteric behavior of enzymes than the alternative models then generally employed. So I set about first casting the basic thermodynamic ideas that Weber discussed into terms that I hoped would be more familiar to enzymologists, employing a variation of the notation proposed by Cleland and widely used by the enzymology community [26, 27]. A key point was to direct attention to the determination of the coupling parameter, which I called “Q.”
This analysis, as it relates to an allosteric enzyme, begins with the single-substrate, single-modifier enzyme mechanism depicted in Scheme 1:
where A is substrate, P is product, X is allosteric ligand, E is free enzyme, XE is enzyme with allosteric ligand bound, EA is enzyme with substrate bound, XEA is the ternary complex with enzyme with both substrate and allosteric ligand bound, K 0a is equal to the Michaelis Constant in the absence of allosteric ligand, \( {\mathrm{K}}_{\mathrm{a}}^{\infty } \) is equal to the Michaelis Constant in the saturating presence of X, K 0ix is equal to the dissociation constant of X in the absence of substrate, and \( {\mathrm{K}}_{\mathrm{ix}}^{\infty } \) is the dissociation constant of X when substrate is saturating. Even a mechanism as simple as this one yields a complex rate equation if only steady state is presumed [28]. However, it is common to presume that the rapid-equilibrium assumption is valid, i.e., that the enzyme-substrate complexes are more likely to dissociate reforming free substrate than to proceed on to form product. This assumption is usually valid for allosteric enzymes, which often have fairly low turnover numbers, and it has been found experimentally to be valid for Fru-6-P binding to PFK [29]. Under conditions of rapid-equilibrium, the rate equation for Scheme 1 is given by:
where V0 equals the maximal velocity in the absence of X, K 0ia equals the dissociation constant for A in the absence of X (which in turn equals K 0a under conditions of rapid-equilibrium). The parameters W and Q are defined by the following two equations:
where \( {\mathrm{V}}^{\infty } \) equals the maximal velocity when X is saturating, \( {\mathrm{K}}_{\mathrm{ia}}^{\infty } \) equals the dissociation constant for substrate when X is saturating, and \( {\mathrm{K}}_{\mathrm{ix}}^{\infty } \) equals the dissociation constant for X when substrate is saturating. W and Q quantitatively describe the nature and magnitude of the allosteric effect on turnover and substrate affinity, respectively, with values greater than one signifying activation and values less than one signifying inhibition. Q defines the effects of PEP and MgADP on PFK since these ligands almost exclusively modulate the affinity PFK displays for its substrate, fructose-6-phosphate. Q is also related to the coupling free energy, or free energy of interaction, between the substrate ligand and the allosteric ligand, ΔGax, as defined by Weber [9–11], as follows:
where R is the gas constant, T is temperature in Kelvin, and the subscripts denote the coupled ligands. Expressing the action of an allosteric ligand in terms of coupling free energy presents two practical advantages: the sign of ΔGax gives the nature of the allosteric effect (positive sign → inhibition, negative sign → activation), and the energetic effects of activation and inhibition are expressed with equal dynamic range.
Determining the coupling between a substrate and an allosteric ligand is straightforward – one simply performs a substrate titration to yield a saturation curve from which an apparent Ka can be determined. This basic experiment is then repeated at varying concentrations of allosteric ligand. The values of Ka as a function of allosteric ligand concentration can then be fit to the functional dependence suggested by Eq. (1), which is given by:
We have found it useful to construct a plot of log (Ka) vs. log [X] to visualize the trend of Ka transitioning from the limiting value at low [X] (K 0ia ) to the limiting value at high [X] (\( {\mathrm{K}}_{\mathrm{ia}}^{\infty } \)) from which the value of log (Qax) can be estimated. The concepts were useful, and could be applied straightforwardly, even on complex, oligomeric, highly cooperative enzymes, such as rat liver PFK, by focusing on the behavior of the half-saturation value, K1/2, which is functionally equivalent to Ka, rather than on the more complex saturation behavior of substrate, and fitting the data to Eq. (5). We demonstrated straightforward utilization of these concepts in the characterization of the pH dependence of the allosteric inhibition of PFK by ATP [30], and subsequently the effects of Fru-2,6-BP [31].
Central to Weber’s point in his papers was that, because the binding energy of a small ligand is derived from multiple interactions each contributing a small amount, it would be unreasonable that the consequences of binding a ligand on the subsequent binding of another ligand at a distant site would be anything other than a fraction of the binding energy, i.e., roughly 1–2 kcal/mole for a single ligand. This magnitude predicted that, even in the case of an inhibitory allosteric ligand, the effects of that ligand should saturate – a consequence of forming the ternary complex with both substrate and inhibitor bound simultaneously to the protein. Although in principal the apparent coupling observed in an oligomeric protein could increase with the number of interacting sites, we were gratified to observe, initially with rat liver PFK and eventually with several other enzymes including at the time mitochondrial isocitrate dehydrogenase (ICDH), that such saturation could be demonstrated – allowing the coupling to be quantified in most cases.
The quantification of the coupling parameter, therefore, provides an unambiguous characterization of both the nature of an allosteric effect. It also allowed for the separation of these issues from the binding affinity of the allosteric ligand, something that becomes conflated when one presumes the nature of the allosteric effect is somehow predetermined by a pre-existing conformational state to which functional attributes are ascribed and the ligand need only bind to that form. This then (and I fear still) prevailing view only leaves the respective binding affinities as the basis of comparison of one inhibitor to another, while clearly the coupling free energy framework allows different ligands to exhibit different efficacies regardless of how their binding affinities compare. Indeed, we have over the ensuing several years made note of several situations where a ligand binds less tightly than its counterpart but actually inhibits more strongly once bound [29, 32].
3 Protein Dynamics and Allosterism
In a classic case of doing the right experiment for the wrong reason, I began to consider how the coupling free energy might vary with temperature. Drawing an analogy to membranes, I wondered whether a temperature might be reached at which the internal organization of the protein’s folded structure might become disorganized – akin to the phase transition in membranes. Although even I didn’t expect such a dramatic change, I wondered whether any discontinuity might be observed in the coupling free energy as a function of temperature. The coupling free energy, after all, was a direct measure of “communication” between two sites on a protein and, particularly if those sites were on opposite sides of the protein, it should be sensitive to any sudden change that might occur. When we proceeded to analyze coupling free energies as a function of temperature, we didn’t observe the expected discontinuities, but upon reflection we felt that we had observed something possibly more significant.
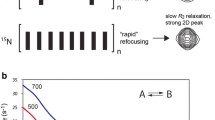
The coupling free energy describing the inhibition of Fru-6-P binding to rat liver PFK by MgATP, as well as coupling free energy describing the activation by ADP of isocitrate binding to beef heart mitochondrial ICDH, instead of exhibiting discontinuities, demonstrated a continuous trend that was quite linear when subjected to a van’t Hoff analysis (Fig. 3) [33]. As Weber had shown, the coupling free energy can be considered to be the standard free energy associated with the disproportionation equilibrium:
Variation with reciprocal temperature of the logarithm of the coupling between isocitrate and ADP binding to isocitrate dehydrogenase (top panel) and between Fru-6-P and MgADP binding to rat liver PFK (bottom panel). The coupling free energy is negative for the data in the top panel and positive for the bottom panel. Yet the positive slope of both lines indicates that the coupling enthalpy is negative for both cases. In PFK, therefore, the positive coupling free energy must be the result of a negative TΔS component that is larger in absolute value than the absolute value of the coupling enthalpy. The significance is discussed in the text. Figure from [33]
Consequently the temperature dependence of the equilibrium constant giving rise to coupling free energy, which we found to be linear when plotted as the logarithm versus reciprocal temperature, can be interpreted in terms of the underlying ΔH and ΔS components that give rise to the coupling free energy. Not surprisingly for both cases, the sign of ΔH and ΔS was the same indicating a substantial degree of compensation when the individual values of ΔH and TΔS are compared to the value of ΔG. The coupling free energy for the activation by ADP of ICDH, which has a negative value by definition according to the sign conventions used by Weber [10, 11] and adopted by us [26], was derived from a negative value of the coupling enthalpy that dominated the contribution of the negative value of TΔS (i.e., ΔH had a greater absolute value than TΔS). This result taken in isolation was not that surprising given a “conformation-centric” view of the structural basis for ligand binding. What was surprising was that the coupling exhibited for the MgATP inhibition of rat liver PFK was entropy dominated, meaning that the positive sign of the coupling free energy was derived from ΔH and TΔS terms that also were negative, just as they had been for ICDH, but unlike that case, for the MgATP–Fru-6-P coupling in PFK the absolute value of TΔS was bigger than that for ΔH giving rise to the positive value for ΔG. We termed this “entropy-dominated coupling” because it was the entropy term that established the sign, and hence the nature, of the allosteric effect. A corollary to this observation was that the sign of the coupling enthalpy was the opposite that for ΔG. Hence, for MgATP inhibition of rat liver PFK, ΔH was negative and thereby actually favoring activation rather than the inhibition that is observed.
Of course, I discussed our observations with Dr. Weber on many occasions, often in his apartment over meals of “microwave pasta” – the result of a process he developed involving a layering of Saran Wrap over the uncooked pasta in water, with only a corner folded back for venting, and then running the microwave a precise amount of time. He delighted in describing this procedure to the uninitiated because it yielded perfect “al dente” pasta every time.
The fact that entropy change could be the dominant, and directing, component of the allosteric effect begged the question of what was causing the entropy change, particularly in the “entropy-dominated” situations. Given that the entropy change pertains to the disproportionation coupling reaction (Scheme 2), it is notable that free ligand is absent from this reaction. Consequently the large hydration changes expected when ligand binding occurs should not be a factor when considering the molecular basis for allosteric coupling. Certainly the possibility exists that the different protein-ligand complexes on each side of the equilibrium have different extents of hydration, but since the filled and unoccupied binding sites are balanced, the likelihood of this mechanism being dominant would seem to be minimized. Instead, the possibility that changes in conformational entropy may be the dominant mechanism at play seemed compelling. To the extent that this occurs, it suggests that conformational changes that might be evident in X-ray crystal structures of various ligated species would not reveal the true conflict that causes an inhibitor to actually antagonize the binding of substrate ligand (and vice versa). Instead, the dynamics of the structure might contain the actual basis for the inhibition. Moreover, since the coupling enthalpy actually favors activation, the crystal structures actually might contain features that would directly mislead one from understanding the basis of inhibition per se.
The other striking feature of our temperature results was that the trend was in fact quite continuous – indeed essentially linear in a van’t Hoff presentation. Although our original data for ICDH and rat liver PFK exhibited positive slopes in the van’t Hoff plots, we have since observed negative slopes as well for other systems. But in almost all cases, linearity is observed over at least a 30° span around 25°C. Since a coupling free energy equal to 0 is indicative of no allosteric effect, with positive signs indicating inhibition and negative values activation, we predicted that it might be possible for temperature alone to cause the sign of the coupling free energy to transition from positive to negative (or vice versa) – hence leading to a temperature-induced inversion of the allosteric effect. Subsequently we published several examples of just this type of cross-over of allosteric effect by modifying only temperature [34, 35].
Of course virtually all of these ideas were explored and matured in my conversations with Dr. Weber over the course of several visits. Ultimately we published our findings in a PNAS paper that was communicated by Dr. Weber [33] in 1989. And I was particularly honored when he chose to include our result and conjectures in his seminal book on “Protein Interactions” published in 1992 [36]. The notion that the transmission of allosteric influence might have an entropic, and hence dynamic, origin was also gaining credence with other investigators [37].
4 Deflating the Two-State Premise
Concomitant with the prediction and the demonstrations of saturation of the allosteric affect, necessary to quantify the coupling free energy, was the obvious incompatibility of these observations with the notion that allosteric enzymes consist of only two conformational states that exclusively bind either substrate or allosteric inhibitor exclusively. Despite Dr. Weber’s eloquent arguments against this simplistic notion as being not just inadequate, but fundamentally misleading with respect to the true nature of proteins and their interactions with other proteins and small molecular weight ligands, the idea, probably because of its simplicity and its appeal to “intuitive common sense,” continued to (and does to this day) dominate both the rhetoric and the thinking about the basis of regulation exhibited by allosteric enzymes. After being emboldened to seek evidence for the role of dynamics in allosteric transmission, we switched much of our attention from rat liver PFK to PFK from prokaryotes, notably E. coli and B. stearothermophilus. These enzymes have several advantages for this objective, being much smaller (subunit molecular weight 34,000), extremely stable obligate homotetramers with no propensity to self-associate further, and much simpler allosteric properties derived from a single allosteric site per subunit to which either the inhibitor phospho(enol)pyruvate (PEP) or the activator ADP can bind. But most compelling for our purposes was that both E. coli PFK and B. stearothermophilus PFK contain only a single tryptophan per subunit. At the time (early 1990s) the fluorescence of many single tryptophan proteins had been characterized using time-resolved techniques in an effort to understand the nature of the internal dynamical properties of proteins. We hoped to utilize similar approaches to examine the relationship between dynamics and allosteric functionality. Eventually this effort would take advantage of the power of site-directed mutagenesis to manipulate the tryptophan position in an effort to sample a wider range of interior environments, but initially we settled for examining the tryptophan in the native position of E. coli PFK. Of course our first experiments involved steady state fluorescence measurements.
Since E. coli PFK could bind either an inhibitor or an activator, and the actions of these ligands seemed to exclusively influence the binding of the substrate, Fru-6-P, the allosteric properties had been viewed as providing a nice example of a straightforward R-T system in which the inhibited form of the enzyme bound the inhibitor PEP well and the substrate Fru-6-P poorly, while the active form of the enzyme bound Fru-6-P and ADP well and PEP poorly. Not only had the kinetic behavior of the enzyme been characterized in this fashion [38], but the previous work on the intrinsic tryptophan fluorescence seemed to support this idea as well [39]. Both ADP and Fru-6-P cause the intrinsic fluorescence of E. coli PFK to diminish to nearly the same level whereas PEP binding causes the intrinsic fluorescence to increase to a small extent. These observations, together with the positive cooperativity the kinetics display with respect to Fru-6-P titrations in the absence of either allosteric ligand, reinforced the notion that apo enzyme exists in a form very similar to the “T” form adopted when PEP binds and that either Fru-6-P or ADP can upon binding induce the “R” form.
We took another look at these results and largely confirmed the effects of these two ligands on quantum yield [40, 41]. The intrinsic fluorescence of the single tryptophan substantially decreased when either Fru-6-P or ADP bound (Fig. 4), and the fluorescence increased to a small but significant extent when PEP bound (Fig. 5). Although we were able to detect a slightly different extent of decrease in quantum yield with ADP binding compared to Fru-6-P binding, that difference was small relative to the overall decrease. Additionally, when Fru-6-P was titrated after PEP saturation, to form the ternary complex, the fluorescence intensity diminished to what was arguably a value comparable to the Fru-6-P binary complex alone. These results were generally consistent with previous observations and in general support of a 2-state model.
Response of intrinsic tryptophan fluorescence emission (A) and polarization (B) to titration of successive equal quantities of ligand to E. coli PFK. Either Fru-6-P (closed squares), MgADP (open triangles), or MgATP (open squares) was added to free enzyme. Subsequently MgADP was added after Fru-6-P (open circles), Fru-6-P was added after MgADP (closed triangles), or MgATP was added after MgADP (plus sign). Note how different the responses are to the binding of Fru-6-P and MgADP when polarization is monitored in contrast to the similar response of the emission. Figure from [40]
Response of intrinsic tryptophan fluorescence emission (A) and polarization (B) to titration of successive equal quantities of ligand to E. coli PFK. Either Fru-6-P (closed squares) or PEP (open circles) was added to free enzyme. Subsequently either PEP was added to enzyme saturated with Fru-6-P (open squares) or Fru-6-P was added to enzyme saturated with PEP (closed circles). Figure from [41]
A very different picture emerged, however, when we looked at the steady state polarization of the tryptophan fluorescence in response to ligand binding. Fru-6-P binding caused a decrease in fluorescence polarization, but ADP did not, clearly demonstrating that Fru-6-P and ADP do not form the same “R” state when assessed in this manner. In addition, when Fru-6-P and ADP both bind simultaneously, the fluorescence polarization of the tryptophan clearly adopts a unique value quite distinct from either of the Fru-6-P or ADP binary complexes.
In contrast to the quantum yield changes, PEP binding caused a decrease in the fluorescence polarization, although to a somewhat lesser extent than did Fru-6-P. The ternary complex formed when Fru-6-P and PEP bind simultaneously exhibits a quantum yield comparable to the Fru-6-P bound form but a fluorescence polarization value comparable to the PEP binary complex. Clearly, when the intrinsic fluorescence of the native tryptophan is thoroughly assessed, one cannot escape the conclusion that Fru-6-P, ADP, and PEP each form binary complexes with unique conformational properties and that the ternary complexes formed by the binding of any two of these ligands (except ADP and PEP which cannot bind simultaneously) produce unique conformations as well.
Moreover, given that the overall size and shape of E. coli PFK do not change upon ligand binding, these results directly suggested that the internal dynamics of the protein, at least in the vicinity of the lone tryptophan per subunit, must be perturbed by ligand binding. We proceeded to examine, via the time-resolved fluorescence properties of this tryptophan, how the motion of this tryptophan changed with ligand binding [41, 42]. In general the data suggested that the amplitude of motion, rather than the rate of motion per se, was perturbed, increasing most notably when the substrate Fru-6-P bound. Thus this prediction made from the temperature dependence data Weber and I had discussed several years previously, also seemed to be born out, although relating the motion changes to a measurable thermodynamic parameter cannot be expected without greater sampling of the overall protein structure.
5 Summary
In the years since these first observations, we have gone on to dissect the coupling relationships in the homotetramer of prokaryotic PFK, study the dynamical properties of allosteric enzymes as they relate to allosteric functionality using an expanding array of techniques including recently NMR, and the implications of ligand-induced quaternary changes as they may relate to enzyme regulatory behavior, using techniques such as fluorescence correlation spectroscopy. In all of these efforts we have been, and continue to be, inspired by Gregorio Weber – by his scientific insights and achievements as well as his approach to science. I am exceedingly grateful that in the beginning he accepted this reserved undergraduate into his research “family.” I am proud to say that in the end he was my mentor and friend.
References
Knopp J, Weber G (1967) Fluorescence depolarization measurements on pyrene butyric-bovine serum albumin conjugates. J Biol Chem 242(6):1353–1359
Knopp JA, Weber G (1969) Fluorescence polarization of pyrenebutyric-bovine serum albumin and pyrenebutyric-human macroglobulin conjugates. J Biol Chem 244(23):6309–6315
Weber G, Bablouzian B (1966) Construction and performance of a fluorescence polarization spectrophotometer. J Biol Chem 241(11):2558–2561
Jameson DM, Spencer RD, Weber G (1976) Construction and performance of a scanning, photon-counting spectrofluorometer. Rev Sci Instrum 47(9):1034–1038
Spencer RD, Weber G (1969) Measurements of subnanosecond fluorescence lifetimes with a cross-correlation phase fluorometer. Ann N Y Acad Sci 158(1):361–376
Clark MG, Kneer NM, Bosch AL, Lardy HA (1974) The fructose 1,6-diphosphatase-phosphofructokinase substrate cycle. A site of regulation of hepatic gluconeogenesis by glucagon. J Biol Chem 249(18):5695–5703
Clark MG, Bloxham DP, Holland PC, Lardy HA (1974) Estimation of the fructose 1,6-diphosphatase-phosphofructokinase substrate cycle and its relationship to gluconeogenesis in rat liver in vivo. J Biol Chem 249(1):279–290
Jameson DM (2014) Introduction to fluorescence. CRC/Taylor and Francis, New York
Weber G (1970) Binding of small ligands by proteins. Hormonal steroids. In: Proceedings of the Third International Congress, Hamburg, September 1970, pp 58–65
Weber G (1972) Ligand binding and internal equilibiums in proteins. Biochemistry 11(5):864–878
Weber G (1975) Energetics of ligand binding to proteins. Adv Protein Chem 29:1–83
Reinhart GD, Lardy HA (1980) Rat liver phosphofructokinase: kinetic activity under near-physiological conditions. Biochemistry 19(7):1477–1484
Aaronson RP, Frieden C (1972) Rabbit muscle phosphofructokinase: studies on the polymerization. The behavior of the enzyme at pH 8, pH 6, and intermediate pH values. J Biol Chem 247(23):7502–7509
Pavelich MJ, Hammes GG (1973) Aggregation of rabbit muscle phosphofructokinase. Biochemistry 12(7):1408–1414
Lad PM, Hill DE, Hammes GG (1973) Influence of allosteric ligands on the activity and aggregation of rabbit muscle phosphofructokinase. Biochemistry 12(22):4303–4309
Reinhart GD, Lardy HA (1980) Rat liver phosphofructokinase: use of fluorescence polarization to study aggregation at low protein concentration. Biochemistry 19(7):1484–1490
Reinhart GD, Lardy HA (1980) Rat liver phosphofructokinase: kinetic and physiological ramifications of the aggregation behavior. Biochemistry 19(7):1491–1495
Furuya E, Uyeda K (1980) An activation factor of liver phosphofructokinase. Proc Natl Acad Sci U S A 77(10):5861–5864
Furuya E, Uyeda K (1980) Regulation of phosphofructokinase by a new mechanism. An activation factor binding to phosphorylated enzyme. J Biol Chem 255(24):11656–11659
Claus TH, Schlumpf J, Pilkis J, Johnson RA, Pilkis SJ (1981) Evidence for a new activator of rat liver phosphofructokinase. Biochem Biophys Res Commun 98(2):359–366
Pilkis SJ, El-Maghrabi MR, Pilkis J, Claus TH, Cumming DA (1981) Fructose 2,6-bisphosphate. A new activator of phosphofructokinase. J Biol Chem 256(7):3171–3174
Van Schaftingen E, Jett MF, Hue L, Hers HG (1981) Control of liver 6-phosphofructokinase by fructose 2,6-bisphosphate and other effectors. Proc Natl Acad Sci U S A 78(6):3483–3486
Richards CS, Uyeda K (1980) Changes in the concentration of activation factor for phosphofructokinase in hepatocytes in response to glucose and glucagon. Biochem Biophys Res Commun 97(4):1535–1540
Hue L, Blackmore PF, Exton JH (1981) Fructose 2,6-bisphosphate. Hormonal regulation and mechanism of its formation in liver. J Biol Chem 256(17):8900–8903
Reinhart GD (1983) Influence of fructose 2,6-bisphosphate on the aggregation properties of rat liver phosphofructokinase. J Biol Chem 258(18):10827–10830
Reinhart GD (1983) The determination of thermodynamic allosteric parameters of an enzyme undergoing steady-state turnover. Arch Biochem Biophys 224(1):389–401
Cleland WW (1963) The kinetics of enzyme-catalyzed reactions with two or more substrates or products. I. Nomenclature and rate equations. Biochim Biophys Acta 67:104–137
Frieden C (1964) treatment of enzyme kinetic data. I. The effect of modifiers on the kinetic parameters of single substrate enzymes. J Biol Chem 239:3522–3531
Tlapak-Simmons VL, Reinhart GD (1994) Comparison of the inhibition by phospho(enol)pyruvate and phosphoglycolate of phosphofructokinase from B. stearothermophilus. Arch Biochem Biophys 308(1):226–230
Reinhart GD (1985) Influence of pH on the regulatory kinetics of rat liver phosphofructokinase: a thermodynamic linked-function analysis. Biochemistry 24(25):7166–7172
Reinhart GD, Hartleip SB (1986) Relationship between fructose 2,6-bisphosphate activation and MgATP inhibition of rat liver phosphofructokinase at high pH. Kinetic evidence for individual binding sites linked by finite couplings. Biochemistry 25(23):7308–7313
McGresham MS, Lovingshimer M, Reinhart GD (2014) Allosteric regulation in phosphofructokinase from the extreme thermophile Thermus thermophilus. Biochemistry 53(1):270–278. doi:10.1021/bi401402j
Reinhart GD, Hartleip SB, Symcox MM (1989) Role of coupling entropy in establishing the nature and magnitude of allosteric response. Proc Natl Acad Sci U S A 86(11):4032–4036
Braxton BL, Tlapak-Simmons VL, Reinhart GD (1994) Temperature-induced inversion of allosteric phenomena. J Biol Chem 269(1):47–50
Tlapak-Simmons VL, Reinhart GD (1998) Obfuscation of allosteric structure-function relationships by enthalpy- entropy compensation. Biophys J 75(2):1010–1015
Weber G (1992) Protein interactions. Chapman and Hall, New York
Cooper A, Dryden DTF (1984) Allostery without conformational change – a plausible model. Eur Biophys J 11:103–109
Blangy D, Buc H, Monod J (1968) Kinetics of the allosteric interactions of phosphofructokinase from Escherichia coli. J Mol Biol 31(1):13–35
Deville-Bonne D, Garel JR (1992) A conformational transition involved in antagonistic substrate binding to the allosteric phosphofructokinase from Escherichia coli. Biochemistry 31(6):1695–1700
Johnson JL, Reinhart GD (1994) Influence of MgADP on phosphofructokinase from Escherichia coli. Elucidation of coupling interactions with both substrates. Biochemistry 33(9):2635–2643
Johnson JL, Reinhart GD (1997) Failure of a two-state model to describe the influence of phospho(enol)pyruvate on phosphofructokinase from Escherichia coli. Biochemistry 36(42):12814–12822
Johnson JL, Reinhart GD (1994) Influence of substrates and MgADP on the time-resolved intrinsic fluorescence of phosphofructokinase from Escherichia coli. Correlation of tryptophan dynamics to coupling entropy. Biochemistry 33(9):2644–2650
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Reinhart, G.D. (2016). Continuing Inspiration: Gregorio Weber’s Influence on Understanding the Basis of Allosteric Regulation of Enzymes. In: Jameson, D. (eds) Perspectives on Fluorescence. Springer Series on Fluorescence, vol 17. Springer, Cham. https://doi.org/10.1007/4243_2016_20
Download citation
DOI: https://doi.org/10.1007/4243_2016_20
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-41326-6
Online ISBN: 978-3-319-41328-0
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)