Abstract
The fluorescence polarization technique that Prof. Weber developed at Cambridge University between the late 1940s and early 1950s has had a tremendous impact on our understanding of the structure and dynamics of macromolecules and in the analysis of proteins interactions and detection of target proteins in biologically complex samples. His decision to develop dimethylaminonaphthalene sulfonyl chloride (Dansyl-Cl) as the first probe for fluorescence polarization studies was brilliant, as its long fluorescence lifetime and well-defined dipole are ideally suited to study protein conjugates as large as 100 kDa. Indeed, after almost 70 years, the Dansyl group is still the probe of choice for in vitro applications of fluorescence polarization. Unfortunately, Dansyl is not very suitable for related studies in living cells, primarily because it requires excitation in the near ultraviolet, while the in vivo labeling of a target protein with Dansyl group is challenging. We have developed a new class of genetically encoded fluorescent protein that may help to overcome these limitations. The lumazine-binding protein (LUMP) harbors a fluorescent probe with a cerulean-colored emission that like Dansyl has a long excited state lifetime (14 ns). Moreover, LUMP has a smaller mass than GFP that allows us to genetically append capture sequences as large as 20 kDa and still generate a fusion protein with sufficient dynamic range in the fluorescence polarization value to quantify the amounts of the free and target-bound states in an equilibrium. In this article, I will compare and contrast key features of Dansyl and LUMP as probes for fluorescence polarization studies and discuss the potential of using LUMP and related encoded proteins to advance the application of fluorescence polarization to analyze target proteins and protein interactions in living cells.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
One of the highlights of my undergraduate experience in the department of biochemistry at Birmingham University (UK) was the final year research project. I was particularly interested in pursuing the project that was offered by Prof. John Teale who had developed a number of thiol-reactive fluorescent dyes that he wanted to use to prepare fluorescent protein conjugates. Recognizing my strong interest in fluorescence, Prof. Teale suggested that I should read papers on fluorescence spectroscopy, including a paper on thiol-reactive fluorophores published by Gregorio Weber [1]. A year after graduating from Birmingham, I applied to PhD programs in the USA – after discussing my choices with Prof. Teale, I decided to go to Urbana where I was lucky enough to end up working in Prof. Weber’s laboratory [2]. I remember soon after joining the group, I had a conversation with Prof. Weber about some of the physical biochemistry courses I had taken at Birmingham with Prof. John Teale – on mentioning Teale’s name, he told me about Teale’s dry humor – the only story I remember was the time when they were in line at the canteen in the Scala Theatre at Sheffield University waiting for the invariable Friday lunch offering of fish and chips, when Teale told Weber “you know Gregorio, ever since I arrived at Sheffield I have become a cod-fearing man.”
I learned a lot of new techniques in the Weber lab, including projects that involved the chemical synthesis of fluorescent probes and site-specific labeling of proteins with reactive fluorescent probes and their applications to study protein dynamics. This early chemical synthesis experience had a great impact on my later research program that over time has led to the development of a wide variety of sensor molecules, including thiol-reactive probes that we have used for quantitative analysis of proteins and their complexes over multiple scales of time and distance [2–5]. I thought I would use the invitation to contribute to this dedicated volume to Gregorio Weber to compare and contrast the properties and performances of synthetic and genetically encoded fluorescent probes for fluorescence polarization-based analysis of macromolecules. The history of this probe development for polarization studies began with 1,8-Dansyl-chloride (Fig. 1a), a probe that Weber designed and synthesized at Cambridge in the latter part of the 1940s. This probe was introduced in the second of his 1952 papers in the Biochemical Journal on the theory and practice of fluorescence polarization [6, 7]. Weber relayed to me the difficulties he had in preparing Dansyl chloride that were finally overcome in a beautifully simple and high-yield reaction that I repeated in my first chemical synthesis in the Weber lab. The reactive sulfonyl chloride of 1,8-dimethylaminonaphthalene sulfonic acid was prepared by grinding solid forms of 1,8-Dansyl sulfonic acid, a light brown flaky material, with phosphorous pentachloride using a mortar and pestle for 15 min. The final step involved solubilizing the liquefied reaction mix in a small volume of acetone, which was poured slowly over a beaker of ice cubes – the product precipitates on the ice as a yellow-colored mass – the yellow precipitate has a mesmerizing brilliance and unique hue.
Molecular structures of probes discussed in this article. (A) 1,8-Dimethylamino-8-naphthalenesulfonylchloride (Dansyl chloride). (B) N-(Iodoacetylaminoethyl)-8-naphthylamine-l-sulfonic acid (1,8-IAEDANS). (C) 1-Pyrenebutyric acid N-hydroxysuccinimide ester. (D) Ribityl-lumazine. (E) Flavin mononucleotide
Dansyl chloride has been used as a label for many other applications in biology and chemistry, including the first fluorescence-based method for protein sequencing. In his recollections of life on the first floor of the MRC building in Cambridge, Brian Hartley described two important discoveries he made using the Dansyl chloride probe he received from Gregorio Weber [8]. The first discovery with Vince Massey was to show Dansyl chloride reacted quickly with the active site serine (Ser-65) in chymotrypsin, which resulted in an orange fluorescent conjugate that was one of the first examples of fluorescent active site titrant [9]. The second application was in protein sequencing [10], where the Dansyl chloride was reacted with the α-amino acid residue in a peptide – the peptide was hydrolyzed and the mixture of amino acids applied to one side of a 2-sided polyamide sheet. The other side of the polyamide sheet was spotted with a solution containing all 20-Dansylated amino acids that were separated using a 2D-chromatographic separation technique [10]. The position of the N-terminal fluorescent amino acid from the peptide was compared to the positions of the 20 known amino acid derivatives of Dansyl that had been separated on the reverse side of the side of the plate. The identity of the N-terminal residue in the peptide was revealed by the overlap of fluorescent spots on each side of the plate.
In this article I will compare and contrast small-molecule synthetic probes and genetically encoded fluorescent proteins as probes for fluorescence polarization measurements. Attention is focused on the properties of 1,8-Dansyl, the prototypical small-molecule probe for fluorescence polarization [7, 11], which is compared and contrasted with genetically encoded fluorescent proteins including GFP and lumazine-binding proteins [12].
1 Design of Small-Molecule Fluorescent Sensors for Target Proteins
Fluorescent sensors including those incorporating 1,8-Dansyl [7] have been widely used to determine the size and shape of macromolecules, to detect target proteins, and to study protein interactions in complex samples [1, 2, 7, 11]. For these latter applications, the fluorescent sensor should exhibit marked differences in one or more properties of the emission between the free and target-bound states. Better still is if these differences are based on an absolute parameter of the fluorescence emission, which we consider as the quantum yield of fluorescence, the lifetime of the excited state, the energy of the emission, or the polarization of the fluorescence. Other absolute measures include changes in the rate of an excited state reaction, including those associated with FRET, excimer formation, and deprotonation. Dansyl probes are highly valued for fluorescence-based investigations of protein interactions. Protein complexation will often result in appreciable changes of the quantum yield, lifetime, and the energy of the emission. This feature arises in part because of the long fluorescence lifetime of 1,8-Dansyl (13–16 ns) [7], which makes it sensitive to dynamic events in its immediate environment, including quenching by nearby residues, while its excited state dipole is sensitive to both general and specific solvent effects [13]. Interestingly, 2,5-Dansyl has a fluorescence lifetime of ~32 ns, which should make it even more sensitive to dynamic quenching events [14].
2 Fluorescence Sensors of Protein Hydrodynamics
Weber developed the theory and experimental technique that relates values of the polarization of the fluorescence to the size and shape of the molecule [6, 7]. Moreover, the increase in the fluorescence polarization value of a sensor on complex formation with a larger target molecule can be predicted accurately using the Perrin–Weber equation [6, 12], a feature that simplifies the design of new FA sensors, especially when compared to that for a FRET-based sensor [13, 14]. Determinations of the fluorescence anisotropy value of appropriately labeled ligands or proteins have been widely used to quantify target proteins or protein interactions [15] – this popularity is a consequence of several factors that include the ability to record accurate measurements of the FA value using steady-state instrumentation under “no-wash” conditions. Before we consider the suitability of small-molecule and genetically encoded fluorescent proteins as probes for fluorescence polarization studies, it may be useful to review some of the essential relationships that relate the fluorescence polarization value to the size (and/or shape) of a probe-labeled protein [6, 7]. I will start by replacing the term fluorescence polarization with fluorescence anisotropy (FA), as the latter is easier to manipulate mathematically. FA values are computed using Eq. (1) from measurements of the polarized components of fluorescence emission of the sensor (I para and I perp) in response to vertically polarized excitation of a solution composed of mixtures of the free and target-bound states of the sensor [2]:
The FA value increases in the sensor-target complex because of its larger volume and slower rate of tumbling in solution [6, 15]. This is shown schematically in Fig. 2. The Perrin–Weber equation [3, 4] expressed in terms of FA values is
where r is the measured FA value and r 0 is the limiting FA value, which depends on molecular properties of the probe molecule and the angle between the absorption and emission transition dipole moments. This limiting value is usually recorded from a dilute solution of the probe in a viscous solvent such as glycerol at −20°C. The excited state lifetime is given by τ f, while τ c is the rotational correlation time, which is the time it takes for a sphere to rotate through 1 radian [12] (Fig. 2) – the rotational correlation time is related to the hydrodynamic volume V according to Eq. (3) [6, 7]:
where η is the solvent viscosity and T is the temperature in Kelvin. Equations (2) and (3) show that a suitable probe or protein conjugate for FA-based analysis of a target protein would have a small molecular volume and a τ f/τ c ratio of unity or less. τ c can also be related to the mass of a spherical protein according to τ c = η MW(ν + h)/RT, where R = 8.314 × 107 erg.mol−1 K−1, η is the viscosity, MW is the molecular weight of the spherical protein, ν is the partial specific volume of 0.74, and h is the degree of hydration 0.3 ml H2O/g protein. We will now consider the suitability of some small-molecule fluorescent probes and larger genetically encoded fluorescent proteins.
3 Small-Molecule Probes for FA-Based Measurements
1,8-Dansyl is well suited for fluorescence polarization studies when used as a fluorescent analogue of a ligand for a target protein or when conjugated chemically to a sensor protein [7]. In particular, 1,8-Dansyl has one of the longest lifetimes (~13 ns) of any small-molecule fluorescent probe, and the direction of the dipole in the molecule is well defined and leads to a relatively uniform anisotropy value at red side of the S0–S1 excitation band (340–350 nm). Another useful feature of Dansyl chloride results from the coupling of the sulfonyl chloride to the ε-amino group on a lysine residue, which produces a short and rigid link that allows the probe to report primarily on the hydrodynamics of the protein molecule [7]. On the other hand, fluorophores having longer and flexible links to the protein allow the probe to experience a range of local and rapid motions that contribute in a complex manner to the depolarization.
The nonlinear relationship between the calculated FA value and the molecular volume (and mass) for fluorescently labeled spherical proteins is revealed by fitting equations (2) and (3) for different fluorescent lifetime values (Fig. 3). The plots clearly reveal the nonlinear relationship between FA and molecular weight. The relationship between the predicted FA value and the mass of a spherical protein conjugated with 1,8-Dansyl is shown in Fig. 3. In particular, the FA curve for a 20 kDa protein chemically coupled to a single 1,8-Dansyl probe is considered, which is easily achieved by treating proteins with a single cysteine residue with the thiol-reactive probe IAEDANS [1] (Fig. 1b). Using a fluorescence lifetime for the Dansyl of 14 ns, one can calculate the FA for a spherical 20 kDa Dansyl conjugate as 0.140 (blue dotted line in Fig. 3). The range of FA values between the free and bound states of this conjugate sensor (0.140 ± 0.001 to the limiting anisotropy of ~0.325 [1]) would allow one to conduct accurate FA-based binding studies with target proteins larger than 80 kDa (Fig. 3). In his classic 1952 Biochemical Journal paper, Weber exploited the FA-properties of 1,8-Dansyl to study the hydrodynamic properties of native and denatured ovalbumin and BSA (35 kDa and 68 kDa) [7]. In addition to providing Dansyl chloride to Hartley, Weber also collaborated with Kenneth Bailey, who with his student T.C. Tsao and G.S. Adair described fluorescence polarization studies on Dansyl-tropomyosin that were performed by Prof. Weber [16]. They reported the tropomyosin molecule has two rotational diffusion constants (ρ) where ρ = 3τ c of 6.3 × 106 s−1 (159 ns) and 1.9 × 108 s−1 (5.3 ns) that correspond to rotations about the short axis and long axis of the molecule, respectively. Separately Tsao acknowledged Prof. Weber in two 1953 studies for his unfailing help in the application of fluorescence polarization to study the size of subunits in myosin [17] and actin [18]. In both studies, he prepared Dansyl conjugates of each protein and used the Perrin plot to determine the size of the subunits. While he was successful in assigning correct molecular weights to the heavy and light subunits of myosin [17], his calculation of the mass of the spherical actin monomer was somewhat high at 70 kDa [18].
It is quite remarkable that in the almost 70 years since Prof. Weber introduced the 1,8-Dansyl probe [7], it is still one of the most suitable probes for FA-based analysis of proteins and their complexes. While pyrene (Fig. 1c) and related symmetrical polyaromatic hydrocarbons can boast even longer excited state lifetimes, they have low limiting anisotropy values, with excitation anisotropy spectra that are often characterized by sharp changes in the anisotropy value [19].
4 Genetically Encoded Fluorescent Proteins for FA-Based Measurements
Unfortunately, 1,8-Dansyl is not a very useful probe for FA-based analysis of proteins in living cells. This limitation arises from the low molar extinction coefficients of Dansyl conjugates, which are on the order of 4–6,000 M−1 cm−1 [7] compared to >100,000 M−1 cm−1 for probes optimized for imaging single molecules [20]. Moreover, excitation of the Dansyl group requires near ultraviolet light (<350 nm), which may result in photocytotoxicity, while also exciting natural fluorescent molecules in cells, including free and protein-bound NADH and flavin mononucleotide. Perhaps the biggest drawback in using small-molecule fluorophores as probes for FA-based analysis of protein interactions in living cells are the difficulties associated with probe delivery and chemical coupling to a target protein in the cytoplasm. It is for this reason perhaps that most studies described to probe protein interactions in living cells use genetically encoded fluorescent proteins with excitation spectra in the visible region of the electromagnetic spectrum. However, GFP-like fluorescent proteins are unsuitable for FA-based imaging of target proteins in living cells for reasons given in the following section.
In spite of the dearth of suitable probes, fluorescence anisotropy holds great promise as a technique to image and quantify specific proteins and their interactions in living cells [11, 15, 21–23]. Unfortunately, the large mass and short lifetime of GFP and related proteins make them unsuitable as probes for FA-based determinations of target proteins. This limitation may be best appreciated with reference to the simulations shown in Fig. 3. This time the predicted FA values for a 20 kDa capture appended to GFP (~30 kDa) is considered. With a mass of ~50 kDa and a fluorescent lifetime of ~2 ns, the predicted steady-state FA value of the free fusion protein in buffer at 20°C is already 0.340, which agrees almost exactly with the experimentally determined FA value for GFP [24]. The small dynamic range in FA values for this fusion protein (0.340–0.390) coupled with the nonlinear response of FA value with mass in this plateau region would make it difficult to record accurately a change in the FA value of GFP in complexes with larger proteins.
5 New Genetically Encoded Proteins for FA-Based Analysis of Proteins
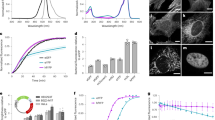
The lumazine-binding protein (LUMP) from Photobacterium leiognathi is a 20 kDa polypeptide composed of two structurally homologous spherical domains (α and β), with the α-subunit binding 6,7-dimethyl-8-(1′-d-ribityl) lumazine (ribityl-lumazine; Figs. 1d and 4) close to its surface with a dissociation constant of 16 nM at 20°C [25–27]. Ribityl-lumazine is synthesized within bacteria and most nonmammalian cells – when these cells are transfected with a gene encoding the α-subunit of LUMP, they produce large amounts of the fluorescent LUMP (Fig. 5a). The absorption spectrum of the lowest energy (S0–S1) transition of purified LUMP is similar to CFP and has a maximum at 420 nm (23,810 cm−1) and a full width half maximum of 3,468 cm−1. LUMP functions as a FRET acceptor in complexes with marine bacterial luciferases, and it is consequently highly optimized as a FRET acceptor probe, shifting the color of the bioluminescence to a cerulean color (Fig. 5a, b) [12, 25, 26]. This optimization also includes respectable fluorescence quantum yield (Φ f = 0.55) and, most importantly, one of the longest excited state lifetimes of any natural fluorescent proteins [12, 26].
High-resolution structure of LUMP showing the surface location of the bound ribityl-lumazine probe [25]
Characterization of LUMP. (a) Photograph recorded on an iPhone showing the cerulean-colored emission from LUMP in a culture of E. coli. (b) Peak-normalized absorption (dashed), excitation spectrum (green) with emission at 470 nm, and emission spectrum of LUMP measured in aqueous buffer. (c) Time-resolved fluorescence intensity decay of LUMP in aqueous buffer. (d) Excitation anisotropy scan with fluorescence emission at 470 nm superimposed on absorbance scan of LUMP. (e) Perrin–Weber plot of inverse anisotropy versus T/η where η is the viscosity in centipoise (cP) in a water/sucrose solution
The FA excitation spectrum of purified LUMP (20 kDa) in a viscous medium (75% sucrose) shows a fairly uniform S0–S1 transition that extends from 380 nm to 480 nm, with a limiting FA value of 0.350 (Fig. 5c). The limiting FA value of LUMP increases to 0.360 when measured at an even higher viscosity [26]. The theoretical maximum FA value of 0.400 is not attained for ribityl-lumazine bound to LUMP presumably because the absorption and emission dipoles are not colinear [25]. The Perrin–Weber plot for LUMP shows a linear plot from which the fluorescence lifetime is calculated as 13.25 ns (Fig. 5d), which is similar to that measured using fluorescence lifetime imaging microscopy (13.6 ns; FLIM; Fig. 5e) [12]. The predicted FA value of LUMP at 20°C is computed using Eq. (2) as 0.133 (τ f = 13.6 ns, r 0 = 0.360 [26]) and assumes τ c = 8.0 ns, which is based on the assumption that τ c increases by 1 ns for every 2.5 kDa increase in mass. The experimentally determined FA value of LUMP is 0.166 ± 0.002, suggesting that the protein molecule is not strictly spherical. The change in FA values between free LUMP at 1 cP (0.166) and its limiting FA value (0.360) is the largest recorded difference in FA values for a genetically encoded fluorescent protein – the corresponding change for GFP would be ~0.050 [24].
The simulations in Fig. 3 illustrate why LUMP and its fusion proteins are suitable probes for FA-based analysis of target proteins [12]. Referring to the purple-colored trace, the predicted FA value for fusion protein composed of LUMP fusion and a 20 kDa capture proteins (40 kDa in total) is ~0.195, which is close to the mid-range of FA values for LUMP – this value could in principle increase to the limiting FA value of 0.360 in complexes with very large proteins. For example, the FA value of this sensor would increase to 0.281 on binding to an 80 kDa target protein and reach 90% of the limiting FA on binding to a 325 kDa protein (τ c = 140 ns).
6 FA-Based Protein Sensors of GTP-Bound Cdc42
A LUMP-based FA sensor for GTP-bound Cdc42 was developed by appending the 32 amino acid GTPase-binding domain (GBD) from kinase ACK1 (human activated Cdc42 kinase 1: residues 448–489) [12] to the N-terminus of LUMP via a flexible six-amino acid linker (GSGSAS; Fig. 6a). LUMP-GBD (25 kDa) binds to Cdc42 specifically with a K d of 23 nM [26]. The FA value of unbound LUMP-GBD is 0.176 ± 0.004 and increases to 0.207 ± 0.002 (Δ ~ 18%) when bound in a stoichiometric complex with GTP-Cdc42. The FA values for the free and bound states of this LUMP sensor agree with those calculated from the Perrin–Weber equation. Thus, using τ f as 13.6 ns, and a limiting FA of 0.360, the free and bound forms of the FA sensor (with masses of 25 kDa and 47 kDa) with calculated τ c values of 10 ns and 19 ns, respectively, would result in calculated FA values of 0.155 and 0.212. Once again these values indicate that the LUMP-GBD molecule is not strictly spherical. Regardless the sensor allows one to conduct accurate titrations of a fixed concentration of LUMP-GBD with increasing amounts of GTP-bound Cdc42 (Fig. 6b), with saturation occurring at equimolar concentrations of Cdc42 and GBD-LUMP (10 μM).
7 Microscope-Based FA Imaging of LUMP Fusion Proteins
Having demonstrated the suitability of LUMP and its fusion proteins as genetically encoded FA sensors of target proteins, we subsequently showed that FA measurements on the same fusion proteins can be carried out using a slightly modified confocal fluorescence microscope (Zeiss 700). In particular, microscope-based imaging of the polarized fluorescence emission of LUMP was used to compute FA images to quantify the distribution of free and bound populations of a LUMP sensor in a sample [28]. A confocal fluorescence microscope was modified to incorporate two film polarizers that were placed in the emission path [12]. The intensity image of LUMP emission shows that the probe is largely localized to the outer surface of the 80 μm NTA-agarose beads, where it presumably binds to the surface coupled NTA. The polarized images of the emission of LUMP in the sample were first registered and then analyzed using Eq. (1) to generate steady-state-polarized emission images of His-tagged LUMP (23 kDa) in a field of nitrilotriacetic acid (NTA)-functionalized agarose beads. The FA values of His-tagged LUMP at the surface of the bead cluster are around 0.310 (Fig. 7b), which is within 14% of the limiting FA value. This result suggests that LUMP molecules are almost completely immobilized when bound to NTA-beads.
An FA image of His-tagged LUMP in solution with agarose beads lacking the NTA-group shows a uniform FA value of 0.185, which corresponds to the unbound LUMP probe. This study highlights an important benefit of using FA images to map the distributions of different molecular forms of the LUMP probe. In particular, since FA values are additive, we can estimate the fraction of LUMP molecules that are free or that interact transiently with the agarose bead in the FA image. Thus, fractional contributions of each species to the total intensity can be calculated according to the relationship (r measured = r 1 f 1 + r 2 f 2), where r 1 is the FA value of LUMP that is transiently immobilized of fractional intensity f 1, r 2 is the FA value of unbound LUMP within the bead of fractional intensity f 2, and f 1 + f 2 = 1. This feature of FA imaging is useful as it can be used to quantify the fractions of two populations of the probe in a sample, whereas the intensity image would indicate the presence of a single and uniform population of LUMP molecules. In this particular study, the FA value of His-tagged LUMP outside of the bead is 0.185 (Fig. 7b). The latter value arises from a mixture of free LUMP (0.185) and LUMP molecules that bind transiently and nonspecifically to the bead (0.310) – an FA value that is obtained from a study of NTA-agarose beads. Using the relationships above, the percentages of free and transiently bound LUMP within agarose beads are calculated as 76% and 24%, respectively.
8 Concluding Remarks
To date it has not been possible to use FA methods to quantify target proteins or their complexes in living cells. This limitation arises because small-molecule fluorophores with long-lived excited states are generally unsuitable for studies in living cells, owing to their requirements for UV excitation and associated phototoxicity and the considerable challenge of directing these probes to target proteins in the cell. Genetically encoded fluorescent proteins offer a chance to overcome these limitations, but their large mass and short lifetimes would severely limit the dynamic range of FA values between the free and bound states of the probe. Interestingly, FA-based measurements of homo-FRET can be used to map changes in the distributions of GFP-fusions with proteins that exist in monomeric or dimeric states [23]. LUMP and related proteins that bind tightly to flavin mononucleotide (Fig. 1e) and combine long-lived excited states with smaller mass than GFP hold great promise in advancing the FA approach to the study of protein complexes in living cells. These imaging studies can be conducted on most commercial confocal fluorescence microscopes with minor modifications. The FA imaging studies on LUMP conducted by our group were carried out using low numerical aperture (NA) objectives that do not result in a “high-NA effect.” In any case, our studies have shown that any high-NA effect within polarized images of fluorescence recorded with high-NA objectives can be corrected for quite easily and used to generate high-resolution FA images of a labeled sample [29].
LUMP is unique among genetically encoded fluorescent proteins in being sensitive changes in protein hydrodynamics and molecular volume. While Dansyl has been a popular probe for FA-based determinations of target proteins in vitro, we believe that LUMP, with its similar fluorescence lifetime, holds even greater promise for quantitative FA analysis of target proteins both in vitro and in vivo. This advantage derives in part from the more favorable excitation condition for LUMP compared to Dansyl. Moreover, the quality of a specific FA sensor can be greatly enhanced by encoding large capture sequences onto the C-terminus of LUMP. In principle it should be possible to generate a library of LUMP-based FA sensors with each member harboring a common LUMP probe and a sequence unique to each protein in an organism. LUMP and related genetically encoded FA sensors [12] also offer an attractive alternative to genetically encoded FRET-based sensors for high-throughput screening, quantitative imaging, and analysis of target molecules in biological systems. FA sensors are far easier to design compared to FRET fusion proteins, as they are composed of a single fluorescent probe linked to a specific capture sequence. Moreover, since FA values are additive, and given the large differences in FA values between the free and target-bound states of LUMP-based FA sensors, one can read out the amounts of the free and bound states of a sensor at any pixel in an FA image.
References
Hudson EN, Weber G (1973) Synthesis and characterization of two fluorescent sulfhydryl reagents. Biochemistry 12(21):4154–4161
Jameson DM (2001) The seminal contributions of Gregorio Weber to modern fluorescence spectroscopy, New trends in fluorescence spectroscopy. Springer, Heidelberg, pp 35–58
Marriott G, Zechel K, Jovin TM (1988) Spectroscopic and functional characterization of an environmentally sensitive fluorescent actin conjugate. Biochemistry 27(17):6214–6220
Sakata T, Yan Y, Marriott G (2005) Family of site-selective molecular optical switches. J Org Chem 70(6):2009–2013
Marriott G, Jovin TM, Yan-Marriott Y (1994) Synthesis and spectroscopic characterization of 1-bromo-(4-bromoacetyl) naphthalene. A thiol-reactive phosphorescent probe. Anal Chem 66(9):1490–1494
Weber G (1952) Polarization of the fluorescence of macromolecules. I. Theory and experimental method. Biochem J 51(2):145
Weber G (1952) Polarization of the fluorescence of macromolecules. II. Fluorescent conjugates of ovalbumin and bovine serum albumin. Biochem J 51(2):155
Hartley B (2004) The First Floor, Department of Biochemistry, University of Cambridge (1952–58). IUBMB Life 56(7):437–439
Richmond V, Hartley BS (1959) A two-dimensional system for the separation of amino acids and peptides on paper. Nature 184:1869–1870
Hartley BS, Massey V (1956) The active centre of chymotrypsin: I. Labelling with a fluorescent dye. Biochim Biophys Acta 21(1):58–70
Jameson DM, Ross JA (2010) Fluorescence polarization/anisotropy in diagnostics and imaging. Chem Rev 110(5):2685–2708
Hoepker AC, Wang A, Le Marois A, Suhling K, Yan Y, Marriott G (2015) Genetically encoded sensors of protein hydrodynamics and molecular proximity. Proc Natl Acad Sci 112(20):E2569–E2574
Chen RF (1967) Fluorescence of dansyl amino acids in organic solvents and protein solutions. Arch Biochem Biophys 120(3):609–620
Herron JN, Voss EW (1981) Characterization of fluorescent 2-dimethylaminonaphthalene-5-sulfonyl-immunoglobulin G conjugates for application in fluorescence polarization studies. J Biochem Biophys Methods 5(1):1–17
Hoepker A, Yan Y, Marriott G (2015) Genetically-encoded sensors of protein hydrodynamics. Oncotarget 6(19):16808
Tsao TC, Bailey K (1953) Aspects of polymerization in proteins of the muscle fibril. Discuss Faraday Soc 13:145–151
Tsao TC (1953) Fragmentation of the myosin molecule. Biochim Biophys Acta 11:368–382
Tsao TC (1953) The molecular dimensions and the monomer-dimer transformation of actin. Biochim Biophys Acta 11:227–235
Knopp J, Weber G (1967) Fluorescence depolarization measurements on pyrene butyric-bovine serum albumin conjugates. J Biol Chem 242(6):1353–1354
Shaner NC, Steinbach PA, Tsien RY (2005) A guide to choosing fluorescent proteins. Nat Methods 2(12):905–909
Suhling K, Siegel J, Lanigan PM, Lévêque-Fort S, Webb SE, Phillips D, Davis DM, French PM (2004) Time-resolved fluorescence anisotropy imaging applied to live cells. Opt Lett 29(6):584–586
Yan Y, Marriott G (2003) Analysis of protein interactions using fluorescence technologies. Curr Opin Chem Biol 7(5):635–640
Gautier I, Tramier M, Durieux C, Coppey J, Pansu RB, Nicolas J-C, Kemnitz K, Coppey-Moisan M (2001) Homo-FRET microscopy in living cells to measure monomer-dimer transition of GFP-tagged proteins. Biophys J 80(6):3000–3008
Volkmer A, Subramaniam V, Birch DJS, Jovin TM (2000) One- and two-photon excited fluorescence lifetimes and anisotropy decays of green fluorescent proteins. Biophys J 78:1589–1598
Chatwell Y et al (2008) Structure of lumazine protein, an optical transponder of luminescent bacteria. J Mol Biol 382:44–55
Lee J, O’Kane DJ, Visser AJWG (1985) Spectral properties and function of two lumazine proteins from photobacterium. Biochemistry 24:1476–1483
Sato Y et al (2010) Crystal structures of the lumazine protein from Photobacterium kishitanii in complexes with the authentic chromophore, 6,7-dimethyl- 8-(1'-d-ribityl) lumazine, and its analogues, riboflavin and flavin mononucleotide, at high resolution. J Bacteriol 192:127–133
Mott HR et al (1999) Structure of the small G protein Cdc42 bound to the GTPase-binding domain of ACK. Nature 399:384–388
Yan Y, Marriott G (2003) Fluorescence resonance energy transfer imaging microscopy and fluorescence polarization imaging microscopy. Methods Enzymol 360:561–580
Acknowledgments
The work was supported by grants awarded to GM including NIH R21CA191067-01. I would like to thank Prof. Yuling Yan for his help in editing the manuscript and Dr. Alexander Hoepker for his valuable discussions on fluorescence.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Marriott, G. (2016). Synthetic and Genetically Encoded Fluorescence Probes for Quantitative Analysis of Protein Hydrodynamics. In: Jameson, D. (eds) Perspectives on Fluorescence. Springer Series on Fluorescence, vol 17. Springer, Cham. https://doi.org/10.1007/4243_2016_17
Download citation
DOI: https://doi.org/10.1007/4243_2016_17
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-41326-6
Online ISBN: 978-3-319-41328-0
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)