Abstract
Every cell within living organisms actively maintains an intracellular Na+ concentration that is 10–12 times lower than the extracellular concentration. The cells then utilize this transmembrane Na+ concentration gradient as a driving force to produce electrical signals, sometimes in the form of action potentials. The protein family comprising voltage-gated sodium channels (NaVs) is essential for such signaling and enables cells to change their status in a regenerative manner and to rapidly communicate with one another. NaVs were first predicted in squid and were later identified through molecular biology in the electric eel. Since then, these proteins have been discovered in organisms ranging from bacteria to humans. Recent research has succeeded in decoding the amino acid sequences of a wide variety of NaV family members, as well as the three-dimensional structures of some. These studies and others have uncovered several of the major steps in the functional and structural transition of NaV proteins that has occurred along the course of the evolutionary history of organisms. Here we present an overview of the molecular evolutionary innovations that established present-day NaV α subunits and discuss their contribution to the evolutionary changes in animal bodies.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Hodgkin and Huxley in the 1940s–1950s suggested the presence of pores that enabled selective permeation of Na+ and K+ ions that was dependent on membrane depolarization and shaped the action potentials (APs). The Na+-based APs were first recorded from the giant axons of the squid Loligo (e.g., Hodgkin and Huxley 1945, 1952). Since then, we have come to recognize voltage-gated Na+ channels (NaVs) as a core element of the nerve impulse that supports essentially all brain function. William A. Catterall and colleagues succeeded in purifying the biochemical component of NaVs from rat brains (e.g., Beneski and Catterall 1980; Hartshorne and Catterall 1981; Hartshorne et al. 1985). Noda et al. (1984) isolated a cDNA encoding the pore-forming α subunit of the NaV from the electric eel, Electrophorus. Thereafter, cDNAs for other NaV α subunits have been cloned from mammals, other vertebrates, and invertebrates. In the fruit fly, Drosophila melanogaster, NaV mutants called para were isolated, and the later studies have clearly linked the genotypes of the NaV α subunit to cellular/whole body-level phenotypes (e.g., Loughney et al. 1989). Then the gene expression of NaV in response to neural inductive signaling was shown to underlie development of membrane excitability in neurons in a simple chordate model, the ascidian Halocynthia roretzi (Okamura et al. 1994). In recent years, Na+ channels derived from marine bacteria were utilized to increase our understanding of the structural and biophysical basis of Na+ ion selectivity and voltage dependence (e.g., Payandeh and Minor 2015; Catterall and Zheng 2015). And very recently, genes from the cockroach Periplaneta and from Electrophorus were utilized to finally resolve the 3D structures of metazoan NaVs (Shen et al. 2017; Yan et al. 2017). This brief history of Na+ channel research reflects the long, but cooperative, struggle to uncover the essence of nature (membrane excitability here) through the use of appropriate biological materials at appropriate times, seemingly making manifest August Krogh’s Principle (Krebs 1975).
The aforementioned analyses were performed using NaVs from a variety of animal species and demonstrated that the amino acid sequences of the channel proteins have changed to varying degrees and have incorporated innovations in accordance with the evolution of the animals harboring the channels. Given the well-known statement by T. Dobzhansky that nothing in biology makes sense except in the light of evolution, the diversity of NaVs provides us with rich insight. Like other gene families, the NaV gene family grew through gene duplication, sequence changes, and natural selection. These processes now enable animals to utilize similar but independent NaVs at different times during development and/or in specific cell types. In this review, we will outline mammalian, vertebrate, chordate, and metazoan NaV α subunit diversity and the cell type-specific usage of different isoforms, and we will introduce some of their molecular innovations that correlate with the evolution of animal lifestyles.
2 Structural Outlines of Voltage-Gated Sodium Channels
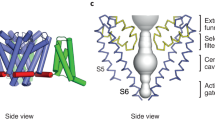
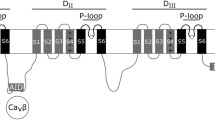
Metazoan NaVs consist of four serially homologous sections, domains I-IV (Fig. 1). Each domain contains six α-helices constituting transmembrane regions S1-S6 linked to each other by extracellular or intracellular loops (Fig. 1a). As a result, the full-length metazoan NaV polypeptide contains a total of 24 transmembrane (TM) regions spanning about 2,000 amino acids. As was proposed for voltage-gated potassium channels (KVs), the S1-S4 segments in each domain function as a voltage sensor (e.g., Catterall 2000). In particular, the S4 segment, in which every third amino acid is a positive charged Arg or Lys residue, is considered essential for sensing membrane voltage. In the resting state, S4 is positioned closer to the intracellular side of the membrane. Upon membrane depolarization, S4 moves in an extracellular direction, and this change of conformation stimulates the channel gate to open (Catterall 2000; Shen et al. 2017; Yan et al. 2017). The S5 and S6 segments and the loop between them (P-loop) from each domain occupy a quarter of the central portion of the protein such that the four domains together form the wall, gate, and ion-selectivity filter of the channel (Catterall 2000; Shen et al. 2017).
Mammalian NaV1 channel α subunits. (a) A schematic image of mammalian NaV1 channels. Diagnostic characteristics of NaV1s are indicated. NaV1s have 24 transmembrane segments and are composed of serially homologous domains I–IV, each of which contains segments S1–S6. S4 segments harbor evenly spaced positively charged residues (+). Amino acids at the inner vertices of the loops between S5 and S6 pore-forming segments (P-loops) constitute an ion-selectivity filter. In the case of NaV1s, the pore signature is Asp/Glu/Lys/Ala (D/E/K/A). The loop between domains II and III contains an ankyrin-binding motif sequence that is reportedly important to localize the NaV1 channels to the axon initial segment and nodes of Ranvier. The sequence between domains III and IV functions as the inactivation latch (inactivation ball), the core of which is represented by the well-conserved Ile-Phe-Met (I-F-M) triplet. (b) A molecular phylogenetic tree of mammalian NaV1 α subunits constructed by the maximum-likelihood method based on gap-free 1,372 amino acid positions using MEGA7. The sequence from the ascidian Halocynthia is used as the out-group. The bootstrap values over 80 are shown. The four groups (NaV1.1/1.2/1.3/1.7, NaV1.4, NaV1.6, NaV1.5/1.8/1.9) are recognized (see text and Table 1)
Mammals, including humans, express nine NaV isoforms (NaV1.1 to NaV1.9). In addition, a protein most similar to NaV1.7, called NaX, has also been identified in mammals (Fig. 1b, Table 1). NaX does not exhibit ion permeability. Instead, it appears to function as a sensor of extracellular Na+ ion and is now thought to be involved in ionic homeostasis in the body (Hiyama et al. 2002, 2004; Hiyama and Noda 2016). The cDNA sequences, predicted amino acid sequences, locations of the respective encoding genes (SCN1A-11A) on the chromosomes, spatiotemporal expression patterns, sensitivity to toxins [e.g., tetrodotoxin (TTX)], single-channel conductances, characteristics of inactivation, and biological functions of these isoforms have all been comprehensively analyzed (some characteristics of mammalian NaVs are listed in Table 1) (Goldin 2001; Catterall et al. 2005). For instance, mammalian NaV1.1, 1.2, and 1.3 are encoded by SCN1A, 2A, and 3A, respectively, and these genes are tandemly arrayed within the genome (on the “q” arm of chromosome 2 in the case of humans) close to the HoxD gene cluster (Table 1). The primary structures of these isoforms are mutually similar, suggesting they emerged through relatively recent tandem gene duplications (Fig. 1b). This tandem cluster also includes the gene for NaV1.7 (SCN9A), which is expressed in the peripheral nervous system (PNS), including the dorsal root ganglia (DRG) (Sangameswaran et al. 1997; Toledo-Aral et al. 1997). The gene encoding NaX (SCN7A) is also found in this cluster. NaV1.7 and NaX constitute a sister clade in the molecular phylogenetic tree, and the duplication of these two genes occurred in parallel with the splits of NaV1.1, 1.2, and 1.3 (Fig. 1b) (Catterall et al. 2005). They commonly show high sensitivity to TTX, with IC50s around 10 nM (summarized in Goldin 2001). In addition to the channels already mentioned, the central nervous system (CNS) expresses NaV1.6, which exhibits characteristic persistent current and a large resurgent current, while the PNS expresses NaV1.8 and 1.9. The heart expresses NaV1.5, encoded by SCN5A, which clusters with SCN10A encoding NaV1.8 and SCN11A encoding NaV1.9, close to the HoxA cluster (Table 1). NaV1.8 and 1.9 are also presumed to have emerged through tandem gene duplication that occurred after the two rounds of whole genome duplication in the ancestor of jawed vertebrates (see below) and share relatively slow activation kinetics (Fig. 1b) (Lai et al. 2004). Along with NaV1.5, NaV1.8 and 1.9 are relatively insensitive to TTX, with IC50 in the range of 1–10 μM or more (Gellens et al. 1992; Akopian et al. 1996; Tate et al. 1998). Skeletal muscles use NaV1.4 for APs, and its gene, SCN4A, is not clustered with other NaV genes and neighbors the HoxB cluster (Table 1). NaV1.4 in skeletal muscle is as sensitive to TTX as NaVs in the CNS (NaV1.1–1.3, and 1.6).
All nine NaVs possess an activation gate that opens in response to membrane depolarization and an ion-selectivity filter that enables selective Na+ permeation. The extracellular fluid around living cells generally contains a high concentration of Na+, while the intracellular fluid has a lower Na+ concentration. NaV gating allows Na+ to flow into the cell and depolarize the membrane, though the channel soon shuts through the process of inactivation. The inactivation function is one that the NaV α subunits themselves possess and enables immediate repolarization of the membrane to sharpen the AP (Hodgkin and Huxley 1952). All known NaVs show some degree of inactivation, which is known to require the linker sequence between domains III and IV (Stühmer et al. 1989), a region called the “inactivation ball” or “inactivation latch” (Fig. 1a). The latch is modeled to fit into the open pore and block ion permeation (ball-and-chain model). Within the sequence of this linker, three consecutive hydrophobic amino acids, Ile-Phe-Met, are well shared by vertebrate NaVs and are totally conserved in all the human NaV isoforms. This triplet, especially the central Phe residue, is essential for inactivation, and thus hydrophobic interaction between the latch and the open pore would be important (West et al. 1992; see also a recent revision to this model proposed from the structural study by Yan et al. 2017).
The P-loop between S5 and S6 in each of the four domains protrudes into the central canal of NaVs, and the inner vertices of the loops are thought to constitute the ion-selectivity filter (Heinemann et al. 1992; Shen et al. 2017). The residues for Na+ ion selectivity in all mammalian NaVs are Asp from domain I, Glu from domain II, Lys from domain III, and Ala from domain IV (asymmetric D/E/K/A signature) (Fig. 1a). By contrast, most CaVs, which have a similar 24-TM conformation, show a symmetric E/E/E/E pore signature (e.g., Hinemann et al. 1992). The 3–4 acidic residues behind the D/E/K/A signature residues (E/E/D/D in all mammalian NaVs) form the “outer ring,” which is also significant for positively charged ion permeability (Catterall 2000). The region around the signature residue in domain I is a definitive binding target for TTX (Noda et al. 1989). This selectivity filter is located midway through the canal, and the outer ring is a bit extracellular to the signature. On the intracellular side is a central cavity enclosed by charged residues, with the predicted activation gate composed of the S6 segments from domains I-IV aligned along the channel fenestration (Catterall 2000; Shen et al. 2017).
These structural features, including the 24-TM segments in the four serially homologous domains, the voltage-sensor domains, D/E/K/A selectivity filter, central cavity, activation gate, and inactivation latch are shared by all mammalian NaV α subunits (e.g., Catterall 2000; Catterall et al. 2005).
3 Historical Origin of Voltage-Gated Sodium Channels and Their Related Proteins
The discovery of NaVs in bacteria (BacNaVs) confirmed that Na+-permeable channel proteins gated in response to changes in membrane potential had already evolved in prokaryotes (Ren et al. 2001; Payandeh and Minor 2015). However, these BacNaVs are composed of homotetrameric 6-TM subunits that contain compact S1-S6 segments (e.g., Payandeh and Minor 2015; Catterall and Zheng 2015). Because a BacNaV can be changed into Ca2+-selective channel through simple mutation(s), it is thought that prokaryotes can also possess voltage-gated Ca2+ channels (Yue et al. 2002). In addition, the KVAP channel, the first voltage-gated ion channel whose structure was resolved, is a K+ channel from prokaryotic archaea (Jiang et al. 2003). It is thus evident that prokaryotes likely made use of a wide variety of voltage-gated, ion-specific 6-TM channels long before the emergence of eukaryotes. The 6-TM segments can be functionally divided into a voltage-sensing unit composed of segments S1-S4 and an ion channel pore domain corresponding to segments S5-S6, including the P-loop in between. Members of the prokaryotic KcsA-related channel and eukaryotic Kir channel families are composed of homotetrameric 2-TM helices, which are thought to represent units of the tetraradial pore domains (e.g., Bichet et al. 2003). The K2P channel family members show a tandem repeat of two pore domains, dimerization of which produces a channel pore of pseudotetrameric symmetry (e.g., Honoré 2007). Recent research has revealed that the voltage-sensor domain can also function independently of the pore domain. For instance, the recently identified voltage-sensing phosphatase (VSP) possesses S1-S4-like segments coupled to phosphatase domain that catalyze membrane phospholipid dephosphorylation in response to changes in membrane potential (Murata et al. 2005). It is also known that the primary structure of a voltage-dependent proton channel, HV1/voltage-sensor-domain-only protein (VSOP), is comparable to segments S1-S4 of 6-TM voltage-gated ion channel subunits (Ramsey et al. 2006; Sasaki et al. 2006).
All of the prokaryotic voltage-gated ion channels identified so far are composed of homotetrameric 6-TM subunits. Eukaryotic voltage-gated K+ channels and Ca2+-permeable Catsper channels are still tetrameric compositions of 6-TM subunits (Liebeskind et al. 2013). In addition to these, eukaryotes express larger voltage-gated ion channel proteins composed of 12-TM or 24-TM segments. The 12-TM channels are two-pore channels (TPCs), which are cation channels functioning within intracellular organelles such as endosomes and lysosomes (Calcraft et al. 2009). The 24-TM ion channel family includes not only NaVs but also L-, T-, and N/P/Q/R-type CaVs (also called CaV1, CaV2, and CaV3, respectively) and cation leak channels (NALCN). It is thought that eukaryotic 24-TM channels harboring four-time serially homologous domains are derived from two rounds of tandem duplication of an original 6-TM factor. In fact, the amino acid sequences of domain I of the NaV and CaV1, 2, and 3 channels are more similar to domain III than to II or IV, while the sequence in domain II is more like that of IV than domain I or III (Strong et al. 1993; Liebeskind et al. 2013). It is also known that the amino acid sequences of these four domains are closer to each other, and to those of the Catsper isoforms, than to the sequences of BacNaVs or KVs. These relationships suggest eukaryotic 24-TM channels were not derived from duplication of a BacNaV, but arose instead through two sequential rounds of duplication of a gene encoding a Catsper-like 6-TM protein; one duplication gave rise to a gene encoding 12-TM segments containing domain I and II, and a second tandem duplication established the present conformation composed of domains I-IV (Liebeskind et al. 2013). This process was significant in that the ion channel molecules got to be formed by a single polypeptide stretch, not by four identical subunits, which would facilitate accumulation of “asymmetric” mutations independently within domains I to IV to make the channel “pseudotetrameric.” This implies that this process would potentiate future molecular evolutionary fine-tuning for specific functions – e.g., Na+ selectivity, fast inactivation, anchoring, etc. (see below).
In addition to the channels mentioned to far, another 24-TM subfamily, NaV2, has been identified in invertebrates, and its members have amino acid sequences similar to NaV1, but exhibits a D/E/E/A pore signature (Salkoff et al. 1987; Sato and Matsumoto 1992; Nagahora et al. 2000; Zhou et al. 2004; Zakon 2012; Gur Barzilai et al. 2012; Moran et al. 2015). Later analyses proved that the members of this family are permeable to Ca2+, and it has recently been proposed that this family be renamed CaV4 (Zhou et al. 2004; Gosselin-Badaroudine et al. 2016). It has also been reported that a mutant NaV1.2 channel of rat giving a D/E/E/A pore signature is permeable not only to Na+ but also to Ca2+ and K+ and that the presence of a physiological concentration of Ca2+ in the extracellular fluid blocks permeation of Na+ through the mutant channel (Heinemann et al. 1992). This nonselective permeation of cations is also observed in a cnidarian NaV family channel having the D/E/E/A pore (called NvNaV2.1), although it does not show the blockade by extracellular Ca2+ (Gur Barzilai et al. 2012). Given its sequence similarity to NaV1 and its functional characteristics, we will refer to this subfamily as NaV2(CaV4) here.
The major diversification events of these families of 24-TM channels of animals, namely, evolutionary splits of the NaV, CaV, and NALCN families, predate the origin of metazoan animals (Fig. 2a). The split of the NaV1/NaV2(CaV4) clade from the CaV1–3 molecular clades occurred before the divergence of animals and choanoflagellates, the protozoan group most closely related to the metazoan clade (Zakon 2012; Liebeskind et al. 2012; Moran et al. 2015). NALCN-like sequences are also found in fungi, and the history of this family can be traced back to the origin of opisthokonta, the monophyletic group containing animals, choanoflagellates, and fungi (Torruella et al. 2011; Liebeskind et al. 2012). Figure 2a shows here that in fact the representative unicellular eukaryotes distantly related with each other, such as the yeast Saccharomyces, the ciliate Paramecium, and green algae Chlamydomonas and Micromonas, possess different types of the 24-TM channel proteins. For instance, Saccharomyces, Chlamydomonas, and Micromonas express a 24-TM channel related to NALCN of animals, while Paramecium, Chlamydomonas, and Micromonas use other types of 24-TM channels closer to NaVs or CaVs of animals (Fig. 2a). This suggests that the gene duplication of NALCN, NaV, and CaV (even between CaV1/2 and CaV3) families preceded the split of Unikonta (including animals, fungi, and amoebozoans) and Bikonta (plants, algae, etc.), namely, had occurred close to or before the origin of eukaryotes (Fig. 2a) (see also the phylogenetic views of eukaryotic lineages in Roger and Simpson 2008; Rogozin et al. 2009; Cavalier-Smith 2010). Despite the deep origin of the voltage-gated 24-TM Ca2+/Na+ channels, it is also known that many eukaryotic groups lack the genes of them, probably because of secondary loss. Brunet and Arendt (2015) argued that the losses of 24-TM NaV/CaV channels that had happened in varied eukaryote lineages are tightly correlated with the absence of flagella in those organisms. The CaV channels of Paramecium and Chlamydomonas are localized in the membrane of their cilia/flagella, in which the channels function to generate APs to change flagellar/ciliary beating waveforms (Machemer and Ogura 1979; Fujiu et al. 2009). The flagellar localization of the 24-TM CaV/NaV channels may be a preadaptation for the later emergence of neurons (Brunet and Arendt 2015).
Molecular phylogeny of eukaryotic 24-TM channels. (a) An unrooted maximum-likelihood tree of 24-TM channels found in green algae (Chlamydomonas reinhardtii, Micromonas commoda), the ciliate (Paramecium tetraurelia), the fungi (Saccharomyces cerevisiae), the apusozoan flagellate (Thecamonas trahens), choanoflagellates (Monosiga brevicollis, Salpingoeca rosetta), and metazoans (Drosophila melanogaster, Homo sapiens). Accession numbers of the sequences are indicated in the tree. The groups of NaV1/NaV2(CaV4), CaV1/2, CaV3, and NALCN and their extended clades are indicated with gray solid and dashed lines. The bootstrap values over 60 are shown. The tree was constructed using the WAG model on MEGA7 from gap-free 847 amino acid positions aligned by MUSCLE program. (b) The pore signatures (at the putative ion-selectivity filter in the P-loops) of the eukaryotic 24-TM channels. The signature sequences were obtained from the alignment used for constructing the tree shown in (a). The species codes (first column) made of the first three and two letters from the genus and species name, respectively, and accession numbers (second column) are shown. The amino acids at the pore signatures are highlighted with colors [Asp (D), magenta; Glu (E), red; Lys (K), blue; Ala (A), yellow; other polar amino acids, green]
While the phylogenetic relationships among the protein sequences from various eukaryotes are based on overall sequence similarities, the ion-selectivity filter signatures (D/E/K/A, E/E/E/E, E/E/D/D, and E/E/K/E in mammalian NaV1, CaV1/2, CaV3, and NALCN, respectively) were established relatively recently (Fig. 2b) (Liebeskind et al. 2011, 2012). This means that the specific ion selectivity of each family may have changed over the course of evolution. For instance, the D/E/K/A pore signature of NaV1 is thought to have emerged at the origin of Bilateria (the clade of animals with bilateral body plans). The evidence is that all NaV-type 24-TM genes found so far in choanoflagellates (the closest relatives to metazoans) (Monosiga and Salpingoeca in Fig. 2) and in ctenophores and placozoans (non-bilaterian metazoans), encode the D/E/E/A filter motif, not D/E/K/A, which implies the NaV2(CaV4)-type pore signature is original (Liebeskind et al. 2011; Zakon 2012; Gur Barzilai et al. 2012; Moran et al. 2015). Cnidarians, another major non-bilaterian metazoan group, express several isoforms of NaV-like 24-TM polypeptides with D/E/E/A (called NaV2.1, 2.3, and 2.4), D/E/E/T (NaV2.2), and D/K/E/A (NaV2.5) pore signatures, and a phylogenetic analysis suggested that the latter two of the isoforms were derived from the first (Fig. 3) (Gur Barzilai et al. 2012; Moran et al. 2015). The cnidarian D/K/E/A channels (NaV2.5) are presumably Na+-selective, representing convergent evolution of Na+-selective channels that was independent from the origin of the D/E/K/A NaV1 subfamily in bilaterians (see below) (Anderson et al. 1993; Gur Barzilai et al. 2012). This molecular parallel evolution may be related to independent acquisitions of large bodies moving fast in cnidarians (e.g., jellyfish) and in bilaterians. On the other hand, the pore signature of cnidarian NALCN-like genes is commonly E/E/E/E, which likely ensures Ca2+ selectivity, while those in all known bilaterians have E/E/K/E at the selectivity filter, making the channels to be cation nonselective (Lu et al. 2007; Liebeskind et al. 2012).
From the evidence summarized above, it can be predicted that at their origin animals possessed two or three CaV subfamily genes, one NaV2(CaV4)/NaV1 subfamily gene and one NALCN-like gene (Moran and Zakon 2014; Moran et al. 2015). Interestingly, none of the channels are Na+-selective, but are Ca2+-preferential. Animals may have evolved as organisms lacking voltage-gated Na+-selective channels, and the true metazoan Na+ channels, NaV1s with the D/E/K/A signature, emerged in a bilaterian ancestor via gene duplication and diversification that at the same time gave rise to the NaV2(CaV4) subfamily.
4 Evolution of Bilaterians and Voltage-Gated Sodium Channel Proteins
Bilateria is composed of animals having bilateral body plans, which are classified into three superphyla, Deuterostomia, Lophotrochozoa, and Ecdysozoa. These animal groups share a basic repertoire of 24-TM ion channel paralogues of three subtypes of CaV (CaV1–3), two of NaV1/NaV2(CaV4), and one of NALCN leak channel. The molecular phylogenetic analysis suggests that the NaV1 subfamily was diverged from the NaV2(CaV4) subfamily and originated in the last common ancestor of bilaterians (Fig. 3) (Liebeskind et al. 2011; Moran et al. 2015). The origin of NaV1 in fact correlated with the development of the D/E/K/A pore signature and therefore represents the occurrence of “true” Na+ selectivity in 24-TM channels (Fig. 3) (Liebeskind et al. 2011). This channel enabled bilateral animals to specifically utilize Na+, the most abundant cation within the environment in which they had adapted. Combined with the function of the Na+ pump (Na,K-ATPase), NaV1 would have conferred a fast electric responsiveness to excitable cells by means of the large driving force across the membrane. NaV1 would also be important in that it could mediate steep depolarization without direct stimulation of the intracellular processes triggered by Ca2+, a major second messenger in the wide variety of unicellular and multicellular organisms. It is also possible the inactivation latch in the loop between domains III and IV developed at the same time (Fig. 3b), which would have facilitated fast recovery to the resting state of membrane potential and thus enabled cells to minimize the changes in intracellular ionic conditions. Importantly, the molecular development of NaV1 would be related not only to fast generation/propagation of APs but also to the occurrence of refractory periods after depolarization. Repetitive excitation of neurons is facilitated by these characteristics of NaV1, which enables the nerves to encode neural signals based on the AP frequencies. Fast neural transmission would have supported evolution of larger bodies, and the ability to inactivate itself can ensure unidirectional flow of signals within the neural network. This would have offered segregation of input (sensory) systems from output (motor) systems, permitting development of the CNS.
Molecular phylogeny of metazoan NaV1/NaV2(CaV4) channels. (a) A maximum-likelihood tree of NaV-related channels from the ctenophore (Mnemiopsis leidyi), the placozoan (Trichoplax adhaerens), the jellyfish (Cyanea capillata), the sea anemone (Nematostella vectensis), squids (Doryteuthis opalescens, Heterololigo bleekeri), the sea slug (Aplysia californica), the annelid worm (Capitella teleta), the fruit fly (Drosophila melanogaster), the cockroach (Blattella germanica), acorn worms (Ptychodera flava, Saccoglossus kowalevskii), the starfish (Acanthaster planci), the sea urchin (Strongylocentrotus purpuratus), ascidians (Ciona intestinalis, Halocynthia roretzi), the larvacean (Oikopleura dioica), the elephant fish chimaera (Callorhinchus milii), the electric eel (Electrophorus electricus), and humans (Homo sapiens). Monophyly of bilaterian NaV1s is evident, while that of NaV2(CaV4) channels is not clear here. The sequences from non-bilaterian metazoans, ecdysozoans, lophotrochozoans, and deuterostomes are labeled with yellow, blue, green, and magenta branches, respectively. NCBI accession numbers and other ID codes are indicated in the tree. The sequences of O. dioica are obtained from OikoBase (http://oikoarrays.biology.uiowa.edu/Oiko/); those of C. intestinalis are from Ghost database (http://ghost.zool.kyoto-u.ac.jp/cgi-bin/gb2/gbrowse/kh/); those of A. planci and P. flava are from the OIST genome browsers (http://marinegenomics.oist.jp/gallery/gallery/index). The bootstrap values over 80 are shown. The tree was constructed using the WAG model on MEGA7 from gap-free 812 amino acid positions aligned by MUSCLE program. (b) The pore signatures and the region corresponding to the inactivation latch in metazoan NaV-related channels. The sequences shown are mostly identical to those in (a). The species codes and color codes are same as in Fig. 2. Sequences corresponding to core triplet of the inactivation latch (I-F-M) are also marked by purple. Cyan residues indicate the amino acids identical to those around the inactivation latch of the mammalian NaV1.6. *1-*6: the genemodel IDs are shown in (a) to identify the sequences in the genome browser of each organism
The evolutionary emergence and divergence of bilaterians represents the geographical event called the “Cambrian explosion.” It is inspiring to consider that the origin of NaV1 is concordant with the evolution of bilaterians. Paleontologists have suggested that predator-prey relationships were established during the Cambrian era, which stimulated increases in body size and complexity and also the sophistication of sensory organs, leading to acceleration of movement and further elaboration of the CNS (Conway-Morris 1986; Gould 1990; Parker 2003). One hypothesis has proposed that development of eyes was a key step in this so-called evolutionary big bang (Parker 2003). Given this context, it would be reasonable to predict that the molecular phylogenetic origin of NaV1 provided a physiological basis for the bilaterian ancestor that potentiated the explosive evolution.
NaV1 originated through gene duplication and diversification that gave rise to the NaV2(CaV4) as well (Fig. 3a). Consequently, most bilaterians express two subtypes of NaV family components, NaV1 and NaV2(CaV4). For example, the genome of Drosophila melanogaster harbors para, which encodes a NaV1-type channel, and DSC1, which encodes a NaV2(CaV4) channel (Salkoff et al. 1987; Ramaswami and Tanouye 1989; Loughney et al. 1989; Hong and Ganetzky 1994; Kulkarni et al. 2002; Zhang et al. 2013). Major invertebrate lineages, such as mollusks, annelids, arthropods, and chordates all express NaV2(CaV4) proteins in addition to NaV1. NaV2(CaV4) family proteins contain voltage sensors – i.e., S4 segments containing evenly spaced, positively charged residues, and their pore signature is generally D/E/E/A. When exogenously expressed in Xenopus oocytes, NaV2(CaV4) family proteins from the cockroach and honeybee (called BSC1 and AmCaV4, respectively) are more permeable to the divalent cations Ca2+ and Ba2+ than to Na+ (Zhou et al. 2004; Gosselin-Badaroudine et al. 2016). These channels are reportedly insensitive to TTX, exhibit relatively slow activation and inactivation, and can be blocked by Cd2+ or Zn2+. Phenotypic analyses of DSC1 gene mutants in Drosophila have suggested its functions in olfaction or odor-responsive behavior and also in stabilizing the performance of neural circuits under stresses (Kulkarni et al. 2002; Zhang et al. 2013).
While bilateral animals share orthologues of NaV1 and NaV2(CaV4), it has also been revealed that some animal lineages have lost either the NaV1 or NaV2(CaV4) subtype, or both. It is well known, for example, that the genome of Caenorhabditis elegans contains neither NaV1 nor NaV2(CaV4) (e.g., Okamura et al. 2005). Whether these paralogues are present or absent is thought to reflect the physical characteristics and lifestyle of each animal group, including body size, locomotion speed, and/or complexity of neural processing. Although the nematode lacks any NaV-class channels, it is not true that this species is “primitive.” The nematode lost this protein because it was not essential for its interstitial life. In fact, nematodes have flourished around the globe without it.
The NaV1 family channels are absent from echinoderms and hemichordates, the group collectively called Ambulacraria, which constitutes the sister clade of the phylum Chordata (Fig. 3) (see also Widmark et al. 2011; Gur Barzilai et al. 2012). This indicates that echinoderms and hemichordates (ambulacrarians) secondarily lost the fast NaV1, while the NaV2(CaV4) with the D/E/E/A pore signature typical for that subfamily remains. This suggests these animal groups are incapable of fast sodium spikes. These animals are small during the larval stage (about 0.1–5.0 mm), and the adults (about 1–10 cm or more sometimes) are generally slow moving. They are not fast predators but protective; echinoderm adults are covered with calcite skeletons, and sometimes also with spines, while hemichordates are generally buried in the seafloor. They develop an ectodermal nerve net over the entire body, and their CNS is relatively rudimentary (Hyman 1955; Holland 2003, 2016; Nomaksteinsky et al. 2009). Their evolutionary status may be regarded like an atavism – i.e., reminiscent of the status of animals before the origin of bilaterians. In other words, they live a “slow life” that is a consequence of the loss of NaV1. On the contrary, the last common ancestor of bilaterians had a NaV1-type channel and lived a “quick life.” Therefore, the long-standing controversy around how the less centralized nervous system seen in echinoderms or hemichordates was integrated into the well-centralized nervous system of vertebrates may not be valid (for reviews, e.g., Holland 2003, 2016). It is noteworthy that recent analyses of the CNS of a polychaete annelid (ragworm) provided surprising evidence of its deep anatomical similarity to the vertebrate CNS (Tessmar-Raible et al. 2007; Tomer et al. 2010; Vergara et al. 2017). A well-centralized nervous system in the last common ancestor of bilaterians would be consistent with the evolution of NaV1.
5 Voltage-Gated Sodium Channels in Chordates
As the bilaterians diversified, the chordate lineage led to vertebrates and two other animal groups, the amphioxi (cephalochordates, lancelets) and tunicates (urochordates). Recent comparative genomic analyses showed that the amphioxi diverged first within these three groups, and tunicates and vertebrates form a sister group (Delsuc et al. 2006; Putnam et al. 2008). Amphioxi and tunicates provided “observation windows” for researchers to investigate past situations before establishment of vertebrate bodies.
Amphioxi inhabit sandy shores and live as filter feeders. They are able to swim out of the sand and dive back into it very quickly. While they develop segmented somites, they lack developed eyes or an expanded brain (Willey 1894). Their repertoire of NaV-related α subunits also appears primitive. An amphioxus likely possesses five genes encoding NaV1/NaV2(CaV4) channel α subunits. At least three of the genes are classified as NaV1 family, and the other two are in the NaV2(CaV4) clade (Fig. 4a). The three NaV1 channels of amphioxi are paralogous with each other and share the D/E/K/A pore signature. On the other hand, the differences in their amino acid sequences are considerable, implying differing functions of the isoforms and situation-dependent differential utilizations (tissue- or life stage-specific expression, etc.). A clearer sign of functional specificity is seen in the NaV2(CaV4) proteins; the one (depicted as NaV2a in Fig. 4) contains a D/D/Q/A, not the D/E/E/A, pore signature at the ion-selectivity filter. Although we do not know the ion permeability of amphioxus NaV2a, it is conceivable the D/D/Q/A signature invests the excitable membranes of this animal with a regulatory option. Examination of these organisms enables us to monitor an “evolutionary experiment” carried out through gene duplication before the emergence of vertebrates (Ohno 1970).
Molecular phylogeny of chordate NaV1 and NaV2(CaV4) channels. (a) A maximum-likelihood tree of NaV-related channels from acorn worms (Ptychodera flava, Saccoglossus kowalevskii), the starfish (Acanthaster planci), the sea urchin (Strongylocentrotus purpuratus), amphioxi (Branchiostoma belcheri and B. floridae), ascidians (Ciona intestinalis, Halocynthia roretzi), the larvacean (Oikopleura dioica), the lamprey (Petromyzon marinus), the elephant fish chimaera (Callorhinchus milii), the whale shark (Rhincodon typus), the coelacanth (Latimeria chalumnae), and humans (Homo sapiens). The clades of NaV1 and NaV2(CaV4) are clearly divided. Vertebrates possess NaV2(CaV4). The sequences from amphioxi, tunicates (ascidians and larvaceans), and vertebrates are labeled with purple, yellow, and red branches, respectively. NCBI accession numbers and other ID codes are mostly indicated in the tree. *1-*9 indicate the genemodel IDs in the genome browser of each organism: *1, KH.C9.462.v3.A.SL1–1; *2, GSOIDP00013476001; *3, KH.C1.1161.v1.A.ND1–1; *4, Harore.CG.MTP2014.S1.g14830; *5, GSOIDP00011229001; *6, KH.C10.502.v2.A.SL1–1; *7, Harore.CG.MTP2014.S25.g02359; *8, KH.C5.200.v1.A.ND1–1; *9, GSOIDP00005042001. The O. dioica sequences are obtained from OikoBase (http://oikoarrays.biology.uiowa.edu/Oiko/); the C. intestinalis sequences are from Ghost database (http://ghost.zool.kyoto-u.ac.jp/cgi-bin/gb2/gbrowse/kh/); the H. roretzi sequences are from Aniseed database (https://www.aniseed.cnrs.fr/); the P. marinus sequences are from the UCSC genome browser gateway (http://genome-asia.ucsc.edu/cgi-bin/hgGateway); and the A. planci and P. flava sequences are from the OIST genome browsers (http://marinegenomics.oist.jp/gallery/gallery/index). The bootstrap values over 90 are shown. The tree was constructed using the WAG model on MEGA7 from gap-free 526 amino acid positions aligned by MUSCLE program. (b) The pore signatures and the regions corresponding to ankyrin-binding motif and the inactivation latch in chordate NaV-related channels. The species codes and color codes are as used in Figs. 2 and 3. Amino acids identical to the mammalian NaV1.6 ankyrin-binding motif are indicated by brown. *1-*9: the genemodel IDs identical to those in (a)
Similar traces are also found in the tunicate lineage, which possess four types of NaV1/NaV2(CaV4) proteins. Ascidians, constituting a representative tunicate class, abundantly distribute along the shores around the world. Their adult form is sessile, while the larvae are in the form of tiny tadpoles that swim in the sea. Ascidians have been utilized as a research model, because of their kinship to vertebrates, but also because of their abundance and the availability of mature gametes among other features. Ascidians have contributed to ion channel studies through the “mosaicism” of their embryogenesis. Historical studies revealed that neuron-like Na+ spikes could be evoked in the neural cell-lineage blastomere of embryos whose cleavage was arrested using an inhibitor of cytokinesis (Takahashi and Yoshii 1981; Takahashi and Okamura 1998). Developmental expression of this Na+ current in the neural cell-lineage blastomere is dependent on a fibroblast growth factor-like inductive signal from a neighboring endomesodermal blastomere, which represents neural induction. This process of differentiation in membrane excitability is firmly correlated with the gene expression of a NaV1 channel, originally called TuNaI (referred to as NaV1a here) (Okado and Takahashi 1988; Okamura et al. 1994; Takahashi and Okamura 1998). This NaV1a α subunit is actually expressed in all known neuronal types (Okamura et al. 1994; Okada et al. 1997). Later studies carried out before and after the genomic sequencing of several species of ascidians revealed that ascidians have four genes encoding Na+ channel α subunits (Nagahora et al. 2000; Okamura et al. 2005; Brozovic et al. 2016). One encodes NaV1a (TuNaI) containing the typical D/E/K/A pore signature, an inactivation latch with the I-F-M triplet between domains III and IV, and a sequence similar to the ankyrin-binding motif found in the loop between domains II and III of vertebrate NaV1s (see below). Another encodes a NaV2(CaV4) subfamily protein containing the typical D/E/E/A pore signature (NaV2, previously called TuNa2), but lacking clear consensus sequences for the inactivation latch and ankyrin-binding motif (Fig. 4) (Nagahora et al. 2000). This gene encoding NaV2(CaV4) is also expressed in some, but not all, neurons in ascidians (Nagahora et al. 2000). The nested patterns of NaV1 and NaV2(CaV4) gene expression are reminiscent of the patterns of para and DSC1 expression in Drosophila embryos (Hong and Ganetzky 1994).
The other two channels in ascidians, called here NaV1b and NaV1c (previously named NaV3 and 4, respectively, in Okamura et al. 2005), are categorized in the NaV1 family, and the tunicate NaV1a, b, and c constitute a clade different from that including the vertebrate NaV1s (Fig. 4a). While the ion-selectivity filter signature of the tunicate NaV1c is the same as that in typical NaV1-type channels (D/E/K/A), NaV1b exhibits a D/E/(K or T or M)/E pore signature (Fig. 4b) (see also Widmark et al. 2011). We do not know the ion permeability of either channel. Temporal expression patterns estimated from the counts of expressed sequenced tags (ESTs) in the ascidian Ciona intestinalis (Satou et al. 2003) suggests the tunicate NaV1a (TuNaI) is expressed in the larval and adult nervous systems which is consistent with in situ analyses of this gene expression pattern (Okamura et al. 1994, 2005; Okada et al. 1997). NaV2 is estimated to be expressed in the nervous systems of larvae and juveniles, and possibly on the juvenile endostyle, which is an organ putatively homologous to the vertebrates’ thyroid gland. The EST counts suggest expression of NaV1b occurs during the larval stage. Our preliminary examination of the spatial expression pattern in C. intestinalis showed that the NaV1b gene is expressed in neurons in the CNS and PNS and in some of the muscle cells in the larva. The EST counts predict a small amount of NaV1c is expressed during the larval stage, though our preliminary in situ hybridization analysis did not detect any clear signal during the larval stage. Also, intriguing is the detection of EST counts for NaV1b and NaV1c in mature hermaphroditic adults producing gametes, not in young immature adults. In the eggs of ascidians, steep membrane depolarization is evoked in response to fertilization, which is known to be mediated by an unknown voltage-gated Na+ channel that is somewhat permeable to Ca2+ along with Na+ (Okamoto et al. 1977; Fukushima 1981; Okamura and Shidara 1987). Thus NaV1b and/or NaV1c may be involved in this process.
Single-channel recordings from cleavage-arrested neuronal blastomeres of the ascidian Halocynthia roretzi support this view. The electrophysiology has uncovered three types of voltage-gated sodium currents that turn over with time after fertilization to matured stage (Okamura and Shidara 1990a, b). The “Type A” Na+ current shows only one decay phase during voltage-dependent inactivation, suggesting that a single type of NaV is responsible for this current. Type A currents are seen in every blastomere within early embryos, and its expression level appears highest at the gastrula stage. This type of Na+ current is identical to that in the fertilization potential of Halocynthia eggs represented by a Na+-dependent AP (Fukushima 1981). The NaV current in Halocynthia eggs is insensitive to TTX but is highly sensitive to scorpion toxin and local anesthetics (Okamoto et al. 1977). These data suggest that the tunicate NaV1b, which has an atypical pore signature and is thus resistant to TTX, is involved in the Type A current. On the other hand, voltage-dependent inactivation of the “Type C” current shows two different, fast and slow, phases of decay (Okamura and Shidara 1987), which is reminiscent of the Nav1.6 channel in mammalian neurons. The Type C is the most predominant in differentiated neuronal blastomeres and is suppressed by microinjection of antisense DNA targeting the Halocynthia NaV1a gene TuNaI (Okamura et al. 1994). During the short period between the disappearance of Type A and appearance of Type C currents during the developmental course of neural-type membrane excitability, an unusual voltage-gated Na+ current, “Type B,” is transiently expressed (Okamura and Shidara 1990a). This current shows persistent gating behavior with multiple short openings (burst activity). At present, the relationship between the classically characterized diversity of voltage-gated Na+ currents and the ascidian NaV isoforms remains unclear, though it appears that NaV1a (TuNa1) carries Type C current. It would be interesting to know whether the tunicate NaV1b or NaV1c carries the Type A current and what underlies the Type B current.
The tree topology shown in Fig. 4 suggests that the ancestor of tunicate NaV1 paralogues became the seed from which there was further molecular evolution of the NaV1 channels in modern vertebrates. The NaV1a of tunicates shares an ankyrin-binding motif sequence with the vertebrate NaV1s (Fig. 1) (Hill et al. 2008), which consists of a dozen amino acids residing in the loop between domains II and III (Fig. 4b), while the NaV1b and NaV1c proteins almost lost it. Similarly, the I-F-M inactivation latch that is conserved in the tunicate NaV1a has been lost in the paralogues, NaV1b and NaV1c (Fig. 4b). Another ankyrin-binding motif similar to that in vertebrate NaV1s is also found in vertebrate KCNQ2/3 (KV7.2/7.3) K+ channels, and these motifs are crucial for ankyrin-G binding and for anchoring of NaV1s and KCNQ2/3s at the axon initial segment (AIS) and nodes of Ranvier in myelinated neurons (Garrido et al. 2003; Lamaillet et al. 2003; Pan et al. 2006; Hill et al. 2008). The sequences of the motif in ascidians’ NaV1a varies somewhat, but ~70% of the amino acids are conserved (Fig. 4b). Even the NaV1 channel in amphioxi, depicted as Branchiostoma NaV1a in Fig. 4, has ~40% identity, though we find no other traces in invertebrate NaV1s (Fig. 4b) (Hill et al. 2008). Myelination and resultant saltatory conduction are regarded as a feature of jawed vertebrates (gnathostomes) (Zalc et al. 2008; Zalc 2016). Despite the absence of nodes of Ranvier in amphioxi and tunicates, the ankyrin-binding motif emerged in these animals and may have initiated interaction with ankyrins within neurons. This may be a key property that the ancestral gene of the tunicate NaV1 paralogues retained, and the reason it was selected as the seed for further evolution in the vertebrate lineage.
The situations seen in amphioxi and tunicates inform us that the isoforms occurring in these so-called “protochordate” organisms through gene duplication differentially evolved, leading to changes, even into the pore signature (as seen in NaV1b of tunicates and NaV2b of amphioxi). The gene duplications can confer specific regulatory options to each of duplicated isoforms as indicated so far (Ohno 1970). What occurred in these organisms is a prelude to what has occurred in the vertebrate lineage: another story of gene duplication and functional differentiation.
6 Evolution of NaV1 Channels in Vertebrates
The monophyletic vertebrate lineage has given rise to agnathans (hagfish and lampreys), cartilaginous fish (sharks, skates, and rays), ray-finned fish (bichirs, sturgeons, gars, bowfin, and teleosts), lobe-finned fish (paraphyletic group of coelacanths and lungfish), and tetrapods (amphibians, reptiles, birds, and mammals) (e.g., Amemiya et al. 2013). During this process of radiation, these organisms inhabited and adapted to various environments in salt and freshwater and in wet and dry terrestrial areas. An ability for predation has been especially well developed in this animal lineage, and several ideas have been proposed in that regard (e.g., Gans and Northcutt 1983). Myelination and saltatory conduction along neuronal axons, as well as the developmental capacities derived from neural crest and placode cells that especially enhanced sensory systems, referred to as “new head,” have enabled vertebrates to become larger and predatory (e.g., Gans and Northcutt 1983; Zalc 2016). The presence of active predators in turn stimulated greater ability to efficiently recognize the predators so as to escape (Parker 2003). Increased complexity of sensory inputs, higher ordered neural processing, and high-speed regulation of locomotion would have strongly supported the radiation of vertebrates under water and on land. NaV1 function was definitely essential to those evolutionary steps.
The lamprey Petromyzon marinus shows multiple types of predicted transcripts encoding NaV1 channel α subunits. Two NaV1 isoforms have previously been identified (Hill et al. 2008; Zakon 2012), but four types, at least, may be there (Fig. 4b). Cartilaginous fish, including the elephant fish (chimaera) Callorhinchus milii, and the whale shark Rhincodon typus also appear to harbor four to five (or more) predicted transcripts for NaV1 isoforms (Fig. 4; some sequences were too short and thus omitted here). Our molecular phylogenetic analysis suggests the NaV1 isoforms in cartilaginous fish well reflect an original state (one gene in each of NaV1.4, NaV1.5/1.8/1.9, NaV1.6, and NaV1.1/1.2/1.3/1.7 groups), before the extensive duplication that occurred in amniotes and in teleosts (Figs. 4 and 5) (see below) (Widmark et al. 2011). It remains difficult, however, to know precise full-length sequence data for these transcripts, and there is not yet sufficient data available to draw firm conclusions.
Molecular phylogeny of vertebrate NaV1s. (a) A maximum-likelihood tree of NaV1 α subunits from the elephant fish chimaera (Callorhinchus milii), the spotted gar (Lepisosteus oculatus), the teleost zebrafish (Danio rerio), the coelacanth (Latimeria chalumnae), the amphibian frog (Xenopus tropicalis), and humans (Homo sapiens). NaV1.4 (magenta), NaV1.6 (green), and NaV1.5/1.8/1.9 (yellow) are monophyletic, while NaV1.1/1.2/1.3/1.7 sequences (blue) does not form a clade in this tree. The branches of mammalian NaV1s are marked with darker colors. NCBI accession numbers are indicated in the tree. The L. oculatus sequences are obtained from Ensembl (http://www.ensembl.org/Lepisosteus_oculatus/Info/Index), and thus sequence IDs are of UniProt. The bootstrap values over 80 are shown. The tree was constructed using the WAG model on MEGA7 from gap-free 1,183 amino acid positions aligned by MUSCLE program. (b) The pore signatures and the regions corresponding to ankyrin-binding motif and the inactivation latch in vertebrate NaV1s. The species codes and color codes are as used in Figs. 2, 3, and 4
Three lamprey NaV1 α subunits for which longer sequence information was found in the database have the D/E/K/A pore signature. On the other hand, at least two of them contain the I-F-M inactivation ball in the loop between domains III and IV, while the third has an I-F-L triplet. The ankyrin-binding motif is conserved to varying degrees (50–100%) in their domain II-III loops (Fig. 4b). The variation in sequence motifs may represent differences of the molecular functions among them, which implies efficient “evolution by gene duplication” working in this gene family from the beginning of the vertebrate lineage (Ohno 1970).
The ankyrin-binding motif may reportedly work to locate and accumulate NaV1s at the AIS in the neurons of lampreys (Hill et al. 2008). True myelin sheaths have not been found along the axons of lampreys, though molecular traces of myelination have been detected (Smith et al. 2013). On the other hand, the proximal portion of neuronal axons in lampreys is thinner than elsewhere along the axon. This narrow initial segment decreases local capacitance and conductance and supports the occurrence of steep APs. Localization of NaV1 at high density in the AIS makes sense for efficient induction of APs (Hill et al. 2008; Kole and Stuart 2012), and full establishment of NaV1 localization at the AIS via its ankyrin-binding motif may have facilitated the increase in body size of the vertebrate ancestor. This AIS machinery may be a preadaptation for the evolution of nodes of Ranvier and saltatory conduction in gnathostomes (jawed vertebrates). KCNQ2/3 (Kv7.2/7.3) channels emerged in gnathostomes and, together with NaV1s, were directed toward the AIS and nodes of Ranvier via their ankyrin-binding motifs, concomitantly with the evolutionary appearance of true myelination (Hill et al. 2008). This innovation would not only change neuronal morphology, but would greatly accelerate the rates of signal relay and processing among sensory organs, neurons themselves, and effectors, which would in turn contribute to the evolution of jaws and the predatory lifestyle.
Interestingly, the genomes of the lamprey, cartilaginous fish, and the coelacanth Latimeria possess the gene for NaV2(CaV4) channel (Fig. 4). The pore signature is D/E/E/G in the Petromyzon NaV2(CaV4), but in cartilaginous fish and the coelacanth, the pore signature is the common D/E/E/A (Fig. 4b). It had been thought that NaV2(CaV4) was present only in invertebrate animals, but it is now recognized that this channel remains in the vertebrate lineage. On the other hand, we did not find NaV2(CaV4) in the genomes of ray-finned fish or amphibians, which suggests the gene encoding this channel was independently lost from the lineages of ray-finned fish and tetrapods. Considering that both “gene loss” events appear to have occurred as the animals left the sea for inland environments, changes in the ionic conditions may have decreased the selection pressure to keep the NaV2(CaV4) gene. However, since the operational principles of this type of channel are still unclear, this issue remains to be determined.
7 Independent Gene Duplications of NaV1 in Teleosts and Amniotes
Recent comparative genome analyses indicate that the last common ancestor of ray-finned fish and tetrapods possessed at least four types of NaV1, each of which was linked to HoxA-D clusters (Widmark et al. 2011; Zakon et al. 2011). The genes encoding NaV1.1, 1.2, 1.3, and 1.7 in amniotes, as well as the NaX shared by therian mammals, emerged from the ancestral gene linked with HoxD cluster through a series of gene duplications, probably in the sequence of [(1.3 (1.2, 1.1)) (1.7, X)] (Fig. 5) (Widmark et al. 2011; Zakon et al. 2011). On the other hand, the gene encoding NaV1.6, a neural NaV1 linked to HoxC, apparently experienced no gene duplications and was single throughout amniote evolution (Fig. 5). In mammals, NaV1.1, 1.2, 1.3, and 1.6 are mainly expressed in CNS neurons and contribute to rapid APs, while NaV1.1 and 1.6 are also expressed to some extent in PNS neurons, and NaV1.7 is selectively expressed in PNS neurons (Table 1) (Goldin 2001). This situation implies that the amniote ancestor utilized NaV1.7 (more precisely the mother gene of 1.7 and 1.1/1.2/1.3) in PNS and NaV1.6 in CNS to independently control the excitability of PNS and CNS neurons, respectively. Later, NaV1.1–1.3, derived from a NaV1.7-like ancestral protein, were recruited for the operation of the CNS in amniotes. The diversity of NaV1.1, 1.2, 1.3, and 1.6 (i.e., the CNS subtypes) enabled independent control of their subcellular distributions in individual neurons, since expression of these subtypes in CNS is not only cell type specific but also cellular compartment specific (Hu et al. 2009; Lorincz and Nusser 2010; Zakon et al. 2011). This diversification of these CNS subtypes would have fine-tuned membrane excitabilities among CNS neurons and led to higher level brain performance in ancestral amniotes, which would contribute to forebrain expansion in amniotes (Zakon et al. 2011).
Like NaV1.6, the amniote NaV1.4 linked to HoxB was not duplicated and remained single. This subtype has presumably remained to work in the skeletal muscle of amniotes as well as anamniotes (Widmark et al. 2011; Zakon et al. 2011). Other subtypes, NaV1.5, 1.8, and 1.9, are clustered with HoxA and were derived via sequential gene duplication, probably in the sequence [1.5 (1.8, 1.9)] (Fig. 5) (Widmark et al. 2011; Zakon et al. 2011). The mammalian (likely all the amniote) NaV1.5 is expressed in the heart (Table 1) (Rogart et al. 1989), as is the NaV1.5 (more precisely the ancestor gene for NaV1.5 and 1.8/1.9) in shark and lungfish (Zakon et al. 2011). The genomes of the lizard Anolis and chick Gallus contain the genes encoding NaV1.8 and 1.9, as well as 1.5, while that of the frog Xenopus does not, suggesting that these innovations occurred with the evolution of amniotes, as in the case of NaV1.1/1.2/1.3/1.7 diversification (Zakon et al. 2011; see also Fig. 5).
It is known that NaV1.8 and NaV1.9 are specialized to work in the nociceptive DRG neurons, a neural crest derivative, in mammals (and likely in all amniotes) (Table 1) (e.g., Lai et al. 2004). The amniote genes encoding NaV1.8 and 1.9 were derived from a NaV1.5-like mother gene and predominantly function for nociception. Unlike other NaV1s, NaV1.8 and 1.9 show slow activation and inactivation (reviewed in Lai et al. 2004), despite a conserved I-F-M inactivation latch in both isoforms (Fig. 5b). The slow kinetics of NaV1.8/1.9 may favor generation of graded membrane potential changes, which could be linearly related to nociceptive input. In fact, these channels, especially NaV1.9, are expressed in small unmyelinated neurons (Lai et al. 2004), which is consistent with the fact that the ankyrin-binding motif is not well conserved in mammalian NaV1.9 (Fig. 5b). This suggests critical aspects of nociceptive properties were innovations in amniotes. Another possible benefit of using multiple NaV1s for nociception is that the slow NaV1.8 and 1.9, fast 1.7, and possibly fast 1.5 may make possible special tuning of the propagation speed of nociceptive signals (Lai et al. 2004). This would enable, for example, adjustment for larger body size or constant/high body temperature in ancestral amniotes. However, answering this question must await testing whether NaV1s expressed in nociceptive peripheral neurons do in fact function in nociceptive neurons in amniotes other than mammals and also await characterization of nociceptive properties in anamniotes.
This expansion of the NaV1 gene family in amniotes, and the additional event needed to derive NaV1.7 and NaX in the therian ancestor, was mediated through tandem gene duplication. This kind of tandem gene duplication was not detected in their neighboring genes nor in the amniote CaV genes. Thus “evolution by gene duplication” in this case apparently occurred specifically to NaV1 family genes of amniotes (Zakon et al. 2011). This diversification pattern is in contrast to what happened in the lineage of ray-finned fish, where another round of whole genome duplication took place in the ancestor of teleosts (e.g., Novak et al. 2006b; Braasch et al. 2016). Indeed, in the zebrafish genome, at least eight genes for NaV1 isoforms have been identified – zscn1Laa/ab, 5Laa/ab, 8aa/ab, and 4aa/ab associated with HoxDa/b, HoxAa/b, HoxCa/b, and HoxBa/b clusters, respectively, although the HoxDb cluster was lost from teleosts (Novak et al. 2006b; Widmark et al. 2011; Zakon et al. 2011, Won et al. 2012). The mode of gene duplication that took place in teleosts was not specific to genes encoding NaV1s; it was global and thus distinct from what happened in amniotes. The gene expression pattern analyses of the zebrafish NaV1 isoforms revealed considerable variation in the expressed isoforms among the CNS, PNS, heart, and skeletal muscles (Novak et al. 2006a; Won et al. 2012). For example, while the zebrafish heart expresses both of the two paralogous NaV1.5 genes (zscn5Laa and ab), PNS cells from the DRG express only one of the paralogues (5Laa) (Novak et al. 2006a; Won et al. 2012). Electrophysiological studies of zebrafish DRG neurons confirmed that a type of Na+ current is slowly inactivating and recorded from smaller neurons (presumably mediated by the NaV1.5 encoded in zscn5Laa), and another Na+ current is rapidly inactivating and recorded from larger neurons (Won et al. 2012). Given that other fast NaV1s are also expressed in the DRG (Won et al. 2012), functional diversification of peripheral neurons and differential utilization of fast and slow NaV1 isoforms may have been established in parallel in teleosts and mammals, although the degree of functional diversification in the DRG neurons (and also in the kinetics of the expressed NaV1s) appears greater in those of mammals.
A remarkable feature of the amniotes’ body is an elaborate forebrain with a large number of neurons. Ancestral amniotes must have developed multiple sensory systems related to their large body size and to terrestrial habitation, which enabled them to utilize complex environmental stimuli for complex motor behaviors (Zakon et al. 2011). “Specific” expansion of the genes encoding NaV1 isoforms in amniotes, namely, the evolution of NaV1.1–1.3 and 1.8/1.9, provided functional options that supported radiation of amniotes. This may have relieved the limitation on body size and facilitated modal innovations in the sensory system and high-speed signal processing in the CNS, which constituted a morphological and physiological framework preadapted for the evolution of humankind.
8 Concluding Remarks
Along the history of voltage-gated Na+ channels, one can see repetitive duplication and diversification of functional units. To establish the 24-TM channel family, two rounds of tandem duplication of a Catsper-like 6-TM unit were required to build a polypeptide composed of serially homologous quadruple domains. The unification of the 24-TM segments made possible asymmetric mutation within a single pseudotetraradial channel such that this structure became a unit for further duplication and diversification that gave rise to various types of voltage-gated channels. NaV channels were established under this molecular evolutionary trend, and the asymmetric pore signatures of NaV1 (D/E/K/A) and NaV2(CaV4) (D/E/E/A), as well as the asymmetric domains that include the inactivation latch and the ankyrin-binding domain emerged. In the lineage leading to NaV channels, some of the genes having emerged via duplication were evolutionary dead ends (e.g., NaV2 paralogues in cnidarians including NaV2.5 with the D/K/E/A pore signature or NaV1b and 1c in tunicates), while others became the seeds for future duplication and diversification. The repertoire of NaV-related channels shrank in some animal lineages but grew in others. Loss of the NaV1 gene in ambulacrarians is an example of the former, while special expansion of NaV1 in amniotes is an example of the latter; and each is in good accord with their specific evolutionary steps for adaptation. The presence/absence of NaV and the multiplication of their isoforms led to fundamental modal shifts in the membrane excitability in animal cells. These fundamental changes expanded/restricted the possibilities for physiological and morphological adaptation of animal bodies, extending the limit on body size, increasing the speed of sensory signal processing and locomotion, facilitating predatory life or habitation on land with an elaborate brain. The evolutionary history of NaVs reflects well the major steps in the broader evolution of organisms on earth, with the former possibly serving as a motive force for the latter. The involvement of NaV2(CaV4) channels in animal evolution was also touched on here. Several ion channel groups appear to have lost their Ca2+ permeability in parallel during the course of vertebrate evolution (Schredelseker et al. 2010; Nishino et al. 2011; Hirai et al. 2017). Further studies of the types of Ca2+-permeable/impermeable NaV-related channels may provide another key to understanding unknown aspects of cellular and organismal adaptations to their environment.
References
Akopian AN, Sivilotti L, Wood JN (1996) A tetrodotoxin-resistant voltage-gated sodium channel expressed by sensory neurons. Nature 379:257–262
Amemiya CT, Alföldi J, Lee AP, Fan S, Philippe H, Maccallum I, Braasch I, Manousaki T, Schneider I, Rohner N, Organ C, Chalopin D, Smith JJ, Robinson M, Dorrington RA, Gerdol M, Aken B, Biscotti MA, Barucca M, Baurain D, Berlin AM, Blatch GL, Buonocore F, Burmester T, Campbell MS, Canapa A, Cannon JP, Christoffels A, De Moro G, Edkins AL, Fan L, Fausto AM, Feiner N, Forconi M, Gamieldien J, Gnerre S, Gnirke A, Goldstone JV, Haerty W, Hahn ME, Hesse U, Hoffmann S, Johnson J, Karchner SI, Kuraku S, Lara M, Levin JZ, Litman GW, Mauceli E, Miyake T, Mueller MG, Nelson DR, Nitsche A, Olmo E, Ota T, Pallavicini A, Panji S, Picone B, Ponting CP, Prohaska SJ, Przybylski D, Saha NR, Ravi V, Ribeiro FJ, Sauka-Spengler T, Scapigliati G, Searle SM, Sharpe T, Simakov O, Stadler PF, Stegeman JJ, Sumiyama K, Tabbaa D, Tafer H, Turner-Maier J, van Heusden P, White S, Williams L, Yandell M, Brinkmann H, Volff JN, Tabin CJ, Shubin N, Schartl M, Jaffe DB, Postlethwait JH, Venkatesh B, Di Palma F, Lander ES, Meyer A, Lindblad-Toh K (2013) The African coelacanth genome provides insights into tetrapod evolution. Nature 496:311–316
Anderson PAV, Holman MA, Greenberg RM (1993) Deduced amino acid sequence of a putative sodium channel from the scyphozoan jellyfish Cyanea capillata. Proc Natl Acad Sci U S A 90:7419–7423
Beneski DA, Catterall WA (1980) Covalent labeling of protein components of the sodium channel with a photoactivable derivative of scorpion toxin. Proc Natl Acad Sci U S A 77:639–643
Bichet D, Haass FA, Jan LY (2003) Merging functional studies with structures of inward-rectifier K+ channels. Nat Rev Neurosci 4:957–967
Braasch I, Gehrke AR, Smith JJ, Kawasaki K, Manousaki T, Pasquier J, Amores A, Desvignes T, Batzel P, Catchen J, Berlin AM, Campbell MS, Barrell D, Martin KJ, Mulley JF, Ravi V, Lee AP, Nakamura T, Chalopin D, Fan S, Wcisel D, Cañestro C, Sydes J, Beaudry FE, Sun Y, Hertel J, Beam MJ, Fasold M, Ishiyama M, Johnson J, Kehr S, Lara M, Letaw JH, Litman GW, Litman RT, Mikami M, Ota T, Saha NR, Williams L, Stadler PF, Wang H, Taylor JS, Fontenot Q, Ferrara A, Searle SM, Aken B, Yandell M, Schneider I, Yoder JA, Volff JN, Meyer A, Amemiya CT, Venkatesh B, Holland PW, Guiguen Y, Bobe J, Shubin NH, Di Palma F, Alföldi J, Lindblad-Toh K, Postlethwait JH (2016) The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat Genet 48:427–437
Brozovic M, Martin C, Dantec C, Dauga D, Mendez M, Simion P, Percher M, Laporte B, Scornavacca C, Di Gregorio A, Fujiwara S, Gineste M, Lowe EK, Piette J, Racioppi C, Ristoratore F, Sasakura Y, Takatori N, Brown TC, Delsuc F, Douzery E, Gissi C, McDougall A, Nishida H, Sawada H, Swalla BJ, Yasuo H, Lemaire P (2016) ANISEED 2015: a digital framework for the comparative developmental biology of ascidians. Nucleic Acids Res 44:D808–D818
Brunet T, Arendt D (2015) From damage response to action potentials: early evolution of neural and contractile modules in stem eukaryotes. Philos Trans R Soc Lond B Biol Sci 371:20150043
Calcraft PJ, Arredouani A, Ruas M, Pan Z, Cheng X, , Hao X, Tang J, Rietdorf K, Teboul L, Chuang K-T, Lin P, Xiao R, Wang C, Zhu Y, Lin Y, Wyatt CN, Parrington J, Ma J, Evans AM, Galione A, Zhu MX (2009) NAADP mobilizes calcium from acidic organelles through two-pore channels. Nature 459:596–600
Catterall WA (2000) From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron 26:13–25
Catterall WA, Zheng N (2015) Deciphering voltage-gated Na+ and Ca2+ channels by studying prokaryotic ancestors. Trends Biochem Sci 40:526–534
Catterall WA, Goldin AL, Waxman SG (2005) International union of pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels. Pharmacol Rev 57:397–409
Cavalier-Smith T (2010) Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. Biol Lett 6:342–345
Conway-Morris S (1986) The community structure of the Middle Cambrian Phyllopod Bed (Burgess Shale). Palaeontology 29:423–467
Delsuc F, Brinkmann H, Chourrout D, Philippe H (2006) Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 439:965–968
Fujiu K, Nakayama Y, Yanagisawa A, Sokabe M, Yoshimura K (2009) Chlamydomonas CAV2 encodes a voltage-dependent calcium channel required for the flagellar waveform conversion. Curr Biol 19:133–139
Fukushima Y (1981) Identification and kinetic properties of the current through a single Na+ channel. Proc Natl Acad Sci U S A 78:1274–1277
Gans C, Northcutt RG (1983) Neural crest and the origin of vertebrates: a new head. Science 220:268–273
Garrido JJ, Giraud P, Carlier E, Fernandes F, Moussif A, Fache MP, Debanne D, Dargent B (2003) A targeting motif involved in sodium channel clustering at the axon initial segment. Science 300:2091–2094
Gellens ME, George AL Jr, Chen L, Chahine M, Horn R (1992) Primary structure and functional expression of the human cardiac tetrodotoxin-insensitive voltage-dependent sodium channel. Proc Natl Acad Sci U S A 89:554–558
Goldin AL (2001) Resurgence of sodium channel research. Annu Rev Physiol 63:871–894
Gosselin-Badaroudine P, Moreau A, Simard L, Cens T, Rousset M, Collet C, Charnet P, Chahine M (2016) Biophysical characterization of the honeybee DSC1 orthologue reveals a novel voltage-dependent Ca2+ channel subfamily: CaV4. J Gen Physiol 148:133–145
Gould SJ (1990) Wonderful life: the Burgess Shale and the nature of history. WW Norton & Co., New York
Gur Barzilai M, Reitzel AM, Kraus JE, Gordon D, Technau U, Gurevitz M, Moran Y (2012) Convergent evolution of sodium ion selectivity in metazoan neuronal signaling. Cell Rep 2:242–248
Hartshorne RP, Catterall WA (1981) Purification of the saxitoxin receptor of the sodium channel from rat brain. Proc Natl Acad Sci U S A 78:4620–4624
Hartshorne RP, Keller BU, Talvenheimo JA, Catterall WA, Montal M (1985) Functional reconstitution of the purified brain sodium channel in planar lipid bilayers. Proc Natl Acad Sci U S A 82:240–244
Heinemann SH, Terlau H, Stühmer W, Imoto K, Numa S (1992) Calcium channel characteristics conferred on the sodium channel by single mutations. Nature 356:441–443
Hill AS, Nishino A, Nakajo K, Zhang G, Fineman JR, Selzer ME, Okamura Y, Cooper EC (2008) Ion channel clustering at the axon initial segment and node of Ranvier evolved sequentially in early chordates. PLoS Genet 4:e1000317
Hirai S, Hotta K, Kubo Y, Nishino A, Okabe S, Okamura Y, Okado H (2017) AMPA glutamate receptors are required for sensory-organ formation and morphogenesis in the basal chordate. Proc Natl Acad Sci U S A 114:3939–3944
Hiyama TY, Noda M (2016) Sodium sensing in the subfornical organ and body-fluid homeostasis. Neurosci Res 113:1–11
Hiyama TY, Watanabe E, Ono K, Inenaga K, Tamkun MM, Yoshida S, Noda M (2002) NaX channel involved in CNS sodium-level sensing. Nat Neurosci 5:511–512
Hiyama TY, Watanabe E, Okado H, Noda M (2004) The subfornical organ is the primary locus of sodium-level sensing by NaX sodium channels for the control of salt-intake behavior. J Neurosci 24:9276–9281
Hodgkin AL, Huxley AF (1945) Resting and action potentials in single nerve fibers. J Physiol 104:176–195
Hodgkin AL, Huxley AF (1952) Currents carried by sodium and potassium ions through the membrane of the giant axon of Loligo. J Physiol 116:449–472
Holland ND (2003) Early central nervous system evolution: an era of skin brains? Nat Rev Neurosci 4:617–627
Holland ND (2016) Nervous systems and scenarios for the invertebrate-to-vertebrate transition. Phil Trans R Soc Lond B 371:20150047
Hong CS, Ganetzky B (1994) Spatial and temporal expression patterns of two sodium channel genes in Drosophila. J Neurosci 14:5160–5169
Honoré E (2007) The neuronal background K2P channels: focus on TREK1. Nat Rev Neurosci 8:251–261
Hu W, Tian C, Li T, Yang M, Hou H, Shu Y (2009) Distinct contributions of NaV1.6 and NaV1.2 in action potential initiation and backpropagation. Nat Neurosci 12:996–1002
Hyman LH (1955) The invertebrates: Echinodermata. The coelomic Bilateria. McGraw-Hill, New York
Jiang Y, Lee A, Chen J, Ruta V, Cadene M, Chait BT, MacKinnon R (2003) X-ray structure of a voltage-dependent K+ channel. Nature 423:33–41
Kole MH, Stuart GJ (2012) Signal processing in the axon initial segment. Neuron 73:235–247
Krebs HA (1975) The August Krogh principle: “For many problems there is an animal on which it can be most conveniently studied”. J Exp Zool 194:221–225
Kulkarni NH, Yamamoto AH, Robinson KO, Mackay TFC, Anholt RR (2002) The DSC1 channel, encoded by the smi60E locus, contributes to odor-guided behavior in Drosophila melanogaster. Genetics 161:1507–1516
Lai J, Porreca F, Hunter JC, Gold MS (2004) Voltage-gated sodium channels and hyperalgesia. Annu Rev Pharmacol Toxicol 44:371–397
Lamaillet G, Walker B, Lambert S (2003) Identification of a conserved ankyrin-binding motif in the family of sodium channel alpha subunits. J Biol Chem 278:27333–27339
Liebeskind BJ, Hillis DM, Zakon HH (2011) Evolution of sodium channels predates the origin of nervous systems in animals. Proc Natl Acad Sci U S A 108:9154–9159
Liebeskind BJ, Hillis DM, Zakon HH (2012) Phylogeny units animal sodium leak channels with fungal calcium channels in an ancient, voltage-insensitive clade. Mol Biol Evol 29:3613–3616
Liebeskind BJ, Hillis DM, Zakon HH (2013) Independent acquisition of sodium selectivity in bacterial and animal sodium channels. Curr Biol 23:R948–R949
Lorincz A, Nusser Z (2010) Molecular identity of dendritic voltage-gated sodium channels. Science 328:906–909
Loughney K, Kreber R, Ganetzky B (1989) Molecular analysis of the para locus, a sodium channel gene in Drosophila. Cell 58:1143–1154
Lu B, Su Y, Das S, Liu J, Xia J, Ren D (2007) The neuronal channel NALCN contributes resting sodium permeability and is required for normal respiratory rhythm. Cell 129:371–383
Machemer H, Ogura A (1979) Ionic conductances of membranes in ciliated and deciliated Paramecium. J Physiol 296:49–60
Moran Y, Zakon HH (2014) The evolution of the four subunits of voltage-gated calcium channels: ancient roots, increasing complexity, and multiple losses. Genome Biol Evol 6:2210–2217
Moran Y, Liebeskind BJ, Zakon HH (2015) Evolution of voltage-gated ion channels at the emergence of Metazoa. J Exp Biol 218:515–525
Murata Y, Iwasaki H, Sasaki M, Inaba K, Okamura Y (2005) Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature 435:1239–1243
Nagahora H, Okada T, Yahagi N, Chong JA, Mandel G, Okamura Y (2000) Diversity of voltage-gated sodium channels in the ascidian larval nervous system. Biochem Biophys Res Commun 275:558–564
Nishino A, Baba SA, Okamura Y (2011) A mechanism for graded motor control encoded in the channel properties of the muscle ACh receptor. Proc Natl Acad Sci U S A 108:2599–2604
Noda M, Shimizu S, Tanabe T, Takai T, Kayano T, Ikeda T, Takahashi H, Nakayama H, Kanaoka Y, Minamino N, Kangawa K, Matsuo H, Raftery MA, Hirose T, Inayama S, Hayashida H, Miyata T, Numa S (1984) Primary structure of electrophorus electricus sodium channel deduced from cDNA sequence. Nature 312:121–127
Noda M, Suzuki H, Numa S, Stühmer W (1989) A single point mutation confers tetrodotoxin and saxitoxin insensitivity on the sodium channel II. FEBS Lett 259:213–216
Nomaksteinsky M, Röttinger E, Dufour HD, Chettouh Z, Lowe CJ, Martindale MQ, Brunet JF (2009) Centralization of the deuterostome nervous system predates chordates. Curr Biol 19:1264–1269
Novak AE, Taylor AD, Pineda RH, Lasda EL, Wright MA, Ribera AB (2006a) Embryonic and larval expression of zebrafish voltage-gated sodium channel α-subunit genes. Dev Dyn 235:1962–1973
Novak AE, Jost MC, Lu Y, Taylor AD, Zakon HH, Ribera AB (2006b) Gene duplications and evolution of vertebrate voltage-gated sodium channels. J Mol Evol 63:208–221
Ohno S (1970) Evolution by gene duplication. Springer, New York
Okada T, Hirano H, Takahashi K, Okamura Y (1997) Distinct neuronal lineages of the ascidian embryo revealed by expression of a sodium channel gene. Dev Biol 190:257–272
Okado H, Takahashi K (1988) A simple “neural induction” model with two interacting cleavage-arrested ascidian blastomeres. Proc Natl Acad Sci U S A 85:6197–6201
Okamoto H, Takahashi K, Yamashita N (1977) One-to-one binding of a purified scorpion toxin to Na channels. Nature 266:465–468
Okamura Y, Shidara M (1987) Kinetic differences between Na channels in the egg and the neutrally differentiated blastomere in the tunicate. Proc Natl Acad Sci U S A 84:8702–8706
Okamura Y, Shidara M (1990a) Changes in sodium channels during neural differentiation in the isolated blastomere of the ascidian embryo. J Physiol 431:39–74
Okamura Y, Shidara M (1990b) Inactivation kinetics of the sodium channel in the egg and the isolated, neutrally differentiated blastomere of the ascidian. J Physiol 431:75–102
Okamura Y, Ono F, Okagaki R, Chong JA, Mandel G (1994) Neural expression of a sodium channel gene requires cell-specific interactions. Neuron 13:937–948
Okamura Y, Nishino A, Murata Y, Nakajo K, Iwasaki H, Ohtsuka Y, Tanaka-Kunishima M, Takahashi N, Hara Y, Yoshida T, Nishida M, Okado H, Watari H, Meinertzhagen IA, Satoh N, Takahashi K, Satou Y, Okada Y, Mori Y (2005) Comprehensive analysis of the ascidian genome reveals novel insights into the molecular evolution of ion channel genes. Physiol Genomics 22:269–282
Pan Z, Kao T, Horvath Z, Lemos J, Sul JY, Cranstoun SD, Bennett V, Scherer SS, Cooper EC (2006) A common ankyrin-G-based mechanism retains KCNQ and NaV channels at electrically active domains of the axon. J Neurosci 26:2599–2613
Parker A (2003) In the blink of an eye. Basic Books, New York
Payandeh J, Minor DL Jr (2015) Bacterial voltage-gated sodium channels (BacNaVs) from the soil, sea, and salt lakes enlighten molecular mechanisms of electrical signaling and pharmacology in the brain and heart. J Mol Biol 427:3–30
Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK, Benito-Gutiérrez EL, Dubchak I, Garcia-Fernàndez J, Gibson-Brown JJ, Grigoriev IV, Horton AC, de Jong PJ, Jurka J, Kapitonov VV, Kohara Y, Kuroki Y, Lindquist E, Lucas S, Osoegawa K, Pennacchio LA, Salamov AA, Satou Y, Sauka-Spengler T, Schmutz J, Shin-I T, Toyoda A, Bronner-Fraser M, Fujiyama A, Holland LZ, Holland PW, Satoh N, Rokhsar DS (2008) The amphioxus genome and the evolution of the chordate karyotype. Nature 453:1064–1071
Ramaswami M, Tanouye MA (1989) Two sodium-channel genes in Drosophila: implications for channel diversity. Proc Natl Acad Sci U S A 86:2079–2082
Ramsey IS, Moran MM, Chong JA, Clapham DE (2006) A voltage-gated proton-selective channel lacking the pore domain. Nature 440:1213–1216
Ren D, Navarro B, Xu H, Yue L, Shi Q, Clapham DE (2001) A prokaryotic voltage-gated sodium channel. Science 294:2372–2375
Rogart RB, Cribbs LL, Muglia LK, Kephart DD, Kaiser MW (1989) Molecular cloning of a putative tetrodotoxin-resistant rat heart Na+ channel isoform. Proc Natl Acad Sci U S A 86:8170–8174
Roger AJ, Simpson AGB (2008) Evolution: revisiting the root of the eukaryote tree. Curr Biol 19:R165–R167
Rogozin IB, Basu MK, Csürös M, Koonin EV (2009) Analysis of rare genomic changes does not support the unikont-bikont phylogeny and suggests cyanobacterial symbiosis as the point of primary radiation of eukaryotes. Genome Biol Evol 1:99–113
Salkoff L, Butler A, Wei A, Scavarda N, Giffen K, Ifune C, Goodman R, Mandel G (1987) Genomic organization and deduced amino acid sequence of a putative sodium channel gene in Drosophila. Science 237:744–749
Sangameswaran L, Fish LM, Koch BD, Rabert DK, Delgado SG, Ilnicka M, Jakeman LB, Novakovic S, Wong K, Sze P, Tzoumaka E, Stewart GR, Herman RC, Chan H, Eglen RM, Hunter JC (1997) A novel tetrodotoxin-sensitive, voltage-gated sodium channel expressed in rat and human dorsal root ganglia. J Biol Chem 272:14805–14809
Sasaki M, Takagi M, Okamura Y (2006) A voltage sensor-domain protein is a voltage-gated proton channel. Science 312:589–592
Sato C, Matsumoto G (1992) Primary structure of squid sodium channel deduced from the complementary DNA sequence. Biochem Biophys Res Commun 186:61–68
Satou Y, Kawashima T, Kohara Y, Satoh N (2003) Large scale EST analyses in Ciona intestinalis: its application as northern blot analyses. Dev Genes Evol 213:314–318
Schredelseker J, Shrivastav M, Dayal A, Grabner M (2010) Non-Ca2+-conducting Ca2+ channels in fish skeletal muscle excitation-contraction coupling. Proc Natl Acad Sci U S A 107:5658–5663
Shen H, Zhou Q, Pan X, Li Z, Wu J, Yan N (2017) Structure of a eukaryotic voltage-gated sodium channel at near-atomic resolution. Science 355:eaal4326
Smith JJ, Kuraku S, Holt C, Sauka-Spengler T, Jiang N, Campbell MS, Yandell MD, Manousaki T, Meyer A, Bloom OE, Morgan JR, Buxbaum JD, Sachidanandam R, Sims C, Garruss AS, Cook M, Krumlauf R, Wiedemann LM, Sower SA, Decatur WA, Hall JA, Amemiya CT, Saha NR, Buckley KM, Rast JP, Das S, Hirano M, McCurley N, Guo P, Rohner N, Tabin CJ, Piccinelli P, Elgar G, Ruffier M, Aken BL, Searle SM, Muffato M, Pignatelli M, Herrero J, Jones M, Brown CT, Chung-Davidson YW, Nanlohy KG, Libants SV, Yeh CY, McCauley DW, Langeland JA, Pancer Z, Fritzsch B, de Jong PJ, Zhu B, Fulton LL, Theising B, Flicek P, Bronner ME, Warren WC, Clifton SW, Wilson RK, Li W (2013) Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet 45(415–421):421e1–421e2
Strong M, Chandy KG, Gutman GA (1993) Molecular evolution of voltage-sensitive ion channel genes: on the origins of electrical excitability. Mol Biol Evol 10:221–242
Stühmer W, Conti F, Suzuki H, Wang X, Noda M, Yahagi N, Kubo H, Numa S (1989) Structure parts involved in activation and inactivation of the sodium channel. Nature 339:597–603
Takahashi K, Okamura Y (1998) Ion channels and early development of neural cells. Physiol Rev 78:307–337
Takahashi K, Yoshii M (1981) Development of sodium, calcium and potassium channels in the cleavage-arrested embryo of an ascidian. J Physiol 315:515–529
Tate S, Benn S, Hick C, Trezise D (1998) Two sodium channels contribute to the TTX-R sodium current in primary sensory neurons. Nat Neurosci 1:653–655
Tessmar-Raible K, Raible F, Christodoulou F, Guy K, Rembold M, Hausen H, Arendt D (2007) Conserved sensory-neurosecretory cell types in annelid and fish forebrain: insights into hypothalamus evolution. Cell 129:1389–1400
Toledo-Aral JJ, Moss BL, He ZJ, Koszowski AG, Whisenand T, Levinson SR, Wolf JJ, Silos-Santiago I, Halegoua S, Mandel G (1997) Identification of PN1, a predominant voltage-dependent sodium channel expressed principally in peripheral neurons. Proc Natl Acad Sci U S A 94:1527–1532
Tomer R, Denes AS, Tessmar-Raible K, Arendt D (2010) Profiling by image registration reveals common origin of annelid mushroom bodies and vertebrate pallium. Cell 142:800–809
Torruella G, Derelle R, Paps J, Lang BF, Roger AJ, Shalchian-Tabrizi K, Ruiz-Trillo I (2011) Phylogenetic relationships within the Opisthokonta based on phylogenomic analyses of conserved single copy protein domains. Mol Biol Evol 29:531–544
Vergara HM, Bertucci PY, Hantz P, Tosches MA, Achim K, Vopalensky P, Arendt D (2017) Whole-organism cellular gene-expression atlas reveals conserved cell types in the ventral nerve cord of Platynereis dumerilii. Proc Natl Acad Sci U S A 114:5878–5885
West JW, Patton DE, Scheuer T, Wang Y, Goldin AL, Catterall WA (1992) A cluster of hydrophobic amino acid residues required for fast Na+-channel inactivation. Proc Natl Acad Sci U S A 89:10910–10914
Widmark J, Sundström G, Ocampo Daza D, Larhammar D (2011) Differential evolution of voltage-gated sodium channels in tetrapods and teleost fishes. Mol Biol Evol 28:859–871
Willey A (1894) Amphioxus and the ancestry of the vertebrates. Macmillan, New York
Won Y-J, Ono F, Ikeda SR (2012) Characterization of Na+ and Ca2+ channels in zebrafish dorsal root ganglion neurons. PLoS One 7:e42602
Yan Z, Zhou Q, Wang L, Wu J, Zhao Y, Huang G, Peng W, Shen H, Lei J, Yan N (2017) Structure of the NaV1.4-β1 complex from electric eel. Cell 170:470–482.e11
Yue L, Navarro B, Ren D, Ramos A, Clapham DE (2002) The cation selectivity filter of the bacterial sodium channel, NaChBac. J Gen Physiol 120:845–853
Zakon HH (2012) Adaptive evolution of voltage-gated sodium channels: the first 800 million years. Proc Natl Acad Sci U S A 109(Suppl 1):10619–10625
Zakon HH, Jost MC, Lu Y (2011) Expansion of voltage-dependent Na+ channel gene family in early tetrapods coincided with the emergence of terrestriality and increased brain complexity. Mol Biol Evol 28:1415–1424
Zalc B (2016) The acquisition of myelin: an evolutionary perspective. Brain Res 1641:4–10
Zalc B, Goujet D, Colman D (2008) The origin of the myelination program in vertebrates. Curr Biol 18:R511–R512
Zhang T, Wang Z, Wang L, Luo N, Jiang L, Liu Z, Wu C-F, Dong K (2013) Role of the DSC1 channel in regulating neuronal excitability in Drosophila melanogaster: extending nervous system stability under stress. PLoS Genet 9:e1003327
Zhou W, Chung I, Liu Z, Goldin A, Dong K (2004) A voltage-gated calcium-selective channel encoded by a sodium channel-like gene. Neuron 42:101–112
Acknowledgments
Drs. Patrick Lemaire, Hiroki Nishida, and Hitoshi Sawada allowed us to utilize genome datasets from ascidians, Halocynthia roretzi and H. aurantium, before the publication of the data.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Nishino, A., Okamura, Y. (2017). Evolutionary History of Voltage-Gated Sodium Channels. In: Chahine, M. (eds) Voltage-gated Sodium Channels: Structure, Function and Channelopathies. Handbook of Experimental Pharmacology, vol 246. Springer, Cham. https://doi.org/10.1007/164_2017_70
Download citation
DOI: https://doi.org/10.1007/164_2017_70
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-90283-8
Online ISBN: 978-3-319-90284-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)