Abstract
Voltage-gated sodium channels initiate and propagate action potentials in excitable cells. They respond to membrane depolarization through opening, followed by fast inactivation that terminates the sodium current. This ON-OFF behavior of voltage-gated sodium channels underlays the coding of information and its transmission from one location in the nervous system to another. In this review, we explore and compare structural and functional data from prokaryotic and eukaryotic channels to infer the effects of evolution on sodium channel structure and function.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
- Activation mechanisms
- Bacterial sodium channels
- Eukaryotic sodium channels
- Evolution of sodium channels
- Gating mechanisms
- Inactivation mechanisms
- Selectivity of sodium channels
1 Introduction
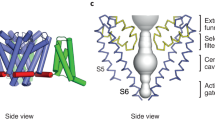
The eukaryotic voltage-gated sodium (Na+) channel is composed of a complex containing a pore-forming α-subunit and up to two β-subunits (Ahern et al. 2016; Catterall 2012; Catterall and Zheng 2015). The pore-forming α-subunit is sufficient to form a functional sodium channel, while the β-subunits are known to modulate the kinetics and voltage dependence of Na+ channel activation and inactivation (Isom et al. 1995; Qu et al. 1995). Nine different isoforms of voltage-gated sodium (NaV) channel α-subunits and four NaV channel β-subunits have been discovered (Catterall et al. 2005). The central nervous system (CNS) expresses four main isoforms: Nav1.1, Nav1.2, Nav1.3, and Nav1.6. Peripheral nervous system (PNS) has three different isoforms Nav1.7, Nav1.8, and Nav1.9. These three subtypes of NaV channels are mainly associated with autonomic regulation and pain sensation. In skeletal muscles, Nav1.4 is the principal NaV channel, while Nav1.5 is the primary NaV channel in cardiac muscle. In each of these channels, the α-subunit is composed of ~2,000 amino acid residues that form four homologous domains (traditionally numbered as I–IV), each with six transmembrane helices (Fig. 1a, numbered S1–S6) (Ahern et al. 2016; Catterall 2012). The fourth transmembrane helix (S4) in each domain has 4–8 positive residues (R or K) that sense changes in voltage, while transmembrane helices S5 and S6 from each domain form the pore-lining residues of the channel (Bezanilla 2000; Catterall 2010). Eukaryotic NaV channels are subject to broad posttranslational modifications including phosphorylation, glycosylation, palmitoylation, ubiquitination, and methylation (Catterall 1986a). This reflects the diversity of eukaryotic NaV channel expression and the role played by posttranslational modification in fine-tuning its function. Recently published structures of NaV channels from American cockroach (NavPas) and electric eel electroplax (Nav1.4) reveal the subunit architecture and structural modules of eukaryotic NaV channels (Shen et al. 2017; Yan et al. 2017).
Overall structures of eukaryotic and prokaryotic sodium channels. (a) Topology of a eukaryotic voltage-gated sodium channel showing four homologous domains. Each domain consists of six segments. Voltage-sensing segments (S1–S4) are shown in gray with S4 segment depicted in marine blue. The pore domain (S5–S6) is shown in light green. Right, topology of a single domain of a bacterial sodium channel. (b) Top view of the overall model of the eukaryotic voltage-gated sodium channel NavPas (left) (5X0M) and bacterial sodium channel NavAb (right) (PDB 3RVZ). Key structural and functional features of NavPas (left) and NavAb (right) channels are color labeled including the voltage sensor domain (S1–S3, green; S4, marine blue), pore domain (light green)
Compared to the complexity of mammalian NaV channels, bacterial NaV channels are composed of four identical subunits of ~250–270 residues. Every subunit has a voltage sensor domain (VSD) and a pore domain (PD) (Payandeh et al. 2011; Ren et al. 2001). They share the major biophysical features with eukaryotic counterparts (Catterall and Zheng 2015). In contrast to eukaryotic channels, bacterial NaV channels lack auxiliary subunits and posttranslational modifications. Crystallization of full-length bacterial NaV channels like NavAb, NavRh, and NavMs and analysis of their structures at high resolution (Payandeh et al. 2011, 2012; Sula et al. 2017; Zhang et al. 2012) make them invaluable models for studying the structural basis of ion conduction, activation, inactivation, and drug interaction. In this review, we use an evolutionary perspective to analyze the differences and similarities between bacterial and eukaryotic NaV channels and how they might have evolved over billions of years.
2 The Voltage-Sensing Module
2.1 The Excitable Membrane and the Voltage Sensor
NaV channels are integral membrane proteins that rely on the potential generated by selective ion conductance and unequal concentrations of ions across the plasma membrane (Hille 2001). This separation of ions across the lipid bilayer plus high resting membrane permeability to K+ results in a membrane potential that ranges from −40 to −95 mV depending on the cell type (measured from inside with respect to the outside). Since the plasma membrane is a lipid bilayer of ~30 Å thickness, this produces an electric field across the plasma membrane that can be as high as 3 × 107 V/m. Voltage-gated ion channels exploit this high electric field by coupling protein conformational changes to physiological changes in the electric field strength. From a structural standpoint, the voltage sensor domains of eukaryotic and bacterial NaV channels are very similar in that there are four voltage-sensing units per channel and each voltage-sensing domain consisting of four transmembrane helices numbered S1–S4 by convention. Transmembrane helix S4 is the key player in sensing the voltage across the cell membrane by placing four to eight positively charged R or K residues across the membrane (Bezanilla 2000; Catterall 2010). These gating charge residues are located at every third position along S4 high order channels (Fig. 2). An important difference between prokaryotic channels and their eukaryotic counterparts is that the number of charged S4 residues is fixed among the four subunits in prokaryotic, homotetrameric channels but can vary a great deal among the subunits and homologous domains of higher-order channels along S4 (Fig. 2). Crystal structures show that the S1, S2, and S3 helices of the voltage sensor module surround the S4 segment and form a gating canal, which facilitates the movement of S4 (Payandeh et al. 2011, 2012; Zhang et al. 2012).
The S1, S2, and S3 helices contain polar or negative residues that create the gating canal that catalyzes the movement of S4 from its inward resting-state position to progressively more activated states upon depolarization (Fig. 2a, b). The positively charged residues in the S4 segments were proposed to form ion pairs with polar/negatively charged amino acid residues present in these surrounding helices (Catterall 1986a, b; Guy and Seetharamulu 1986). S1 has an N residue (N25 in NavAb numbering) that is highly conserved in bacterial and eukaryotic sodium channels (Fig. 1S). Another less conserved residue is E32 (NavAb numbering), which is present in some bacterial NaV channels and in two or three domains of eukaryotic NaV channels. The S2 segment has two negative (or one negative and one polar) residues separated by nine, mostly hydrophobic, residues. On the N-terminal side, either D, E, or N is present, and on the C-terminal, E is always present. In-between these two negative residues, a set of hydrophobic residues (including F56 in NavAb numbering) serve as a hydrophobic plug that prevents ionic leak through VSD. This hydrophobic constriction site (HCS, Fig. 2) is conserved among voltage sensors from many different proteins, including Kv, Nav, Cav, proton channels, and voltage-dependent phosphatase (VSP) enzymes (Li et al. 2014; Payandeh et al. 2011; Tao et al. 2010; Wu et al. 2016). In the case of segment S3, E80 (NavAb numbering) is a very conserved residue in bacterial and eukaryotic NaV channels. The high sequence identity of residues within VSD indicates that gating has been conserved through evolution (Figs. 1S and 2a).
The prokaryotic voltage sensor shares structural features with eukaryotic channels. (a) Sequence alignment of S4 segment of NavAb with Nav1.4 showing the conservation of residues discussed in the text. (b) Structural model of NavAb in orthogonal views, highlighting residues and structural regions discussed in the text. Helices are shown as ribbon models, while important side chains are shown as sticks. R1–R4 refer to voltage-sensing arginine residues; ENC, HCS, and INC refer to extracellular negative cluster, hydrophobic constriction site, and intracellular negative cluster, respectively. (c) Structure-based alignment of the voltage sensor of NavAb and Domain II of the recently solved NavPas structure. Alignment was performed using the entire voltage sensor sequence of each respective protein. (d) Cartoon model highlighting the variability of gating charge positions in NavPas when compared to NavAb. The gray model represents the S4 helix of NavAb, while the blue helices represent those of NavPas. Gating charges are shown as yellow boxes, the HCS is shown as an orange box, and lipids are drawn in a cartoon format. We have assigned a gating charge position to each S4 helix, shown in the line below the diagram and meant to highlight the different position of gating charges in DI–DIV on NavPas
2.2 A Conserved Mechanism of Activation
There is a consensus among voltage-gated ion channel biophysicists that the electromechanical coupling between the voltage sensor module and the activation gate of the pore domain follows a sliding-helix mechanism (Vargas et al. 2012). The S4 segment serves as the main voltage sensor by virtue of its high concentration of positively charged residues (Catterall 1986a). Mutagenesis studies of these residues resulted in reduction of the steepness of voltage-dependent gating and a shift of its voltage dependence (Logothetis et al. 1992; Papazian et al. 1991; Stuhmer et al. 1989). The S1–S3 segments form the structure of the gating pore through which S4 moves from its resting to the activated state (Catterall 2010). The shape of the voltage-sensing module looks like an hour glass, with a large extracellular aqueous cleft (~10 Å) and a smaller intracellular cleft (Fig. 2b). The hydrophobic constriction site (HCS, Fig. 2b) is present between these two aqueous clefts. Very highly conserved residues (I in S1, F in S2, and V or I in S3) form this hydrophobic seal, which prevents ionic leak through voltage-sensing module.
The activated and resting states of voltage sensors of sodium channels have been probed with gating pore current studies (Gamal El-Din et al. 2010; Gamal El-Din et al. 2014; Sokolov et al. 2005, 2008), disulfide cross-linking experiments (DeCaen et al. 2008, 2009, 2011), and substituted cysteine accessibility methods (Larsson et al. 1996; Yang et al. 1996). These studies showed that the R3 or R4 gating charges seal the gating pore at the hydrophobic constriction site during activation, probably by interacting with nearby negatively charged or hydrophilic residues in the S2 segment (E, D, or N). Crystal structures of many voltage sensors showed that either R3 or R4 has proximity to the outermost residue in the extracellular negative cluster (N49 in NavAb or D48 in NavRh) on the S2 segments (Payandeh et al. 2011; Zhang et al. 2012). Interestingly, computational models using the Rosetta method combined with disulfide cross-linking revealed the existence of three different activated states of the bacterial sodium channel NaChBac (Yarov-Yarovoy et al. 2012). In activated state I, gating charges R1 and R2 interact with the external negative cluster, while R3 is located between N49 and F56. In activated state II, R1 is solvated in the extracellular aqueous cleft, while R2 interacts with E43 and R3 interacts with N49. In activated state III, R1 is still solvated, R2 and R3 interact with E43 and N49, respectively, and R4 passes through the hydrophobic constriction site and is located just on the extracellular side of F56.
Lack of a crystal structure of a NaV channel in the resting state has placed primary emphasis on experimental and computational methods to assist in seeing the unseen. Fluorometric studies have shown that the electric field is focused around the part of the S4 segment that interacts with the hydrophobic constriction site (Ahern and Horn 2005; Asamoah et al. 2003). The substituted cysteine accessibility method (SCAM), in which the gating charge of interest is mutated to cysteine and perfusion of cysteine-reactive agent (MTSET, MTSES, or MTSEA) probes its aqueous accessibility, showed that R1 is not accessible from either side in mammalian or bacterial NaV channels (Blanchet and Chahine 2007; Yang and Horn 1995), which means that it is buried within the gating canal. R2, on the other hand, was accessible from both sides in the resting state, indicating that it can move through the hydrophobic constriction site in a pair of structurally similar resting states. Mutation of R1, R2, or both to neutral, shorter, and uncharged residues causes a leak current, the gating pore current, through the mutated voltage-sensing module. R1 and R2 gating pore currents are active in the resting state and shut off in the activated state (Gamal El-Din et al. 2010, 2014; Sokolov et al. 2005; Starace and Bezanilla 2001, 2004).
Based on the available data regarding resting and activated states of the voltage sensor, we can now consider the current evidence regarding the gating transition between these states. When the potential difference across the plasma membrane is reduced during depolarization, the voltage-sensing modules undergo a conformation transition from the resting state to the activated state. During this transition, sequential formation and breakage of salt bridges and hydrogen-bonding interactions between arginine residues (R1–R4) on S4 and the negatively charged and polar residues on neighboring segments S1–S3 mediate low-energy passage of the S4 gating charges through the hydrophobic constriction site, as posited in the sliding helix or helical screw models (Catterall 1986a, b; Guy and Seetharamulu 1986). This transition of gating charges from resting to activated state is equivalent to moving ~8–14 elementary charges across the membrane electric field (Bezanilla 2000; Catterall 2010; Gamal El-Din et al. 2008; Hirschberg et al. 1995). Gating pore current and disulfide-locking experiments tracked this journey of the S4 segment from resting to activated states (Gamal El-Din et al. 2010, 2014; DeCaen et al. 2008, 2009, 2011). There is agreement that the S4 helix slides ~10 Å outward through the gating pore formed by the S1, S2, and S3 segments, accompanied by ~30° rotation and a sideways tilt at the pivot point in the hydrophobic constriction site (Vargas et al. 2012; Yarov-Yarovoy et al. 2012). All the resting-state models show the positive gating charge residues along S4 are in position to form salt bridges with acidic residues in the S1–S3 helices or interact with the aqueous regions of the lipid head groups. These conclusions agree with the experimental data on sodium channels obtained from the disulfide cross-linking experiments (DeCaen et al. 2008, 2009, 2011; Yarov-Yarovoy et al. 2012) and measurements of gating pore currents (Gamal El-Din et al. 2014).
Even though the overall mechanism of S4 activation is very similar between the bacterial and eukaryotic NaV channels, the details of this transition differ in several ways. First, the position of the resting state of each voltage sensor relative to the activation gate is different (Gosselin-Badaroudine et al. 2012). Second, the voltage sensors of eukaryotic NaV channels are in a different axial position in the plasma membrane relative to the prokaryotic voltage sensor and often in different positions relative to each other (Shen et al. 2017; Yan et al. 2017). These differential positions may enable these channels to activate faster and tune individual voltage sensors for specific aspects of sodium channel function (Fig. 2d). The result of these different resting and activated states may be stabilization of the channel in various conformations over a specific range of membrane potentials as required for the physiological niche for each NaV channel.
3 The Selectivity Filter: From Symmetry to Asymmetry
It is intuitive that ion channels could attract cations with a cluster of negative charges (Hille 2001), and, indeed, the structure of the selectivity filter of the prokaryotic NaV channels showed just such a mechanism (Payandeh et al. 2011). These channels are considered the ancestors of eukaryotic NaV and CaV channels, and each contains a quartet of negatively charged E residues arranged around the selectivity filter, confirming this idea (Payandeh et al. 2011; Ren et al. 2001). The selectivity filter itself is formed of four identical segments, located in the “turn” in the “P1 helix-turn-P2 helix” structure from each domain. The selectivity filter of bacterial NaV channels has the signature sequence motif TLX 1 SWX 2 , where X 1 is the site of high-field-strength anionic site in most channels. The only exception is NavMs, where X 2 is the high-field-strength site instead of X 1 . The selectivity filter in NavAb has the sequence (TLESWSM). It has been suggested that the carboxylate groups of the four E residues (one from each domain) form an extracellular symmetric acidic coordination site (SiteHFS), while the backbone carbonyls of the four L and T residues form the central and inner coordination sites (SiteCEN and SiteIN), respectively. The crystal structure of NavMs showed three Na+ at almost the same sites proposed for NaVAb (Naylor et al. 2016). Ordered water molecules were observed in the NaVAb and NaVMs structures in the lower part of the selectivity filter. Molecular dynamics studies elucidated the catalytic mechanism of Na+ conduction (Chakrabarti et al. 2013; Ulmschneider et al. 2013). Remarkably, highly degenerate but energetically favorable dunking motions, in which one to three of the carboxylate side chains at SiteHFS bend at a single torsion angle and move inward accompanying partially, dehydrated Na+ (Chakrabarti et al. 2013).
While mammalian NaV channels are virtually impermeable to Ca2+ under physiological conditions, bacterial NaV channels show a significant permeability for Ca2+, as demonstrated by permeability ratios of PCa/PNa of 0.15, 0.14, 0.27, and 0.2 for NaChBac, NavRh, NavAb, and NavMs, respectively (Naylor et al. 2016; Tang et al. 2014; Yue et al. 2002; Zhang et al. 2012). This indicates that ancient bacterial NaV channels might have served as a conductance pathway for both Na+ and Ca2+. Over billions of years of evolution, the homomeric structure of eukaryotic NaV channels was converted into a single polypeptide chain by covalently linking the C- and N-termini of two neighboring subunits. This resulted in loss of the fourfold symmetry of the pore and selectivity filter, as well as the signature motif at SiteHFS. This stepwise evolution is reflected in the many different signature motifs at SiteHFS in invertebrate sodium channels: EEEE, EEKE, EEAA, DEAA, etc. (Stephens et al. 2015). The signature motif EEEE has remained the same for Cav channels, the position of the carbon backbone stayed the same, but the geometry of the locations of the E residues side chains changed (Wu et al. 2016) (Fig. 3). On the other hand, the evolution of bacterial to eukaryotic NaV channels involved changes in the chemical signature of residues that select Na+ to give much higher selectivity. K appeared in Domain III as the residue conferring higher selectivity for Na+ and preventing the permeability of Ca2+ (Favre et al. 1996). Studies that target this residue in NaV channels by mutating it to E rescued CaV conductance (Heinemann et al. 1992). Overall, the structure of the carbon backbone of the high-field-strength site of the cockroach NavPas channel resembles that of the quartet of E residues in NavAb, though the exact placement of the side chains is uncertain (Fig. 3b).
The selectivity filters of NavAb, Cav1.1, and NavPas. (a) Comparing the selectivity filters from a view above the plane of the membrane. Each selectivity filter is shown in stick format, with the residue equivalent to HFS2 (E177 in NavAb) highlighted. NavAb is shown in gray, Cav1.1 in orange, and NavPas in light blue. (b) Side view of NavAb and NavPas selectivity filters highlighting the position of residues important for selectivity (E177 in NavAb, D375, E701, K1061, A1353 in NavPas) and overall structure of the selectivity filter (W179 in NavAb and W377 and W702 in NavPas are highlighted)
Two amino acids have been conserved throughout the evolution process, W179 and T175 (NaVAb numbering). T175 forms a hydrogen bond with W179 of a neighboring subunit. This bond is a key interaction site to connect adjacent subunits at the selectivity filter and to stabilize the overall structure of the selectivity filter (Payandeh et al. 2011).
The apparent similarity of Na+ and Ca2+ permeation through eukaryotic ion channels indicate that they have evolved from the same ancestor. In both cases, Na+ and Ca2+ have to be partially hydrated in order to move through the relatively wide selectivity filter (Hille 1971; Naylor et al. 2016; Payandeh et al. 2011; Tang et al. 2014; Wu et al. 2016) (Fig. 3). This is fundamentally different from K+ channels where K+ ions pass completely dehydrated through the selectivity filter (Zhou et al. 2001). Moreover, in both NaV and CaV channels, there is an extracellular cluster of negatively charged residues that recruit cations to the selectivity filter. In contrast, KV channels use only interactions with backbone carbonyls to select and conduct K+ (Naylor et al. 2016; Wu et al. 2016). Another major difference between permeation in eukaryotic versus bacterial NaV and CaV channels on one hand and KV channels on the other hand is flexibility versus rigidity of the selectivity filter. The finding that the size of the selectivity filter of NaV and CaV channels is much wider than KV channels indicates that the side chains of the surrounding residues have more degrees of freedom. Classic studies of CaV channels suggested that carboxylate side chains of the EEEE locus move independently to form a high-affinity binding site for incoming Ca2+ (Sather and McCleskey 2003; Yang et al. 1993). Molecular dynamics studies of NavAb showed that the E side chains of SiteHFS undergo a “dunking” motion with each permeating Na+ (Chakrabarti et al. 2013). In contrast, the relative tightness of the KV channel selectivity filter requires backbone carbonyls to select the right ions, while the side chains of the selectivity filter residues point away from the lumen of the pore (Morais-Cabral et al. 2001; Zhou et al. 2001).
4 Inactivation Evolved from Bacterial to Eukaryotic Sodium Channels
In contrast to the conserved mechanism of activation between bacterial NaV channels and their eukaryotic counterparts, inactivation mechanisms have evolved over billions of years to become faster and more efficient as required for the rapid information processing in excitable cells of multicellular organisms. Eukaryotic NaV channels have two main types of inactivation: fast inactivation that occurs on a time scale of 1–5 ms (Hodgkin and Huxley 1952a, c, d) and slow inactivation that occurs on a time scale of hundreds of milliseconds to seconds (Adelman and Palti 1969; Palti and Adelman 1969; Rudy 1975, 1981). Mechanistically, fast inactivation takes place when a short peptide located in the segment between domains III and IV (IFM) binds to the intracellular mouth of the pore, which stops ion conduction (Catterall 2012). Slow inactivation, on the other hand, is believed to result from collapse of the pore of the NaV channel (Payandeh et al. 2012; Zhang et al. 2012).
4.1 Slow Inactivation of Eukaryotic Sodium Channels
When NaV channels are exposed to prolonged depolarizations, either by applying one long positive pulse or a series of high-frequency repetitive ones, they stop Na+ conduction by entering the slow-inactivated state (Ulbricht 2005; Vilin and Ruben 2001). Just as entry into this inactivated state takes much longer than fast inactivation, recovery from it also proceeds very slowly with time constants in the range of hundreds of milliseconds to minutes, compared to 20 - 30 ms for fast inactivation. During this time, excitable membranes become refractory to further stimulation. Slow inactivation seems to be an ancient mechanism that NaV channels used to protect cells against high stress conditions that generate highly repetitive stimuli.
Studies of slow inactivation of eukaryotic NaV channels using site-directed mutagenesis methods implicated amino acid residues in the selectivity filter and surrounding S5 and S6 segments in conformational changes that accompany slow inactivation (Balser et al. 1996; Benitah et al. 1996, 1997; Vilin and Ruben 2001). Mutation of W402 located in the selectivity filter of Domain I of Nav1.4 dramatically reduces slow inactivation (Balser et al. 1996). Four charged residues within the selectivity filter of rNav1.4 (E403/DI, E758/DII, D1214/DIII, and D1532/DIV) move toward each other during establishment of slow inactivation (Xiong et al. 2003), as indicated by disulfide bond formation between substituted cysteine residues. Similarly, F1236C was accessible only from the outside during short depolarizing pulses, which did not induce slow inactivation but became inaccessible during long depolarizing pulses (6 s), which induce slow inactivation (Ong et al. 2000).
4.2 Studies of Slow Inactivation of Bacterial Sodium Channels
Bacterial NaV channels lack the equivalent linker between Domain III and IV that is responsible for fast inactivation in eukaryotic NaV channels and thus have only slow inactivation (Pavlov et al. 2005). The crystal structure of NavAb showed that its selectivity filter is rigidly anchored by a hydrogen bond (~ 3.0 Ǻ) between T175 (equivalent to T in domains I, III, and IV in NaV1.4) and W179 (equivalent to W402 in NaV1.4) in neighboring subunits [(Payandeh et al. 2011, 2012); Fig. 4]. However, the crystal structure of an inactivated state of NavAb/WT showed that two of the key T175–W179 interactions have become very weak, forming hydrogen bonds with a length ~3.8 Ǻ compared to ~3 Ǻ in case of pre-open state (Payandeh et al. 2012). This suggests that the hydrogen bond to W at this outer side of the selectivity filter is keeping the pore open for permeation of hydrated Na+. In the inactivated structure of NavAb, S180 has flipped into a different conformation and interacts with the carboxylate side chain of E177 of a neighboring subunit (Payandeh et al. 2012). This movement led to the asymmetric collapse of two of the four pore-lining S6 segments of NavAb. In order to have a highly conductive channel, the geometry at the selectivity filter must be perfectly sized to coordinate and conduct a hydrated square-planar Na+, as in the NaVAb-I217C structure (Chakrabarti et al. 2013; Payandeh et al. 2011). Therefore, the change in size and shape of the selectivity filter in the slow-inactivated state of NaVAb/WT (Payandeh et al. 2012) is likely to make it poorly conductive.
The selectivity filter changes conformation as sodium channels inactivate. (a–c) Space filling (top row) and stick models (bottom row) are shown for pre-open NavAb (gray, pdb 3RVY), inactivated NavAb (purple, pdb 4EKW AB tetramer), and NavRh (green, pdb 4dxw) highlighting conformational changes as the channel moves into the inactivated state. Residues and hydrogen bonds discussed in the text are highlighted in the stick figures
Structure-function studies suggested that both activation and inactivation of bacterial NaV channels occur via twisting and bending movements of the S6 pore-lining helix at a glycine hinge, such as G219 in NaChBac (Zhao et al. 2004b). The same mechanism has been proposed for the bacterial KcsA and MthK channels (Cuello et al. 1998; Jiang et al. 2002a, b; Perozo et al. 1999). In NaChBac, mutation of the neighboring T residue (T220) also greatly slows inactivation during the pulse (Lee et al. 2012; Zhao et al. 2004a, b). Crystal structures of NavMs show a kink at T219 that is equivalent to the glycine hinge in NaChBac (McCusker et al. 2012). Recently, the same kink in NavAb has been observed, indicating a similar mechanism of twisting and bending movements of the S6 pore-lining helix (Lenaeus et al. 2017). Mutation of T206 at the bending point in the NaVAb S6 segment causes a negative shift of the voltage dependence of activation and is also required for early voltage-dependent inactivation (Gamal El-Din et al. 2017). These results suggest that this segment of the S6 segment undergoes two conformational changes, one that initiates opening of the activation gate and a second that begins the process of inactivation.
The amino acid residues that move during slow inactivation of mammalian NaV channels are also intimately involved in inactivation of bacterial NaV channels. For example, residues E403 in Domain I, E758 in Domain II, D1214 and F1236 in Domain III, and D1532 in Domain IV of NaV1.4 move when the channel is subject to high-frequency depolarizing pulses. E403 and E758 are equivalent to S180, F1236 is equivalent to I176, and D1532 is equivalent to M181 in NavAb, which all move during the transition from the pre-open to the slow-inactivated state (Payandeh et al. 2011, 2012). Evidently, the molecular machinery for slow inactivation has been conserved from bacteria to mammals, even as the rate and timing of inactivation have evolved.
4.3 Evolution of Fast Inactivation in Eukaryotic Sodium Channels
In contrast to the conserved evolution of the slow inactivation from bacterial to the mammalian sodium channel, current inactivation during application of a depolarizing stimulus has evolved tremendously over billions of years by addition of a fast inactivation using the linker connecting Domain III to Domain IV to act as a hinged lid and block the pore from the intracellular side. The need for efficient information processing in the mammalian nervous system might be the driving force behind the evolution of the mechanisms of inactivation during the pulse.
A typical mammalian NaV channel in the nerve or muscle opens very rapidly upon depolarization and closes within 1–2 ms, even if the stimulus is still on. In their seminal work on action potential initiation and propagation, Hodgkin and Huxley (1952b, c, d) described the Na+ channel conductance as being proportional to m 3 h, where m and h are gating particles responsible for activation and inactivation, respectively (Hille 2001). This was the first indication that the fast inactivation process is voltage dependent, which predicted that there is a differential functionality of voltage sensor modules in eukaryotic NaV channels. Currently, there is a consensus that S4 segments of domains I, II, and III are mainly responsible for activation, while Domain IVS4 is primarily responsible for initiating and maintaining fast inactivation (Capes et al. 2013; Chanda and Bezanilla 2002; Kuhn and Greeff 1999; Rogers et al. 1996; Sheets et al. 1999).
The structural motif that is responsible for translating the electrical signal sensed by the S4 segment in Domain IVS4, and thus initiating fast inactivation, is the intracellular linker between domains III and IV (Vassilev et al. 1988, 1989). Cutting this motif (Stuhmer et al. 1989) or mutating the critical hydrophobic residues (IFM) abolishes inactivation during the pulse (Kellenberger et al. 1997; Patton et al. 1992; West et al. 1992). The inactivation gate NMR structure shows that it has an alpha-helix backbone capped at its N-terminus by two turns, with the hydrophobic IFM residues located in this turn motif. Structural studies confirmed the physiological results that the IFM triad and adjacent threonine residue are essential components of the latch (Rohl et al. 1999). This structure is seen exactly in the corresponding segment in the cryo-EM structure of NaV1.4 (Yan et al. 2017).
5 Modulation of Sodium Channels by Their C-Terminal Tail
In eukaryotic NaV channels, the intracellular extensions of S6 segments begin the large C-terminal segments following each homologous domain. They are ~250 residues in length and they constitute the major targets for channel regulation. The C-terminal domains regulate fast inactivation (Cormier et al. 2002; Deschenes et al. 2001; Mantegazza et al. 2001; Nguyen and Goldin 2010) as well as slow inactivation that is modulated by interaction of calmodulin with a conserved site in the proximal segment (Gabelli et al. 2016). The newly published cryo-EM structure of a NaV1.4 channels reveals extensive interactions between the C-terminal domain and Domain III–IV linker, which may explain the ability of the C-terminal tail to modulate fast inactivation (Shen et al. 2017) (Fig. 5).
The interaction between the C-terminal domain and the pore domain is different in prokaryotic channels and NavPas. Cartoon models of prokaryotic channels NavAb (closed state, gray), NavMs (open state, light green), and NavPas (uncertain state, blue). In each case, a cartoon model is shown with overlying space-filling figure to highlight the interactions between C-terminal and membrane-spanning domains. Each channel shows the pore and voltage-sensing domains in a single color, then the associated C-terminal domain in orange to allow for contrast. The NavPas model also shows the DIII–DIV linker associated with inactivation in pink. The implications related to inactivation are discussed in the text
Most bacterial NaV channels have a short (~40 residues) C-terminal tail. The crystal structure of the tail was resolved for NavMs, NavAe, NavSulP, and NaVAb (Bagneris et al. 2013; Irie et al. 2012; Lenaeus et al. 2017; Shaya et al. 2014). There is a striking similarity among these structures, in which the C-terminal tails from four subunits form a four-helix bundle (the neck) proximal to the activation gate, which terminates in a four-stranded coiled coil. While there is a consensus about the similarity of C-terminal crystal structure, there are differences in its function. Originally, it was proposed that C-terminal is essential for tetramer formation (Powl et al. 2010; Shaya et al. 2014), but subsequent reports argued that it is only important for stabilizing subunit-subunit interactions (Mio et al. 2010). Indeed, many bacterial NaV channels are functional even after deleting the entire C-terminal (Arrigoni et al. 2016; Irie et al. 2012; Lenaeus et al. 2017). Arrigoni et al. showed that the proximal part of the C-terminal (the neck domain) is subject to temperature-dependent unfolding transitions during gating, while the coiled-coil part stays intact (Arrigoni et al. 2016). They found that only the neck has a major effect on channel activation profile, with a minor role for the coiled coil. Their experiments demonstrated little effect of the C-terminal tail on inactivation. In contrast, C-terminal truncations modulate early voltage-dependent inactivation of NaVAb during single depolarizations (Gamal El-Din et al. 2017). Progressive deletions from the C-terminus cause graded increases in the rate of the early voltage-dependent inactivation process, without major effects on the rate of activation. This reflects a loss of voltage dependence, as if truncation of the C-terminal tail removes a voltage-dependent brake on the early phase of the inactivation process. A similar effect has been shown previously in the KcsA channel where truncation of the C-terminal tail enhanced inactivation kinetics (Cuello et al. 1998). In contrast, deleting the C-terminal tail of NaVSulP slowed its early phase of inactivation during single depolarizations, and point mutations that disrupt the four-helix bundle also caused that effect (Irie et al. 2012). These results suggest that the C-terminal tail can modulate the voltage dependence and kinetics of the early phase of voltage-dependent inactivation of different NaV channels in different directions, depending on the molecular context.
The fine-tuning of inactivation during the pulse via the C-terminal tail has been preserved over billions of years of evolution, as truncation or mutation of C-terminal residues of eukaryotic NaV channels affects inactivation kinetics. Truncation of 129 residues of the distal C-terminus of Nav1.3 accelerates fast inactivation by approximately fourfold (Nguyen and Goldin 2010). The C-terminus has a strong influence on kinetics and voltage dependence of inactivation of brain (Nav1.2), cardiac (Nav1.5), and skeletal muscle (Nav1.4) channels and is primarily responsible for their differing rates of channel inactivation (Deschenes et al. 2001; Mantegazza et al. 2001).
The interplay between the early and late phases of inactivation in bacterial NaV channels is quite evident. This explains the recent results showing that truncation of the C-terminal tail of NaVAb/WT also removes the late, use-dependent phase of inactivation (Gamal El-Din et al. 2013, 2017). Cutting as few as ten residues from the distal end of the tail was both sufficient to abolish use-dependent inactivation at low frequency (0.2 Hz); however, at higher frequency (1 Hz), ~ 20–30% use-dependent inactivation occurs during the first few pulses, and then the peak current stays constant for up to 50 pulses. Overall, the movement of S6 segments seems to be finely tuned by the C-terminal tail in eukaryotic or bacterial NaV channels.
6 Conclusion
As this brief review shows, sodium channels are ancient membrane signaling proteins with many functional properties conserved from bacteria to human. The simple bacterial sodium channels have provided an invaluable resource for high-resolution structural analysis of sodium channel function. The recent addition of cryo-EM structures of eukaryotic sodium channels at near-atomic resolution shows striking conservation of the core structures of sodium channels over this full length of evolutionary time. Looking ahead, the combination of structural studies of bacterial and eukaryotic sodium channels seems likely to reveal the mechanisms of their core functions in atomic detail and their complex regulation in eukaryotes with increasing resolution. It is an exciting time in this field of research!
References
Adelman WJ Jr, Palti Y (1969) The effects of external potassium and long duration voltage conditioning on the amplitude of sodium currents in the giant axon of the squid, Loligo pealei. J Gen Physiol 54:589–606
Ahern CA, Horn R (2005) Focused electric field across the voltage sensor of potassium channels. Neuron 48:25–29
Ahern CA, Payandeh J, Bosmans F, Chanda B (2016) The hitchhiker’s guide to the voltage-gated sodium channel galaxy. J Gen Physiol 147:1–24
Arrigoni C, Rohaim A, Shaya D, Findeisen F, Stein RA, Nurva SR, Mishra S, McHaourab HS, Minor DL Jr (2016) Unfolding of a temperature-sensitive domain controls voltage-gated channel activation. Cell 164:922–936
Asamoah OK, Wuskell JP, Loew LM, Bezanilla F (2003) A fluorometric approach to local electric field measurements in a voltage-gated ion channel. Neuron 37:85–97
Bagneris C, Decaen PG, Hall BA, Naylor CE, Clapham DE, Kay CW, Wallace BA (2013) Role of the C-terminal domain in the structure and function of tetrameric sodium channels. Nat Commun 4:2465
Balser JR, Nuss HB, Chiamvimonvat N, Pérez-García MT, Marban E, Tomaselli GF (1996) External pore residue mediates slow inactivation in mu-1 rat skeletal muscle sodium channels. J Physiol 494:431–442
Benitah JP, Tomaselli GF, Marban E (1996) Adjacent pore-lining residues within sodium channels identified by paired cysteine mutagenesis. Proc Natl Acad Sci U S A 93:7392–7396
Benitah JP, Ranjan R, Yamagishi T, Janecki M, Tomaselli GF, Marban E (1997) Molecular motions within the pore of voltage-dependent sodium channels. Biophys J 73:603–613
Bezanilla F (2000) The voltage sensor in voltage-dependent ion channels. Physiol Rev 80:555–592
Blanchet J, Chahine M (2007) Accessibility of four arginine residues on the S4 segment of the Bacillus halodurans sodium channel. J Membr Biol 215:169–180
Capes DL, Goldschen-Ohm MP, Arcisio-Miranda M, Bezanilla F, Chanda B (2013) Domain IV voltage-sensor movement is both sufficient and rate limiting for fast inactivation in sodium channels. J Gen Physiol 142:101–112
Catterall WA (1986a) Molecular properties of voltage-sensitive sodium channels. Annu Rev Biochem 55:953–985
Catterall WA (1986b) Voltage-dependent gating of sodium channels: correlating structure and function. Trends Neurosci 9:7–10
Catterall WA (2010) Ion channel voltage sensors: structure, function, and pathophysiology. Neuron 67:915–928
Catterall WA (2012) Voltage-gated sodium channels at 60: structure, function and pathophysiology. J Physiol 590:2577–2589
Catterall WA, Zheng N (2015) Deciphering voltage-gated Na+ and Ca2+ channels by studying prokaryotic ancestors. Trends Biochem Sci 40:526–534
Catterall WA, Goldin AL, Waxman SG (2005) International Union of Pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels. Pharmacol Rev 57:397–409
Chakrabarti N, Ing C, Payandeh J, Zheng N, Catterall WA, Pomes R (2013) Catalysis of Na+ permeation in the bacterial sodium channel NaVAb. Proc Natl Acad Sci U S A 110:11331–11336
Chanda B, Bezanilla F (2002) Tracking voltage-dependent conformational changes in skeletal muscle sodium channel during activation. J Gen Physiol 120:629–645
Cormier JW, Rivolta I, Tateyama M, Yang AS, Kass RS (2002) Secondary structure of the human cardiac Na+ channel C terminus: evidence for a role of helical structures in modulation of channel inactivation. J Biol Chem 277:9233–9241
Cuello LG, Romero JG, Cortes DM, Perozo E (1998) pH-dependent gating in the Streptomyces lividans K+ channel. Biochemistry 37:3229–3236
DeCaen PG, Yarov-Yarovoy V, Zhao Y, Scheuer T, Catterall WA (2008) Disulfide locking a sodium channel voltage sensor reveals ion pair formation during activation. Proc Natl Acad Sci U S A 105:15142–15147
DeCaen PG, Yarov-Yarovoy V, Sharp EM, Scheuer T, Catterall WA (2009) Sequential formation of ion pairs during activation of a sodium channel voltage sensor. Proc Natl Acad Sci U S A 106:22498–22503
DeCaen PG, Yarov-Yarovoy V, Scheuer T, Catterall WA (2011) Gating charge interactions with the S1 segment during activation of a Na+ channel voltage sensor. Proc Natl Acad Sci U S A 108:18825–18830
Deschenes I, Trottier E, Chahine M (2001) Implication of the C-terminal region of the α-subunit of voltage-gated sodium channels in fast inactivation. J Membr Biol 183:103–114
Favre I, Moczydlowski E, Schild L (1996) On the structural basis for ionic selectivity among Na+, K+, and Ca2+ in the voltage-gated sodium channel. Biophys J 71(6):3110–3125
Gabelli SB, Yoder JB, Tomaselli GF, Amzel LM (2016) Calmodulin and Ca(2+) control of voltage gated Na(+) channels. Channels (Austin) 10:45–54
Gamal El-Din TM, Grogler D, Lehmann C, Heldstab H, Greeff NG (2008) More gating charges are needed to open a Shaker K+ channel than are needed to open an rBIIA Na+ channel. Biophys J 95:1165–1175
Gamal El-Din TM, Heldstab H, Lehmann C, Greeff NG (2010) Double gaps along Shaker S4 demonstrate omega currents at three different closed states. Channels (Austin) 4(2):93–100
Gamal El-Din TM, Martinez GQ, Payandeh J, Scheuer T, Catterall WA (2013) A gating charge interaction required for late slow inactivation in the bacterial sodium channel NaVAb. J Gen Physiol 142:181–190
Gamal El-Din TM, Scheuer T, Catterall WA (2014) Tracking S4 movement by gating pore currents in the bacterial sodium channel NaChBac. J Gen Physiol 144:147–157
Gamal El-Din TM, Lenaeus MJ, Ramanadane K, Zheng N, Catterall WA (2017) Control of slow, use dependent inactivation of NaVAb by its C-terminal tail. Biophys J 112(3, Suppl 1):105a. https://doi.org/10.1016/j.bpj.2016.11.597
Gosselin-Badaroudine P, Delemotte L, Moreau A, Klein ML, Chahine M (2012) Gating pore currents and the resting state of Nav1.4 voltage sensor domains. Proc Natl Acad Sci U S A 109:19250–19255
Guy HR, Seetharamulu P (1986) Molecular model of the action potential sodium channel. Proc Natl Acad Sci U S A 83(2):508–512
Heinemann SH, Terlau H, Stuhmer W, Imoto K, Numa S (1992) Calcium channel characteristics conferred on the sodium channel by single mutations. Nature 356:441–443
Hille B (1971) The hydration of sodium ions crossing the nerve membrane. Proc Natl Acad Sci U S A 68:280–282
Hille B (2001) Ionic channels of excitable membranes. Sinauer Associates, Sunderland, MA
Hirschberg B, Rovner A, Lieberman M, Patlak J (1995) Transfer of twelve charges is needed to open skeletal muscle Na+ channels. J Gen Physiol 106:1053–1068
Hodgkin AL, Huxley AF (1952a) Currents carried by sodium and potassium ions through the membrane of the giant axon of Loligo. J Physiol 116:449–472
Hodgkin AL, Huxley AF (1952b) The dual effect of membrane potential on sodium conductance in the giant axon of Loligo. J Physiol 116:497–506
Hodgkin AL, Huxley AF (1952c) A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol 117:500–544
Hodgkin AL, Huxley AF (1952d) The components of membrane conductance in the giant axon of Loligo. J Physiol 116:473–496
Irie K, Shimomura T, Fujiyoshi Y (2012) The C-terminal helical bundle of the tetrameric prokaryotic sodium channel accelerates the inactivation rate. Nat Commun 3:793
Isom LL, Ragsdale DS, De Jongh KS, Westenbroek RE, Reber BF, Scheuer T, Catterall WA (1995) Structure and function of the beta 2 subunit of brain sodium channels, a transmembrane glycoprotein with a CAM motif. Cell 83:433–442
Jiang Y, Lee A, Chen J, Cadene M, Chait BT, MacKinnon R (2002a) Crystal structure and mechanism of a calcium-gated potassium channel. Nature 417:515–522
Jiang Y, Lee A, Chen J, Cadene M, Chait BT, MacKinnon R (2002b) The open pore conformation of potassium channels. Nature 417:523–526
Kellenberger S, West JW, Scheuer T, Catterall WA (1997) Molecular analysis of the putative inactivation particle in the inactivation gate of brain type IIA Na+ channels. J Gen Physiol 109:589–605
Kuhn FJ, Greeff NG (1999) Movement of voltage sensor S4 in domain 4 is tightly coupled to sodium channel fast inactivation and gating charge immobilization. J Gen Physiol 114:167–183
Larsson HP, Baker OS, Dhillon DS, Isacoff EY (1996) Transmembrane movement of the Shaker potassium channel S4. Neuron 16:387–397
Lee S, Goodchild SJ, Ahern CA (2012) Local anesthetic inhibition of a bacterial sodium channel. J Gen Physiol 139:507–516
Lenaeus MJ, El-Din TMG, Ing C, Ramanadane K, Pomes R, Zheng N, Catterall WA (2017) Structures of closed and open states of a voltage-gated sodium channel. Proc Natl Acad Sci U S A 114:E3051–E3060
Li Q, Wanderling S, Paduch M, Medovoy D, Singharoy A, McGreevy R, Villalba-Galea CA, Hulse RE, Roux B, Schulten K et al (2014) Structural mechanism of voltage-dependent gating in an isolated voltage-sensing domain. Nat Struct Mol Biol 21:244–252
Logothetis DE, Movahedi S, Satler C, Lindpaintner K, Nadal-Ginard B (1992) Incremental reductions of positive charge within the S4 region of a voltage-gated K+ channel result in corresponding decreases in gating charge. Neuron 8:531–540
Mantegazza M, Yu FH, Catterall WA, Scheuer T (2001) Role of the C-terminal domain in inactivation of brain and cardiac sodium channels. Proc Natl Acad Sci U S A 98:15348–15353
McCusker EC, Bagneris C, Naylor CE, Cole AR, D’Avanzo N, Nichols CG, Wallace BA (2012) Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing. Nat Commun 3:1102
Mio K, Mio M, Arisaka F, Sato M, Sato C (2010) The C-terminal coiled-coil of the bacterial voltage-gated sodium channel NaChBac is not essential for tetramer formation, but stabilizes subunit-to-subunit interactions. Prog Biophys Mol Biol 103:111–121
Morais-Cabral JH, Zhou Y, MacKinnon R (2001) Energetic optimization of ion conduction rate by the K+ selectivity filter. Nature 414:37–42
Naylor CE, Bagneris C, DeCaen PG, Sula A, Scaglione A, Clapham DE, Wallace BA (2016) Molecular basis of ion permeability in a voltage-gated sodium channel. EMBO J 35:820–830
Nguyen HM, Goldin AL (2010) Sodium channel carboxy terminal residue regulates fast inactivation. J Biol Chem 285(12):9077–9089
Ong BH, Tomaselli GF, Balser JR (2000) A structural rearrangement in the sodium channel pore linked to slow inactivation and use dependence. J Gen Physiol 116:653–662
Palti Y, Adelman WJ Jr (1969) Measurement of axonal membrane conductances and capacity by means of a varying potential control voltage clamp. J Membr Biol 1:431–458
Papazian DM, Timpe LC, Jan YN, Jan LY (1991) Alteration of voltage-dependence of Shaker potassium channel by mutations in the S4 sequence. Nature 349:305–310
Patton DE, West JW, Catterall WA, Goldin AL (1992) Amino acid residues required for fast Na+-channel inactivation: charge neutralizations and deletions in the III-IV linker. Proc Natl Acad Sci U S A 89:10905–10909
Pavlov E, Bladen C, Winkfein R, Diao C, Dhaliwal P, French RJ (2005) The pore, not cytoplasmic domains, underlies inactivation in a prokaryotic sodium channel. Biophys J 89:232–242
Payandeh J, Scheuer T, Zheng N, Catterall WA (2011) The crystal structure of a voltage-gated sodium channel. Nature 475:353–358
Payandeh J, Gamal El-Din TM, Scheuer T, Zheng N, Catterall WA (2012) Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature 486:135–139
Perozo E, Cortes DM, Cuello LG (1999) Structural rearrangements underlying K+-channel activation gating. Science 285:73–78
Powl AM, O'Reilly AO, Miles AJ, Wallace BA (2010) Synchrotron radiation circular dichroism spectroscopy-defined structure of the C-terminal domain of NaChBac and its role in channel assembly. Proc Natl Acad Sci U S A 107:14064–14069
Qu Y, Isom LL, Westenbroek RE, Rogers JC, Tanada TN, McCormick KA, Scheuer T, Catterall WA (1995) Modulation of cardiac Na+ channel expression in Xenopus oocytes by beta 1 subunits. J Biol Chem 270:25696–25701
Ren D, Navarro B, Xu H, Yue L, Shi Q, Clapham DE (2001) A prokaryotic voltage-gated sodium channel. Science 294:2372–2375
Rogers JC, Qu Y, Tanada TN, Scheuer T, Catterall WA (1996) Molecular determinants of high affinity binding of alpha-scorpion toxin and sea anemone toxin in the S3-S4 extracellular loop in domain IV of the Na+ channel alpha subunit. J Biol Chem 271:15950–15962
Rohl CA, Boeckman FA, Baker C, Scheuer T, Catterall WA, Klevit RE (1999) Solution structure of the sodium channel inactivation gate. Biochemistry 38:855–861
Rudy B (1975) Proceedings: slow recovery of the inactivation of sodium conductance in Myxicola giant axons. J Physiol 249:22–24
Rudy B (1981) Inactivation in Myxicola giant axons responsible for slow and accumulative adaptation phenomena. J Physiol 312:531–549
Sather WA, McCleskey EW (2003) Permeation and selectivity in calcium channels. Annu Rev Physiol 65:133–159
Shaya D, Findeisen F, Abderemane-Ali F, Arrigoni C, Wong S, Nurva SR, Loussouarn G, Minor DL Jr (2014) Structure of a prokaryotic sodium channel pore reveals essential gating elements and an outer ion binding site common to eukaryotic channels. J Mol Biol 426:467–483
Sheets MF, Kyle JW, Kallen RG, Hanck DA (1999) The Na channel voltage sensor associated with inactivation is localized to the external charged residues of domain IV, S4. Biophys J 77:747–757
Shen HZ, Zhou Q, Pan XJ, Li ZQ, Wu JP, Yan N (2017) Structure of a eukaryotic voltage-gated sodium channel at near-atomic resolution. Science 355(6328). pii: eaal4326
Sokolov S, Scheuer T, Catterall WA (2005) Ion permeation through a voltage-sensitive gating pore in brain sodium channels having voltage sensor mutations. Neuron 47:183–189
Sokolov S, Scheuer T, Catterall WA (2008) Depolarization-activated gating pore current conducted by mutant sodium channels in potassium-sensitive normokalemic periodic paralysis. Proc Natl Acad Sci U S A 105:19980–19985
Starace DM, Bezanilla F (2001) Histidine scanning mutagenesis of basic residues of the S4 segment of the Shaker K+ channel. J Gen Physiol 117:469–490
Starace DM, Bezanilla F (2004) A proton pore in a potassium channel voltage sensor reveals a focused electric field. Nature 427:548–553
Stephens RF, Guan W, Zhorov BS, Spafford JD (2015) Selectivity filters and cysteine-rich extracellular loops in voltage-gated sodium, calcium, and NALCN channels. Front Physiol 6:153
Stuhmer W, Conti F, Suzuki H, Wang X, Noda M, Yahadi N, Kubo H, Numa S (1989) Structural parts involved in activation and inactivation of the sodium channel. Nature 339:597–603
Sula A, Booker J, Ng LC, Naylor CE, DeCaen PG, Wallace BA (2017) The complete structure of an activated open sodium channel. Nat Commun 8:14205
Tang L, Gamal El-Din TM, Payandeh J, Martinez GQ, Heard TM, Scheuer T, Zheng N, Catterall WA (2014) Structural basis for Ca2+ selectivity of a voltage-gated calcium channel. Nature 505:56–61
Tao X, Lee A, Limapichat W, Dougherty DA, MacKinnon R (2010) A gating charge transfer center in voltage sensors. Science 328:67–73
Ulbricht W (2005) Sodium channel inactivation: molecular determinants and modulation. Physiol Rev 85:1271–1301
Ulmschneider MB, Bagneris C, McCusker EC, Decaen PG, Delling M, Clapham DE, Ulmschneider JP, Wallace BA (2013) Molecular dynamics of ion transport through the open conformation of a bacterial voltage-gated sodium channel. Proc Natl Acad Sci U S A 110:6364–6369
Vargas E, Yarov-Yarovoy V, Khalili-Araghi F, Catterall WA, Klein ML, Tarek M, Lindahl E, Schulten K, Perozo E, Bezanilla F et al (2012) An emerging consensus on voltage-dependent gating from computational modeling and molecular dynamics simulations. J Gen Physiol 140:587–594
Vassilev PM, Scheuer T, Catterall WA (1988) Identification of an intracellular peptide segment involved in sodium channel inactivation. Science 241:1658–1661
Vassilev P, Scheuer T, Catterall WA (1989) Inhibition of inactivation of single sodium channels by a site-directed antibody. Proc Natl Acad Sci U S A 86:8147–8151
Vilin YY, Ruben PC (2001) Slow inactivation in voltage-gated sodium channels: molecular substrates and contributions to channelopathies. Cell Biochem Biophys 35:171–190
West JW, Patton DE, Scheuer T, Wang Y, Goldin AL, Catterall WA (1992) A cluster of hydrophobic amino acid residues required for fast Na+-channel inactivation. Proc Natl Acad Sci U S A 89:10910–10914
Wu J, Yan Z, Li Z, Qian X, Lu S, Dong M, Zhou Q, Yan N (2016) Structure of the voltage-gated calcium channel CaV1.1 at 3.6 a resolution. Nature 537:191–196
Xiong W, Li RA, Tian YL, Tomaselli GF (2003) Molecular motions of the outer ring of charge of the sodium channel: do they couple to slow inactivation? J Gen Physiol 122:323–332
Yan Z, Zhou Q, Wang L, Wu J, Zhao Y, Huang G, Peng W, Shen H, Lei J, Yan N (2017) Structure of the Nav1.4-beta1 complex from electric eel. Cell 170(3):470–482.e11
Yang N, Horn R (1995) Evidence for voltage-dependent S4 movement in sodium channels. Neuron 15:213–218
Yang J, Ellinor PT, Sather WA, Zhang JF, Tsien RW (1993) Molecular determinants of Ca2+ selectivity and ion permeation in L-type Ca2+ channels. Nature 366:158–161
Yang N, George AL Jr, Horn R (1996) Molecular basis of charge movement in voltage-gated sodium channels. Neuron 16:113–122
Yarov-Yarovoy V, Decaen PG, Westenbroek RE, Pan CY, Scheuer T, Baker D, Catterall WA (2012) Structural basis for gating charge movement in the voltage sensor of a sodium channel. Proc Natl Acad Sci U S A 109:E93–E102
Yue L, Navarro B, Ren D, Ramos A, Clapham DE (2002) The cation selectivity filter of the bacterial sodium channel, NaChBac. J Gen Physiol 120:845–853
Zhang X, Ren W, DeCaen P, Yan C, Tao X, Tang L, Wang J, Hasegawa K, Kumasaka T, He J et al (2012) Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature 486:130–134
Zhao Y, Scheuer T, Catterall WA (2004a) Reversed voltage-dependent gating of a bacterial sodium channel with proline substitutions in the S6 transmembrane segment. Proc Natl Acad Sci U S A 101:17873–17878
Zhao Y, Yarov-Yarovoy V, Scheuer T, Catterall WA (2004b) A gating hinge in Na+ channels; a molecular switch for electrical signaling. Neuron 41:859–865
Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R (2001) Chemistry of ion coordination and hydration revealed by a K+ channel- Fab complex at 2.0 a resolution. Nature 414:43–48
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
1 Electronic Supplementary Material
Supplementary Fig. 1
(PDF 377 kb)
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Gamal El-Din, T.M., Lenaeus, M.J., Catterall, W.A. (2017). Structural and Functional Analysis of Sodium Channels Viewed from an Evolutionary Perspective. In: Chahine, M. (eds) Voltage-gated Sodium Channels: Structure, Function and Channelopathies. Handbook of Experimental Pharmacology, vol 246. Springer, Cham. https://doi.org/10.1007/164_2017_61
Download citation
DOI: https://doi.org/10.1007/164_2017_61
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-90283-8
Online ISBN: 978-3-319-90284-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)