Abstract
Diffusion-weighted magnetic resonance imaging (DWI) is the use of specific MRI sequences, which uses the diffusion of Hydrogen atoms to generate contrast and it allows the mapping of the diffusion process of molecules in vivo and reflects interactions with macromolecules, fibers, and membranes among other. Hydrogen atom diffusion patterns (quantification of anisotropy) can reveal microscopic details about tissue architecture, either normal or in a diseased state. A special kind of DWI, diffusion tensor imaging (DTI), has been used extensively to map white matter tractography in the brain. Tractography is a procedure that is used to highlight neural tracts (axon), its fibers position estimation in brain areas has broad potential implications in cognitive neuroscience fields. An algorithm based on diffusion tensor Image is developed and implemented in order to evaluate brain connectivity in different regions of interest. The major objective of this work is represent two-dimensional and three-dimensional connectivity between areas thereby show the potential of the DTI. Results shows how Connectivity Matrix provides statistical data on the pattern of anatomical relationships, this connectivity pattern is formed by synapses that represent the cross correlations and the flow of information.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Overall, the nervous system comprises, into two parts: the Central Nervous System (CNS) and the Peripheral Nervous System (SNP). The central nervous system consists of the brain and spinal cord. The brain has long connections between them that make up the white matter and gray matter (Fig. 1).
White matter is 60% of the brain, they are myelinated nerve fibers (axons) protected by a myelin sheath that provides electrical isolation processes that allow nerve signals to be transmitted faster [1, 2]. Neural networks created in white matter, help in the process of memory, learning, cognitive processes and the development of intelligence [3,4,5].
In neuroscience, a tractography is a non-invasive, in vivo 3D modeling technique used to visually represent nerve tracts using data collected by diffusion MRI (MRI-DWI). It provides the necessary information to analyze of nerve fibers (tracts) qualitatively and quantitatively, reproducing their volume and direction. Tracts are not directly identifiable; hence, experts must use image processing techniques to detect them, see Fig. 4.
DTI (Diffusion Tensor Imaging) Tractography uses diffusion tensor data and allows 3D visualization of specific white matter tracts.. Diffusion tensor by white matter can detect the orientation of axons and the magnitude or degree of anisotropy [6,7,8,9]. Diffusion is also known as Brownian motion, defined through eq. 1 as follows:
where r2 refers to the mean square displacement of molecules, t is the diffusion time, and D is the diffusion constant, a constant of proportionality for the particular substance being measured, in this case the hydrogen atom.
The Diffusion tensor vector matrix is a 3 × 3 matrix. Can relate to signal strength weighted by diffusion, S, and the intensity of the signal b 0 s / mm2, S0, described with Eq.2:
where
where bij comprises the direction and magnitude of the applied gradient vector diffusion (Gx, Gy, Gz).
Isotropy refers to the freedom with which water molecules move through tissues. More obstacles decrease isotropy (anisotropy), but when water molecules move freely, isotropy levels are high. Isotropy values range from 0 (no isotropy, much hindered movement) to 1 (total isotropy, free movement). Anisotropy can be measured in multiple ways, including fractional anisotropy (FA). An anisotropy of 0 corresponds to a perfect sphere, while 1 would be an ideal linear diffusion. Well-defined tracts have a FA greater than 0.2, there are few regions with higher FA (i.e. 0.9). Each isotropy is linked to a predominant orientation axis (predominant direction of diffusion).
DTI evaluates two parameters, among others:
Fractional anisotropy (FA), measures the direction of diffusion and detects white matter lesions
Average diffusivity, measures the extent of diffusion and is sensitive to ultrastructural damage in white matter.
Our algorithm provides a post-processing directional information of the image to obtain a matrix relationship in a brain atlas. This characteristic is important, since the brain is not formed by modular structures that function in isolation but rather by distributed neural networks that connect different regions and integrate them into broad functional networks (i.e. neural connectivity), which we call neuronal connectivity, where, the integrity and correct functioning of the white matter (axons, tracts) is essential for the normal development of any cognitive process. Tracts allow communication between gray matter and other body parts. They transmit information from different parts of the body to the cerebral cortex and enable different brain regions involved in cognition to connected (i.e. brain connectivity). Tracts are not directly involved in the cognitive process itself, but they still play role by allowing the different brain regions involved in cognition to remain connected. Consequently, information flows at an appropriate speed. In conclusion, changes in the white matter can cause changes in cognitive ability [10, 11].
Brain connectivity refers to a pattern of anatomical relationships between areas within a nervous system. The units correspond to individual neurons, neuronal populations, or anatomically separate brain regions. In fact, intuitive solutions to problems arise when the brain gets more intense connectivity between its two hemispheres. Cooperation between both hemispheres increases mental performance, that is, the ability to actively support the memory-learning process for longer, with greater productivity and more efficiently. Analyzing brain connectivity, including differentiating connectivity by gender [12], is crucial to understanding neurons and their information process in the network [13,14,15].
The aim of this paper is to represent two-dimensionally and three-dimensional brain connectivity in all brain areas through an algorithm developed in MatLab (https://www.mathworks.com/).
2 Methodology
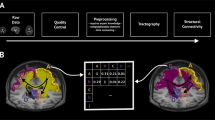
We followed the methodology depicted in Fig. 2 to create connectivity matrix that provides statistical data on structural relationship In order to carry out the connectivity matrix that provides statistical data on the structural relationship between two brain areas of interest (i.e. pattern of anatomical relationships between different units within a nervous system).
2.1 Brain atlas
Brain atlases are one of the most effective ways to accurately locate anatomical and functional structures in patients.
Brain atlas used is the AAL (Automated Anatomical Labeling) with 116 structures [16, 17]. In order to separate structures we proceed as shown in Fig. 3, the intensity of each pixel in the original image (116 intensity) 116 will separate structures. Intensity levels determine the structure to which it belongs.
2.2 Tractography
Tractography determines the directional pattern of diffusion tensor imaging using an “anisotropic map” on a 3D coordinate system in which the color represents the direction, given the difficulty of representing 2D grayscale images. See Fig. 4.
Red indicates directions in the X axis: right to left or left to right.
Green indicates directions in the Y axis: posterior to anterior or from anterior to posterior.
Blue indicates directions in the Z axis: foot-to-head direction or vice versa.
To separate tracts we proceed with the reading of trk file (trackvis, with the implementation of an algorithm in MatLab), where you can see that our algorithm obtains the same results as the commercial as shown in Fig. 5.
2.3 Algorithm
A network is defined by a set of nodes and links between pairs of nodes. Large brain networks whereas links represent the anatomical, functional, or effective connections, according to the data set. Our algorithm implemented with MatLab looks for the areas and ROIs involved as connectivity between tracts. This process can be defined as follows:
- 1.
trk file properties retrieved; the trk file is a binary file, with the first 1000 bytes of the header and the rest as the body.
- 2.
Each scalar properties tract are defined from the trk file, in order to differentiate them from the matrix.
- 3.
Tract structures are loaded. T = tract number
- 4.
The total size of the tract is read.
- 5.
ROIs (areas of interest, brain atlas) R = number of ROI is read
- 6.
Tx vs Rx is evaluated
- 7.
Connectivity matrix is obtained
Table 1 shows the obtained connectivity matrix representing Fig. 6. These results were validated with Trackvis with identical results. UCLA_Multimodal_Connectivity_Package https://www.ccn.ucla.edu/wiki/index.php/UCLA_Multimodal_Connectivity_Package
3 Results
Our connectivity matrix provides statistical data on structural relationship between two areas of interest (pattern relationships between different units within a nervous system). This research proposes a tool to help medical professionals quantitatively validate the results of a therapy treatment or cognitive-motor treatment. As seen in Fig. 6, our algorithm calculate the points of the structure tract so that they can validate the area of interest (ROI),
The first part of the results shows the connectivity among the 116. The second part shows the segmentation results of specific areas (ROI1, ROI2, ROI3, ROI4, ROI5) as chosen by users, and the connectivity among them.
3.1 First part
Our algorithm shows the connectivity matrix between each and every areas of the AAL brain atlas. The area less connected to other areas is 108 Cerebelum_10_R, whereas the most connected area is 31 Cingulum_Ant_L.
The connectivity matrix shows areas from the brain atlas are more connected to another. As depicted in Fig. 7 and according to our matrix results, area 108 has the least connectivity with the other areas, namely 3 of 116 (98,100,106). Conversely, area 31 showed the highest connectivity with other areas, namely 23 of 116.
Areas with more tracts involved in connectivity are shown in Fig. 8. As can be observed, 99. Cerebelum_6_L, provides the greatest total number of tracts involved. Conversely, Cerebelum_3_L, provides the least total number of tracts involved.
3.2 Second part
Our algorithm can group interest areas designed by the user and not necessarily a square matrix, i.e.:
There can be no connectivity between ROI1 - ROI2 –ROI3-ROI5 with t_4. See Fig. 9 and Table 2.
The greatest connectivity according to this example is ROI_2 with t_5. See Fig. 9 and Table 2.
Table 2 and Fig. 9 introduce our conectivity matrix. As Fig. 9 despicts, t_5 is the hihest connectivity tract, where 90% of connectivity is in ROI_2 and 10% is in ROI_4.
4 Conclusion
Brain signal and image acquisition techniques have evolved, thus becoming essential tools for diagnosis and research in neurosciences. Some of the most important applications in connectome have been developed in the field of Education, neuronal connectivity and the changes experienced by the brain through experience are part of brain plasticity or synaptic plasticity (neuronal connectivity) that makes learning possible.
The structural connectome provides relevant information on experience and training-related changes in the brain. Much of what is currently in know of brain connectivity and operation in distributed neural networks has come from the hand of neuroimaging, specifically DTI technologies. DTI-based tracktography makes great contributions to the study of the structure and functionality of axons.
Tracts allow communication between gray matter and other body parts, it sends information from different parts of the body to the cerebral cortex. Even though tracts are not directly involved in the cognitive process itself, they allow different brain regions involved in cognition remain connected. Consequently, information flows rapidly. Track integrity and its proper operation is essential for the normal development of cognitive functions (e.g. attention, memory, ultimately, for any cognitive process).
DTI applies to any condition that is likely to create an anomaly in the tracts, such as multiple sclerosis, Alzheimer, mild cognitive impairment or simply normal aging. Tractography by diffusion tensor calculus is the only method available today to evaluate cerebral white matter tracts in vivo. This field (tractography) opens a range of possibilities, especially for mental disorders that have traditionally been considered as lacking a structural brain base. Analyzing of the effects of brain damage on cognition helps to understand these anatomical and functional relationships, noting the many ways that can disintegrate cognition. Finally, neuroimaging technologies remain promising alternatives for solving important questions in neuroscience.
References
Blumenfeld H. Areas of the CNS made up mainly of myelinated axons are called white matter, de Neuroanatomy through clinical cases (2nd ed.), Sunderland, Sinauer Associates Inc, 2010, p. 21.
R. D. Fields, «White Matter Matters,» Sci Am, vol. 1, n° 298, pp. 54 - 61 , 2008.
S. Farquharson, «White matter fiber tractography: why we need to move beyond DTI,» J Neurosurg, vol. 118, n° 6, pp. 1367-1377, 2013.
D. K. Jones, «White matter integrity, fiber count, and other fallacies: the do's and don'ts of diffusion MRI,» Neuroimage, vol. 1, n° 73, pp. 239-254, 2013.
L. Penke, «Brain white matter tract integrity as a neural foundation for general intelligence,» Mol Psychiatry, vol. 17, n° 10, p. 1026, 2012.
Duque A, «Anatomía de la sustancia blanca mediante tractografía por tensor de difusión,» Radiología, vol. 50, n° 2, pp. 99-111, 2008.
M. F. Glasser, «DTI tractography of the human brain's language pathways,» Cereb Cortex, vol. 18, n° 11, pp. 2471-2482, 2008.
M. Catani, «A diffusion tensor imaging tractography atlas for virtual in vivo dissections,» Cortex, vol. 44, n° 8, pp. 1105-1132., 2008.
P. Mukherjee , «Diffusion Tensor MR Imaging and Fiber Tractography: Theoretic Underpinnings,» AJNR Am J Neuroradiol, vol. 29, n° 1, pp. 632-641, 2008.
G. Kocevar, «Brain structural connectivity correlates with fluid intelligence in children: A DTI graph analysis,» Intelligence, vol. 72, n° 1, pp. 67-75, 2019.
F. Roman, «Enhanced structural connectivity within a brain sub-network supporting working memory and engagement processes after cognitive training,» Neurobiol Learn Mem, vol. 141, n° 1, pp. 33-43, 2017.
M. Ingalhalikar, «Sex differences in the structural connectome of the human brain,» Proc Natl Acad Sci, vol. 111, n° 2, pp. 823-828, 2014.
P. Hagmann, «Understanding diffusion MR imaging techniques: from scalar diffusion-weighted imaging to diffusion tensor imaging and beyond,» Radiographics, vol. 26, n° 1, pp. 205-223, 2006.
R. D. Rafal, «Connectivity between the superior colliculus and the amygdala in humans and macaque monkeys: virtual dissection with probabilistic DTI tractography,» J Neurophysiol, vol. 114, n° 3, pp. 1947-1962, 2015.
Verly M. Microstructural organization of the language connectome in typically developing left-handed children: a DTI tractography study, de ISMRM, Singapore, 2016.
N. D. Institute, Neurofunctional Imaging, Université de Bordeaux, Group (GIN-IMN), [En línea]. Available: http://www.gin.cnrs.fr/en/tools/aal-aal2/. [Último acceso: 01 01 2019].
N. Tzourio-Mazoyer, «Automated Anatomical Labeling of Activations in SPM Using a Macroscopic Anatomical Parcellation of the MNI MRI Single-Subject Brain,» NeuroImage, vol. 15, n° 1, pp. 273-289, 2002.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical approval
This paper does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hernandez, N.R., Montelongo, R.H., Bernal, J.M.C. et al. Development and implementation of algorithms with diffusion tensor images to evaluate brain connectivity. Health Technol. 10, 471–478 (2020). https://doi.org/10.1007/s12553-019-00376-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12553-019-00376-7