Abstract
Purpose of Review
The diagnosis of invasive fungal disease remains challenging in the clinical laboratory. In this paper, the use of matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) for the identification of filamentous fungi as well as its application for antifungal resistance testing and strain typing is evaluated.
Recent Findings
Most studies report very high accuracy for the identification of filamentous fungi by MALDI-TOF MS. Its cost effectiveness, short analysis time, and low error rate and the fact that it can also discriminate between closely related and cryptic species make it appropriate for implementation in the clinical routine. Two drawbacks remain in the availability of extended reference spectra databases and the fact that this technique can only be applied on isolates.
Summary
More work on (simultaneous) antifungal susceptibility testing and strain typing is needed. The application of MALDI-TOF MS directly on clinical specimens would further improve the diagnosis of invasive fungal disease and improve its successful management.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fungi play an important role in a variety of ecosystems and are widely and abundantly present in the human environment. Some of these naturally occurring fungi are able to cause opportunistic invasive fungal disease (IFD) in immunocompromised hosts. With the growing number of these patients at risk, IFD represent a real challenge in the clinical laboratory, as moreover, the mortality rates linked to IFD are particularly high. Yet, IFD are still underrepresented in research projects and surveillance programs, and epidemiological data that could give an idea of the prevalence of these infections is very poor. However, Brown and his colleagues have estimated from extrapolated data that more people die from IFD than from tuberculosis or malaria worldwide [1]. More innocent fungal infections, that are however much more prevalent, are superficial infections of skin and nails. These infections, mostly caused by dermatophytes, are estimated to affect 25% of the world’s population [2]. Species-level identification is critical for correct patient treatment, and it is generally accepted that a rapid diagnosis, coupled to the early onset of the appropriate treatment, leads to a better patient outcome [3].
Though important advances have been made over the past decades at the level of identification of filamentous fungi, it remains quite a challenge. Whereas historically, strains have been classified based upon their morphological characteristics, the implementation of molecular methods have unraveled an important genetic variability within the sometimes very homomorphic groups. Multi-locus gene sequencing is indeed now considered as the gold standard for the identification of filamentous fungi. This costly and time-consuming approach is not always realistic in the clinical laboratory setting, and as it did for the identification of bacteria and yeasts, matrix-assisted laser desorption/ionization-time of flight mass spectroscopy (MALDI-TOF MS) is now doing its entry for the identification of filamentous fungi in the clinical routine.
The MALDI-TOF MS Technology
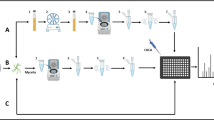
MALDI-TOF MS exists already for over three decades and was developed initially as a soft ionization mass spectrometry approach for the analysis of large biomolecules in 1987. It was even honored with a Nobel prize in 2002 for John B. Fenn and Koichi Tanaka. The technology has since then been evolving and is used since the beginning of this century for the identification of micro-organisms [4]. Whereas the identification of bacteria and also yeasts by MALDI-TOF MS is getting widely integrated in the clinical routine since over a decade, it is now becoming more and more accepted that MALDI-TOF MS could also be a powerful alternative for the identification of filamentous fungi [4, 5•, 6, 7]. The principle is quite simple: a protein extract of the micro-organism is deposited on a metal plate together with a matrix solution. Irradiation with a laser beam leads to the ionization of the material, which is subsequently accelerated through an electric field, allowing the separation of the molecules according to their mass to charge ratio (m/z) in a vacuum tube. The whole process generates a spectrum, reflecting the protein content of the sample, and in the case of micro-organisms, this spectrum is shown to be species-specific. A comparison of the obtained spectrum with a reference spectra database eventually allows the identification of the micro-organism [4]. The obtained spectrum should be considered as a fingerprint of the strain, but the peaks in the spectrum remain unidentified. Different MALDI-TOF MS platforms for microbial identification are currently available: the Bruker BioTyper (Bruker Daltonics GmbH, Bremen, Germany), the Andromas (Andromas SAS, Paris, France), the Axima-iD Plus (Shimadzu/AgnosTec, Duisburg, Germany), and the Vitek MS system in combination with the SARAMIS database (bioMérieux, Marcy l’Etoile, France). Though the technology is similar in all four platforms, the data processing and the algorithms used for the comparison to the database are quite different and often remain a black box for the user [4, 5•, 8].
The Andromas system expresses the similarity of the unknown spectra with the reference spectra as a percentage. The results are subdivided in three categories: “good identification” when the similarity is over 65%, “identification to be confirmed” when between 60 and 64%, and “no identification” when under 60%. In the Biotyper software, similarity of the spectra with reference spectra of the database is expressed as log scores between 0 and 3. A log score above 2.3 indicates “highly probable species-level identification,” while a log score between 2 and 2.299 indicates secure genus level identification and probable species level identification. When the log score is between 1.7 and 1.99, a “probable genus level identification” is indicated, and with a score below 1.7, the identification should be interpreted as not reliable. However, it is not clear whether these guidelines apply for all types of micro-organisms, as in different reports, deviations of these guidelines did not seem to have an effect on the sensitivity and specificity of the results. The SARAMIS database was initially developed by AgnosTec but was then purchased by Biomérieux and incorporated in the Vitek MS platform. Also in this system, the similarity of the spectra is expressed as a percentage, and a value over 60% is considered reliable [4, 8]. Several studies have attempted to compare the performance of the available MALDI-TOF MS diagnostic platforms, either focusing on one type of pathogen or on a variety of less commonly encountered organisms, and the main conclusion seems to be that all systems perform quite well and that they all have advantages and disadvantages, depending on the type of organism studied [5•, 9, 10, 11, 12•].
MALDI-TOF MS for the Identification of Filamentous Fungi
The application of MALDI-TOF MS for the identification of filamentous fungi in the clinical routine has been hampered by several factors. Firstly, though some studies reported the successful use of the “direct deposit” method, a specific sample preparation procedure is recommended to obtain high quality and reproducible spectra, mainly due to the fact that fungi have a thorough cell wall [13, 14]. Secondly, fungal cultures are heterogenic. Not only do they consist of different cell types such as mycelium, spores, and fructification organs, there is also a significant difference between the center of a colony and the periphery. At the center, older cells produce secondary metabolites and pigments, while the younger undifferentiated mycelium is at the periphery of the colony. This age effect was very nicely demonstrated by Coulibaly et al. in a study on Pseudallescheria/Scedosporium spp. where it was shown that the spectra of the same colony varied in function of age [15]. To minimize the effect of growth conditions, Bruker Daltonics proposed a protein extraction protocol from 24 to 48 h old liquid and rotating cultures and dedicated a special database to it. This database and the procedure based upon liquid cultures were evaluated by the group of Michael Hombach in Zurich. Though the reported correct species identification levels of 54.2 and 79% (using collection material and clinical isolates respectively) seem not convincing, the authors attributed this to the limitation of the database, as the majority of the unidentified strains were not recognized because the species was lacking from the database and only a little amount of false identifications were encountered [16]. This brings us to the third and major drawback for fungal identification by MALDI-TOF MS, namely, the lack of extended reference spectra databases. Whereas the commercially available databases are quite elaborate for bacteria and yeasts, they are very limited for fungi, which explains why the vast majority of the publications on fungal identifications use an in-house created database [5•].
Performance of MALDI-TOF MS for Fungal Identification
First of all, many reports are available on the performance level of MALDI-TOF MS for the identification of filamentous dermatophyte and non-dermatophyte fungi. The success rate of the fungal identification has been extensively reviewed by Cassagne et al. [5•].
Though the percentages of correct identifications are generally high and the amount of false identifications are often close to zero, they can vary between the different reports, depending mainly on the used method, the database, and on the variety of the test panel evaluated, making it difficult to compare the results. For instance, Nenoff et al. have reported the very successful identification of dermatophytes by MALDI-TOF MS using the Saramis software. The success rate of 98.8% correct identifications seems convincingly high, but this may be explained by the fact that the authors have used the same strains for constructing the database as for challenging it [17].
Lau et al. constructed a reference spectra database based on 249 reference isolates, and challenged this database with a test panel of 421 clinical isolates. 88.9% of the isolates were correctly identified up to the species level, whereas no false identifications were encountered [18]. Cassagne et al. developed a database of 143 reference spectra, and their evaluation of 177 isolates led to an 87% accuracy rate and only two misidentifications [19]. Both studies used an extraction protocol and the Bruker BioTyper. Other reports using the Bruker BioTyper system reported success rates in the same range of 89 and 87% when looking at a variety of species [20, 21]. Becker et al. have evaluated the performance of their in-house created database with a test panel of 490 clinical isolates, leading to a success rate of 95.4% [22]. For this study, the authors did not use the cut-off values as recommended by the manufacturer, instead they considered the result valid if three out of the four technical repeats (i.e., spots) gave the same identification. With only one false identification, the specificity in this study remained very high [22]. Indeed, several studies have reported deviations from the cut-off values in the manufacturer’s instructions without impacting the specificity of the method, underlining the robustness of the technology [22–23].
In other reports, several groups have focused on specific types or genera of (closely related) organisms, and most of them have focused on Aspergillus spp. Iriart et al. evaluated the use of the Vitek MS system, without applying a protein extraction protocol, for the identification of Aspergillus strains. Out of the 44 Aspergillus isolates, 36 were correctly identified (81.8%) and eight remained unidentified. Though in this study, the percentage of correct species identifications was even lower than in their routine identification test, it is worthwhile mentioning that also in this case, none of the strains were misidentified [24]. Oliveira et al. have reported the 100% successful distinction of Sporothrix species using an in-house created database with the SARAMIS software [25]. Two studies have evaluated the performance of the Andromas system for Aspergillus identification. Bille et al. have tested a panel of 64 Aspergillus strains and achieved an overall success rate of 98.4%, using a formic acid extraction procedure, but they only looked at a limited number of different species (6) and the vast majority were Aspergillus fumigatus [26]. In the study by Alanio et al., the test panel included 24 different Aspergillus species, but no protein extraction protocol was applied and 98.6% of correct identifications up to the species level were obtained and no false identifications were encountered [13].
Whereas for evaluating the performance of the Bruker BioTyper system for fungal identification, most of the studies have reported a full extraction protocol; De Carolis et al. have shown that Aspergillus, Fuisarium, and Mucorales species were successfully identified using a direct deposit method (96.8% correct identifications when present in the database) [14, 18,19,20,21,22]. Triest et al. have reported a success rate of 91% for the identification of a wide variety of closely related Fusarium species using an in-house created database and a formic acid extraction procedure [23]. Dolatabadi et al. have reported the successful distinction of Mucorales species by MALDI-TOF MS using 38 isolates and a formic acid extraction procedure. Though generating distinct clades using a PCA dendrogram, the identification performance was not reported by the authors [27]. Also for dermatophytes, which usually represent an even higher diagnostic challenge as the taxonomy of this group is very complex and the isolates are often atypical, MALDI-TOF MS seems to perform very well in the clinical setting. Identification rates reported are very high, up to 100% using in-house created databases [5, 28,29,30].
It is clear from these reports that the good performance of MALDI-TOF MS, and more specifically, the very limited amount of false identifications that are encountered, makes this technique suitable for the clinical laboratory, also for filamentous fungi, and CLSI guidelines (CLSI M-58 Ed1) are now available for its implementation in the medical laboratory. MALDI-TOF MS is more effective for species identification as compared to the classical microscopic approaches and is a cost-effective alternative for multi-locus gene-sequencing [22]. However, it is also important to strive for standardized cultivation and sample preparation methods when implementing the method in a clinical laboratory, i.e., one approach for all types of filamentous fungi. To improve identification scores, the sample preparation procedure of the unknown sample are preferably the same as the method used to generate the spectra of the database. Lastly, making the generated spectra publically available would be an important step forward to overcome the problem of the availability of the databases. However, taken the previous point into consideration, for the successful use of such publically available databases, complete transparency about the used protocols to construct the spectra is a prerequisite.
Promising Perspectives for the Future
One limitation of the MALDI-TOF MS technology remains the fact that it is only applicable on isolates. This is inherent to the technology that produces a spectrum reflecting the complete content of the sample, a fingerprint, but that is not detecting any specific molecules. It is well known that cultures are often false negative and it has been estimated that positive blood cultures are only obtained in about 50% of the cases of invasive candidiasis and that the success rate is even lower in invasive infections with filamentous fungi [31]. For bacteria and yeasts, several reports have shown the MALDI-TOF MS based identification of pathogens directly on clinical specimens or blood cultures [4]. The direct identification of filamentous fungi such as Fusarium spp. from positive blood cultures has also been recently described for the first time, but further work is needed [32••]. Alternative MS approaches targeting specific molecules have been evaluated. Triest et al. have developed a method using LC-ESI-MS/MS to detect species-specific peptides in mixed cultures. This could be a promising approach to detect the organisms directly in clinical specimens [33••]. In another attempt, serum disaccharide has been targeted as an early but pan-fungal marker to diagnose invasive candidiasis, aspergillosis, and mucormycosis [31]. Indeed, the detection of species-specific molecules using specialized mass spectrometers can overcome certain limitations of MALDI-TOF MS, as these specific molecules could theoretically be detected in any matrix and independently of culture conditions. However, this highly specialized equipment currently requires specialized analysts and is not accessible in the clinical microbiology laboratory.
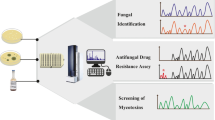
Several studies have also been undertaken to use MALDI-TOF MS for antifungal susceptibility testing in yeasts and for susceptibility to caspofungin in Candida and Aspergillus, a proof of concept delivered by De Carolis et al. [34,35,36]. A combined approach for identification and simultaneous antifungal susceptibility testing as it was described for Mycobacteria could indeed significantly decrease turnaround time in the clinical routine [37••]. Decreasing the turnaround time will positively affect patient outcome as it allows an evidence-based treatment strategy. Shorter hospital stays and improved prognosis will in its turn also have financial implications.
As MALDI-TOF MS differentiates closely related species that are not picked up by the conventional methods, it represents a true added value in evaluating the real species diversity and appearance of emerging species in the clinical routine and to help us to generate epidemiological insights. Gautier et al. showed that the number of species encountered in the clinical routine increased from 16 to 42 different species of filamentous fungi [38]. Strain typing is another promising possibility of the technology that could be of major importance in outbreak prevention and management. Currently used typing methods are complex and often not available in routine clinical laboratories, leading to a delay in the appropriate management of hospital-acquired fungal infections. The report of Dhieb et al. on the typing of Candida glabrata seemed promising; other attempts have failed to cluster genetically related isolates [39–40]. In a recent report, the successful identification and clustering of the emerging and particularly important pathogen Candida auris has been shown using the VITEK MS [41].
Conclusion
MALDI-TOF MS has revolutionized the clinical laboratory, and its successful application for the identification of filamentous fungi has numerously been reported. What is particularly interesting when looking at the different reports is that the level of false identifications is close to zero, making it a robust and reliable technology. The true added value lies in the fact that MALDI-TOF MS can differentiate closely related species that are missed by conventional methods, creating new epidemiological insights and a better view on the emerging problems. Promising future developments include antifungal resistance testing and strain typing, but the developments in these fields are delayed as compared to their advances for bacteria and yeasts and more work is needed. One disadvantage of the technique remains the fact that it can only be applied on isolates, and different strategies are investigated to overcome this, though it needs to be seen whether MALDI-TOF MS will turn out to be the appropriate method for this, or whether more specialized MS approaches will need to come into play. Either way, the future is challenging.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, White TC. Hidden killers: human fungal infections. Med Mycol. 2012;4(165):165rv13. doi:10.1126/scitranslmed.3004404.
Havlickova B, Czaika VA, Friedrich M. Epidemiological trends in skin mycoses worldwide. Mycoses. 2008;51(Suppl. 4):2–15.
Perfect JR. Fungal diagnosis: how do we do it and can we do better? Curr Med Res Opin. 2013;29(Suppl 4):3–11.
Clarck AE, Kaleta EJ, Arora A, Wolk DM. Matrix-assisted laser desorption ionization-time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clin Micr Rev. 2013;26(3):547–603.
• Cassagne C, Normand AC, L’Ollivier C, Ranque S, Piarroux R. Performance of MALDI-TOF MS platforms for fungal identification. Mycoses. 2016;59(11):678–90. This review paper provides an excellent overview on the identification efficacy of yeasts, filamentous fungi and dermatophytes.
van Veen SQ, Claas EC, Kuijper EJ. High-throughput identification of bacteria and yeast by matrix-assisted laser desorption ionization-time of flight mass spectrometry in conventional medical microbiology laboratories. J Clin Microbiol. 2010;48(3):900–7.
Marklein G, Josten M, Klanke U, Müller E, Horré R, Maier T, Wenzel T, Kostrzewa M, Bierbaum G, Hoerauf A, Sahl HG. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for fast and reliable identification of clinical yeast isolates. J Clin Microbiol. 2009;47(9):2912–7.
Sanguinetti M, Posteraro B. Identification of molds by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2016;55(2):369–79.
Bader O, Weig M, Taverne-Ghadwal L, Lugert R, Gross U, Kuhns M. Improved clinical laboratory identification of human pathogenic yeasts by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Microbiol Infect. 2011;17(9):1359–65.
Martiny D, Busson L, Wybo I, El Haj RA, Dediste A, Vandenberg O. Comparison of the Microflex LT and Vitek MS systems for routine identification of bacteria by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2012;50(4):1313–25.
Lacroix C, Gicquel A, Sendid B, Meyer J, Accoceberry I, François N, Morio F, Desoubeaux G, Chandenier J, Kauffmann-Lacroix C, Hennequin C, Guitard J, Nassif X, Bougnoux ME. Evaluation of two matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) systems for the identification of Candida species. Clin Microbiol Infect. 2014;20(2):153–8.
• Lévesque S, Dufresne PJ, Soualhine H, Domingo MC, Bekal S, Lefebvre B, Tremblay C. A side by side comparison of Bruker Biotyper and VITEK MS: utility of MALDI-TOF MS technology for microorganism identification in a public health reference laboratory. PLoS One. 2015;10(12):e0144878. doi:10.1371/journal.pone.0144878. Excellent comparison between two available platforms for a wide variety of species.
Alanio A, Beretti JL, Dauphin B, Mellado E, Quesne G, Lacroix C, Amara A, Berche P, Nassif X, Bougnoux ME. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for fast and accurate identification of clinically relevant Aspergillus species. Clin Microbiol Infect. 2011;17(5):750–5.
De Carolis E, Posteraro B, Lass-Flörl C, Vella A, Florio AR, Torelli R, Girmenia C, Colozza C, Tortorano AM, Sanguinetti M, Fadda G. Species identification of Aspergillus, Fusarium and Mucorales with direct surface analysis by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Microbiol Infect. 2012;18(5):475–84.
Coulibaly O, Marinach-Patrice C, Cassagne C, Piarroux R, Mazier D, Ranque S. Pseudallescheria/Scedosporium complex species identification by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Med Mycol. 2011;49(6):621–6.
Schulthess B, Ledermann R, Mouttet F, Zbinden A, Bloemberg GV, Böttger EC, Hombach M. Use of the Bruker MALDI Biotyper for identification of molds in the clinical mycology laboratory. J Clin Microbiol. 2014;52(8):2797–803.
Nenoff P, Erhard M, Simon JC, Muylowa GK, Herrmann J, Rataj W, Gräser Y. MALDI-TOF mass spectrometry—a rapid method for the identification of dermatophyte species. Med Mycol. 2013;51(1):17–24.
Lau AF, Drake SK, Calhoun LB, Henderson CM, Zelazny AM. Development of a clinically comprehensive database and a simple procedure for identification of molds from solid media by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2013;51(3):828–34.
Cassagne C, Ranque S, Normand AC, Fourquet P, Thiebault S, Planard C, Hendrickx M, Piarroux R. Mould routine identification in the clinical laboratory by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. PLoS One. 2011;6(12):e28425.
Ranque S, Normand AC, Cassagne C, Murat JB, Bourgeois N, Dalle F, Gari-Toussaint M, Fourquet P, Hendrickx M, Piarroux R. MALDI-TOF mass spectrometry identification of filamentous fungi in the clinical laboratory. Mycoses. 2014;57(3):135–40.
Normand AC, Cassagne C, Ranque S, L’ollivier C, Fourquet P, Roesems S, Hendrickx M, Piarroux R. Assessment of various parameters to improve MALDI-TOF MS reference spectra libraries constructed for the routine identification of filamentous fungi. BMC Microbiol. 2013;13:76.
Becker PT, de Bel A, Martiny D, Ranque S, Piarroux R, Cassagne C, Detandt M, Hendrickx M. Identification of filamentous fungi isolates by MALDI-TOF mass spectrometry: clinical evaluation of an extended reference spectra library. Med Mycol. 2014;52(8):826–34.
Triest D, Stubbe D, De Cremer K, Piérard D, Normand AC, Piarroux R, Detandt M, Hendrickx M. Use of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of molds of the Fusarium genus. J Clin Microbiol. 2015;53(2):465–76.
Iriart X, Lavergne RA, Fillaux J, Valentin A, Magnaval JF, Berry A, Cassaing S. Routine identification of medical fungi by the new Vitek MS matrix-assisted laser desorption ionization-time of flight system with a new time-effective strategy. J Clin Microbiol. 2012;50(6):2107–10.
Oliveira MM, Santos C, Sampaio P, Romeo O, Almeida-Paes R, Pais C, Lima N, Zancopé-Oliveira RM. Development and optimization of a new MALDI-TOF protocol for identification of the Sporothrix species complex. Res Microbiol. 2015;166(2):102–10.
Bille E, Dauphin B, Leto J, Bougnoux ME, Beretti JL, Lotz A, Suarez S, Meyer J, Join-Lambert O, Descamps P, Grall N, Mory F, Dubreuil L, Berche P, Nassif X, Ferroni A. MALDI-TOF MS Andromas strategy for the routine identification of bacteria, mycobacteria, yeasts, Aspergillus spp. and positive blood cultures. Clin Microbiol Infect. 2012;18(11):1117–25.
Dolatabadi S, Kolecka A, Versteeg M, de Hoog SG, Boekhout T. Differentiation of clinically relevant Mucorales Rhizopus microsporus and R. arrhizus by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS). J Med Microbiol. 2015;64(7):694–701.
Packeu A, De Bel A, l’Ollivier C, Ranque S, Detandt M, Hendrickx M. Fast and accurate identification of dermatophytes by matrix-assisted laser desorption ionization-time of flight mass spectrometry: validation in the clinical laboratory. J Clin Microbiol. 2014;52(9):3440–3.
L’Ollivier C, Cassagne C, Normand AC, Bouchara JP, Contet-Audonneau N, Hendrickx M, Fourquet P, Coulibaly O, Piarroux R, Ranque S. A MALDI-TOF MS procedure for clinical dermatophyte species identification in the routine laboratory. Med Mycol. 2013;51(7):713–20.
Theel ES, Hall L, Mandrekar J, Wengenack NL. Dermatophyte identification using matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2011;49(12):4067–71.
Mery A, Sendid B, François N, Cornu M, Poissy J, Guerardel Y, Poulain D. Application of mass spectrometry technology to early diagnosis of invasive fungal infections. J Clin Microbiol. 2016;54(11):2786–97.
•• de Almeida JN Jr, Sztajnbok J, da Silva AR Jr, Vieira VA, Galastri AL, Bissoli L, Litvinov N, Del Negro GM, Motta AL, Rossi F, Benard G. Rapid identification of moulds and arthroconidial yeasts from positive blood cultures by MALDI-TOF mass spectrometry. Med Mycol. 2016;54(8):885–9. Paper describes promising new application of MALDI-TOF MS directly on clinical specimens.
•• Triest D, Hendrickx M, Piérard D, Piarroux R, Fraselle S, De Cremer K. Proof-of-concept study of a new LC-ESI-MS/MS-basd assay to identify Aspergillus spp. in artificially mixed samples using species/genus-specific proteotypic peptides. Mycol Progress. 2017; doi:10.1007/s11557-017-1273-5. Paper describes promising new MS/MS approach for the detection of species specific peptides.
Marinach C, Alanio A, Palous M, Kwasek S, Fekkar A, Brossas JY, Brun S, Snounou G, Hennequin C, Sanglard D, Datry A, Golmard JL, Mazier D. MALDI-TOF MS-based drug susceptibility testing of pathogens: the example of Candida albicans and fluconazole. Proteomics. 2009;9(20):4627–31.
Vella A, De Carolis E, Vaccaro L, Posteraro P, Perlin DS, Kostrzewa M, Posteraro B, Sanguinetti M. Rapid antifungal susceptibility testing by matrix-assisted laser desorption ionization-time of flight mass spectrometry analysis. J Clin Microbiol. 2013;51(9):2964–9.
De Carolis E, Vella A, Florio AR, Posteraro P, Perlin DS, Sanguinetti M, Posteraro B. Use of matrix-assisted laser desorption ionization-time of flight mass spectrometry for caspofungin susceptibility testing of Candida and Aspergillus species. J Clin Microbiol. 2012;50(7):2479–83.
•• Ceyssens PJ, Soetaert K, Timke M, Van den Bossche A, Sparbier K, De Cremer K, Kostrzewa M, Hendrickx M, Mathys V. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for combined species identification and drug sensitivity testing in mycobacteria. J Clin Microbiol. 2017;55(2):624–34. In this paper the simultaneous identification and antifungal susceptibility testing of Mycobacteria by MALDI-TOF MS is described. This is also promising for fungi.
Gautier M, Ranque S, Normand AC, Becker P, Packeu A, Cassagne C, L’Ollivier C, Hendrickx M, Piarroux R. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry: revolutionizing clinical laboratory diagnosis of mould infections. Clin Microbiol Infect. 2014;20(12):1366–71.
Dhieb C, Normand AC, Al-Yasiri M, Chaker E, El Euch D, Vranckx K, Hendrickx M, Sadfi N, Piarroux R, Ranque S. MALDI-TOF typing highlights geographical and fluconazole resistance clusters in Candida glabrata. Med Mycol. 2015;53(5):462–9.
Dhieb C, Normand AC, L’Ollivier C, Gautier M, Vranckx K, El Euch D, Chaker E, Hendrickx M, Dalle F, Sadfi N, Piarroux R, Ranque S. Comparison of MALDI-TOF mass spectra with microsatellite length polymorphisms in Candida albicans. J Mass Spectrom. 2015;50(2):371–7.
Girard V, Mailler S, Chetry M, Vidal C, Durand G, van Belkum A, Colombo AL, Hagen F, Meis JF, Chowdhary A. Identification and typing of the emerging pathogen Candida auris by matrix-assisted laser desorption ionisation time of flight mass spectrometry. Mycoses. 2016;59(8):535–8.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The author declares no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Advances in Diagnosis of Invasive Fungal Infections
Rights and permissions
About this article
Cite this article
Hendrickx, M. MALDI-TOF MS and Filamentous Fungal Identification: A Success Story?. Curr Fungal Infect Rep 11, 60–65 (2017). https://doi.org/10.1007/s12281-017-0277-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12281-017-0277-6