Abstract
Seed germination is regulated by several environmental factors, such as moisture, oxygen, temperature, light, and nutrients. Light is a critical regulator of seed germination in small-seeded plants, including Arabidopsis and lettuce. Phytochromes, a class of photoreceptors, play a major role in perceiving light to induce seed germination. Classical physiological studies have long suggested the involvement of gibberellin (GA) and abscisic acid (ABA) in the phytochrome-mediated germination response. Recent studies have demonstrated that phytochromes modulate endogenous levels of GA and ABA, as well as GA responsiveness. Several key components that link the perception of light and the modulation of hormone levels and responsiveness have been identified. Complex regulatory loops between light, GA and ABA signaling pathways have been uncovered.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Upon the completion of embryogenesis, the embryo within a seed stops growth and becomes dormant. The dormant seeds do not germinate and can survive under changing temperature and moisture conditions for a long time. Dormancy breaks down after a period of storage and seeds germinate under a certain condition. Seeds are equipped with sophisticated sensors and machineries to monitor and determine whether the surrounding environment is suitable for germination and subsequent plant growth. For small-seeded plants including Arabidopsis, lettuce, tomato and tobacco, light is an important regulator of seed germination. If germinated deep underground, the seedlings with small nutrient reserves cannot reach the surface of soil due to lack of energy. Thus, light sensing by the seed is thought to play a role in detecting its position. Phytochromes are the major class of photoreceptors responsible for this process. Light signals received by phytochromes are converted to internal cues, which in turn regulate physiological processes in seeds. Gibberellin (GA) and abscisic acid (ABA) are the internal signals that play central roles in the regulation of seed germination; GA induces, whereas ABA inhibits, seed germination. Recent studies have begun to reveal a tight interaction between light, GA and ABA signaling pathways in seeds at the molecular level.
In this review, we first describe a brief overview of each pathway, and then discuss how light and hormone signals interact to control seed germination. Here, we mainly focus on Arabidopsis, but also briefly discuss distinct as well as common mechanisms in lettuce and Arabidopsis during light-induced seed germination.
Phytochromes
Plants have at least four different types of photoreceptors; the red (R)/far-red (FR) light photoreceptor phytochromes, the UV-A/blue light photoreceptor cryptochromes, the blue light receptor phototropins, and the newly discovered blue light photoreceptors zeitlupes (Bae and Choi 2008). Phytochromes were originally discovered based on the photo-reversible germination of lettuce seeds. Irradiation of dark-imbibed seeds with a R light pulse induces germination and a subsequent FR light pulse cancels the effect of R light (Borthwick et al. 1952). Two forms of phytochromes, Pr and Pfr (the R light and FR light absorbing forms, respectively) exist in plants. Pr is an inactive form and is converted to active Pfr by R light. Pfr is reversibly converted to Pr by FR light (Fig. 1). This is the typical photo-reversible regulation of light response by phytochromes. The conformational change is accompanied by the reversible targeting of Pfr into the nucleus, where it binds to interacting proteins and regulates various physiological processes including seed germination.
a Modes of phytochrome mediated seed germination in Arabidopsis. PhyB, the major phytochrome accumulated in seeds, mediates typical R/FR photoreversible response (LFR) to promote seed germination in the early phase of seed imbibition. After a prolonged imbibition in the dark, PhyA is accumulated in the seeds and promote seed germination in response to either very low fluence of wide range of wavelength from UV-A to FR (VLFR) or relatively longer irradiation of FR light (FR-HIR). Phytochromes are dimeric proteins typically consisting of two identical apoproteins. b Light treatments for PhyA- and PhyB-dependent germination assay in Arabidopsis seeds. For PhyB-dependent germination assay, R and FR light pulses (10−7~10−4 mol m−2) are irradiated shortly after imbibition (~1 h) to reversibly control germination. Germination (radicle emergence) is normally observed within 24 h after PhyB activation. For PhyA-dependent germination assay, a FR light pulse is first irradiated to inactivate PhyB at the beginning of imbibition. After a long period of dark incubation (~48 h), FR light (~4 h, >10−2 mol m−2) is irradiated to activate PhyA. Seed germination can be also observed within 24 h of PhyA activation
Phytochromes are encoded by a multigene family in most plant species. In Arabidopsis, there are five PHY genes encoding phytochrome apoproteins (PHYA to PHYE) (Mathews 2006). PhyB (holoprotein), mediating R/FR photoreversible low fluence responses (LFR), promotes seed germination in response to low R light during initial stage (up to several hours) of seed imbibitions, whereas PhyA, mediating photo-irreversible very low fluence response (VLFR) and FR high irradiance response (FR-HIR), promotes seed germination in response to very low fluence of relatively wide spectra of light and continuous FR light after longer imbibition period in the absence of active PhyB in the dark (Shinomura et al. 1994, 1997; Fig. 1). In addition, PhyE has been shown to mediate LFR and FR-HIR once seeds are imbibed. The typical R/FR photo-reversible germination in lettuce corresponds to LFR.
GA pathways
Endogenous hormone levels are determined by the rates of their formation and conversion into inactive forms. Here, we refer to biosynthesis as the production of bioactive forms of a hormone, deactivation as conversion of bioactive forms (and their precursors) to the inactive (or less active) forms, and metabolism as both biosynthesis and deactivation.
GAs are a group of diterpenoids that are biosynthesized from geranylgeranyl diphosphate (GGDP) (Fig. 2). Among more than 100 GAs identified from plants, only a small number of them, such as GA1 and GA4, act as bioactive hormones that control seed germination, leaf expansion, stem elongation and flowering (Yamaguchi 2008). The late steps in the GA biosynthetic pathway are catalyzed by GA 20-oxidase (GA20ox) and GA 3-oxidase (GA3ox), both of which are 2-oxoglutarate-dependent dioxygenases (2ODDs) (Fig. 2). GA20ox and GA3ox are each encoded by small multigene families (Fig. 2). Accumulating evidence indicates that these 2ODDs are the primary targets for the regulation of bioactive GA levels (Yamaguchi 2008). GAs are deactivated by multiple classes of enzymes. The best-characterized GA deactivation enzyme is GA 2-oxidase (GA2ox), which is also a class of 2ODDs (Fig. 2). Recently, studies have shown that GA 16, 17-epoxydase (GA16,17ox) (CYP714D1/ELONGATED UPPERMOST INTERNODE) from rice and GA methyltransferases (GAMT1 and GAMT2) from Arabidopsis also act as GA-deactivating enzymes (Yamaguchi 2008; Fig. 2).
The GA metabolic pathway. In germinating Arabidopsis seeds, GA4 is the major bioactive GA. Inactivated GAs are shown in grey letters. Enzymes, genes (locus) and AGI codes are shown. See Yamaguchi (2008) for details. 2ox, GA 2-oxidase; 3ox, GA 3-oxidase; 20ox, GA 20-oxidase; 16,17ox, GA 16α,17-epoxidase, GAMT, GA methyltransferase; GGDP, geranylgeranyl diphosphate; CPS, ent-copalyl diphosphate synthase; KS, ent-kaurene synthase; KO, ent-kaurene oxidase; KAO, ent-kaurenoic acid oxidase. *16,17ox and GAMT also use other GAs as substrates, which are not shown for simplification
Several components in the GA signaling pathways have been identified. DELLA proteins (named after an amino acid sequence in a conserved domain) are members of the GRAS protein family acting as negative regulators in the GA signaling pathway (Sun and Gubler 2004). A soluble GA receptor, GIBBERELLIN INSENSITIVE DWARF 1 (GID1) in rice and its Arabidopsis orthologs, AtGID1a, b and c, interact with DELLA proteins in the presence of bioactive GAs (Ueguchi-Tanaka et al. 2007). The GA-GID1-DELLA complex is recognized by an F-box protein (SLEEPY1 [SLY1] in Arabidopsis and GID2 in rice) (McGinnis et al. 2003; Sasaki et al. 2003) for ubiquitination of the DELLA protein, which is then degraded through the 26S proteasome pathway. This results in de-repression (activation) of the GA signaling pathway (Fig. 3; Ueguchi-Tanaka et al. 2007; Hirano et al. 2008; Schwechheimer 2008). Genetic evidence has proven the role of these three components in regulating seed germination in Arabidopsis. Loss-of-function mutations in DELLA genes lead to increased germination capacity (Lee et al. 2002; Cao et al. 2005), whereas mutants defective in the SLY1 F-box protein or the AtGID1 receptors are non-germinating (Steber et al. 1998; Griffiths et al. 2006; Iuchi et al. 2007; Willige et al. 2007).
Model for GA action through degradation of DELLA proteins. a In the absence of GA, DELLA proteins are stable and negatively regulate the GA response pathway. b Once GA binds the GA receptor GID1, the GID1-GA complex interacts with DELLA protein. The GA-GID1-DELLA complex is recognized by an F-box protein (SLY1 in Arabidopsis and GID2 in rice) for ubiquitination (Ub) in the SCF complex (containing Skp1, Cullin and Rbx1). The ubiquitinated DELLA protein is degraded through the 26S proteasome, which results in de-repression of the GA response pathway
ABA pathways
ABA is a sesquiterpenoid derived from carotenoids and is involved in the induction of stress-responsive genes, stomatal closure, seed development, dormancy and germination (Nambara and Marion-Poll 2005). The ABA metabolic pathway is outlined in Fig. 4. Mutants defective in an earlier step of carotenoid synthesis exhibit an albino phenotype in addition to ABA deficiency. In contrast, mutants defective in zeaxanthin epoxidase (ZEP) show typical phenotypes due to ABA deficiency. There is evidence that the step catalyzed by 9-cis epoxycarotenoid dioxygenase (NCED) is limiting the level of ABA in the biosynthesis pathway (Nambara and Marion-Poll 2005). It has recently been reported that ABA is also produced via hydrolysis of ABA glucose ester by ABA glucosiodase, AtBG1, in Arabidopsis (Lee et al. 2006; Fig. 4). A major route for ABA deactivation is hydroxylation of the C-8′ position to produce 8′-hydroxy ABA by ABA 8′-hydroxylase, a class of P450 s designated as CYP707As. 8′-hydroxy ABA is subsequently converted to phaseic acid and dihydrophaseic acid (Nambara and Marion-Poll 2005; Fig. 4).
The ABA metabolic pathway. Enzymes, genes and AGI codes are shown. See Nambara and Marion-Poll (2005); Lee et al. (2006) and North et al. (2007) for details. ABA8ox, ABA 8′-hydroxylase; ABAG, ABA glucosidase; ABA-GE, ABA glucose ester; ABAO, abscisic aldehyde oxidase; DPA, dihydro phaseic acid; MOSU, molybdenum cofactor sulfurase; NCED, nine-cis-epoxycarotenoid dioxigenase; NSY, neoxanthin synthase; PA, phaseic acid; XO, xanthoxin oxidase; ZEP, zeaxanthin epoxidase
Three possible ABA receptors, the RNA binding protein FCA, the Mg-chelatase H subunit CHLH/ABAR/GUN5 and the G protein-coupled receptor GCR2, have been reported to date (Wang and Zhang 2008). Among these, CHLH/ABAR/GUN5 appears to function in seeds because a loss-of-function mutation in this gene results in ABA-insensitive germination (Shen et al. 2006). ABA signaling has been extensively reviewed elsewhere (Finkelstein et al. 2002, 2008; Hirayama and Shinozaki 2007) and will not be discussed here in detail. Future work will need to elucidate how ABA signaling components are tied to regulate germination.
Regulation of GA metabolism by light
Light promotes germination of small-seeded plants, such as Arabidopsis and lettuce via phytochromes (Shinomura 1997). R right that induces germination can be substituted by an application of bioactive GA to lettuce seeds (Kahn et al. 1957). In addition, R light cannot induce seed germination of severe GA-deficient mutants, such as ga1-3, in Arabidopsis (Oh et al. 2006). These results suggest that phytochromes promote germination by regulating GA levels in seeds. Light-regulation of GA metabolism has been studied in detail under PhyB-dependent or related germination conditions in Arabidopsis and lettuce seeds, where endogenous levels of bioactive GAs increase after irradiation with a R-light pulse and the increase is suppressed by a FR-light pulse given immediately after the R-light pulse (Toyomasu et al. 1993; Oh et al. 2006; Seo et al. 2006; Fig. 5). Consistent with the change in GA levels, expression of GA biosynthetic genes encoding GA3ox (AtGA3ox1 and AtGA3ox2 in Arabidopsis and LsGA3ox1 in lettuce) is induced by R light and the induction is canceled by FR light (Toyomasu et al. 1998; Yamaguchi et al. 1998; Fig. 5). In contrast, transcript levels of a GA deactivating gene GA2ox (AtGA2ox2 in Arabidopsis and LsGA2ox2 in lettuce) are decreased by R light treatment (Nakaminami et al. 2003; Oh et al. 2006; Seo et al. 2006; Yamauchi et al. 2007; Fig. 5). Light-regulation of GA3ox and GA2ox genes are also observed during PhyA-dependent germination (Oh et al. 2006). These data demonstrate that bioactive GA levels are regulated by phytochromes through reciprocal regulation of GA3ox and GA2ox genes.
Regulation GA and ABA metabolism during phytochrome-dependent germination in Arabidopsis. a Light treatments for PhyB-dependent germination. b Germination profiles. c Expression profiles of GA biosynthetic genes (AtGA3ox1 and AtGA3ox2) and a GA deactivating gene (AtGA2ox2). d Endogenous levels of bioactive GAs. e Expression profile of an ABA biosynthetic gene (AtNCED6) and an ABA deactivating gene (CYP707A2). f Endogenous ABA levels. DS, dry seeds. [Modified from Seo et al. (2006).]
Regulation of ABA metabolism by light
ABA treatment inhibits germination of lettuce seeds induced by R light or by exogenous application of bioactive GAs (Khan 1968; Sankhla and Sankhla 1968). More recently, ABA-deficient mutant seeds have been shown to partially germinate even in the absence of inductive light in Arabidopsis (Seo et al. 2006). Thus, endogenous ABA levels might also be controlled by light. In fact, R light decreases ABA levels through LFR in Arabidopsis and lettuce seeds (Toyomasu et al. 1994; Seo et al. 2006; Oh et al. 2007; Fig. 5). In Arabidopsis seeds, ABA levels decrease at the initial phase of imbibition regardless of light treatment. However, the rates of decrease at the later phase of imbibition are modulated by light; FR light-treated seeds contain higher levels of ABA than do FR/R light-treated seeds (Seo et al. 2006; Fig. 5). Consistent with the change in ABA levels, expression levels of ABA biosynthetic genes encoding ZEP (AtZEP/AtABA1 in Arabidopsis) and NCED (AtNCED6 and AtNCED9 in Arabidopsis and LsNCED2 and LsNCED4 in lettuce) are decreased by R light treatment (Seo et al. 2006; Oh et al. 2007; Sawada et al. 2008a; Fig. 5). In contrast, transcript levels of ABA deactivating genes encoding CYP707A (CYP707A2 in Arabidopsis and LsABA8ox4 in lettuce) are elevated by R light treatment (Seo et al. 2006; Oh et al. 2007; Sawada et al. 2008a; Fig. 5). In lettuce, a decrease in ABA levels is accompanied by an increase in phaseic acid levels upon R light treatment (Sawada et al. 2008a). These results indicate that ABA concentrations in seeds are regulated by phytochrome in a manner opposite to GA concentrations.
Interaction between GA and ABA metabolism
Unlike wild type, ABA-deficient mutants including aba2, aao3 and nced6 partially germinate even in the absence of R light treatment (Seo et al. 2006). Likewise, treatment with fluridone (an ABA biosynthesis inhibitor) increases the germination frequency in the absence of R-light. The aba1 mutation allows ga1 mutant seeds to germinate under continuous white light, suggesting that GA biosynthesis is not required for germination in an ABA-deficient mutant background (Koornneef et al. 1982). In comparison, the germination of aba2 seeds under the non-inductive light condition is inhibited by a GA biosynthesis inhibitor paclobutrazol, suggesting that de novo GA biosynthesis causes the germination (Seo et al. 2006). In fact, the levels of GA4 and expression of GA biosynthetic genes (AtGA3ox1 and AtGA3ox2) are elevated in the aba2 mutant compared to those in wild type. Furthermore, the PhyB-mediated induction of these GA biosynthetic genes is partly suppressed in an ABA over-accumulating cyp707a2 mutant (Seo et al. 2006). Altogether, these data show that bioactive GA levels are negatively regulated by ABA during seed germination (Seo et al. 2006).
There is also evidence that GA negatively regulates endogenous ABA concentrations. In Arabidopsis, higher ABA levels, activation of ABA biosynthetic genes and suppression of an ABA deactivating gene are observed in the GA deficient mutant ga1 in comparison with wild type (Oh et al. 2007). Moreover, an application of bioactive GA to ga1 seeds under non-inductive light condition decreases and increases expression levels of ABA biosynthetic and ABA deactivating genes, respectively (Oh et al. 2007). Similarly, treatment of lettuce seeds with bioactive GA decreases ABA content and LsNCED4 transcript levels in the absence of Pfr (Toyomasu et al. 1994; Sawada et al. 2008a). Zentella et al. (2007) reported that XERICO, one of the early GA- and DELLA-responsive genes, regulated ABA levels in seeds. XERICO encodes a protein containing a RING (really interesting new gene)-H2 type zinc finger motif. Overexpression of this gene causes accumulation of ABA and enhanced drought tolerance in vegetative tissues presumably through an increased ABA biosynthesis (Ko et al. 2006). Chromatin immunoprecipitation (ChIP) analysis indicates that DELLA proteins repress the expression of XERICO by directly associating with its promoter (Zentella et al. 2007). These results indicate that the repression of XERICO by DELLA proteins is an important mechanism by which GA regulates ABA metabolism. The mutual negative regulation between GA and ABA might contribute to an efficient change in the balance of these antagonistic hormones in response to an external cue (Fig. 6).
Interaction of ABA and GA metabolism. a Pfr formed by R light increases GA levels and decreases ABA levels. R-light induced GA accumulation also contributes to reducing ABA levels. b In the absence of Pfr, ABA levels are higher because of the loss of negative regulation by Pfr. Increased ABA levels are required to reduce GA levels. Arrows indicate positive regulation and T-bars indicate negative regulation. Grey elements denote inactive regulation or low abundance in each condition
Factors that link light signal to GA and ABA signals
PIL5
Phytochromes modulate GA and ABA metabolism through one of their interacting proteins, PIL5 (PIF3–LIKE 5). PIL5 is a phytochrome-interacting basic helix-loop-helix (bHLH) protein that belongs to the Arabidopsis bHLH subfamily 15 together with PIF3 (PHYTOCHROME INTERACTING FACTOR 3), PIF4, PIL6, and 10 other bHLH proteins (Toledo-Ortiz et al. 2003; Yamashino et al. 2003). PIL5 preferentially interacts with the Pfr form of PhyA and PhyB, and the interaction induces the degradation of PIL5 protein through the 26S proteasome (Oh et al. 2004, 2006). Loss-of-function pil5 mutant seeds exhibit constitutive germination even under non-inductive light condition, whereas the PIL5 overexpressor requires the higher fluence of light for seed germination. Taken together, the results indicate that phytochromes promote seed germination by activating the degradation of PIL5 (Oh et al. 2004).
Gene expression analysis and quantification of hormone levels indicate that PIL5 regulates both GA and ABA levels by modulating expression of their metabolic genes (Fig. 7). Expression of two GA biosynthetic genes (AtGA3ox1 and AtGA3ox2) is activated, whereas a GA deactivating gene (AtGA2ox2) is suppressed, in the pil5 mutant (Penfield et al. 2005; Oh et al. 2006). Consistent with gene expression patterns, endogenous GA4 levels are increased in the pil5 mutant compared to wild type (Oh et al. 2006; Fig. 7). ABA biosynthetic genes (AtABA1, AtNCED6 and AtNCED9) are highly expressed in FR light-irradiated seeds but repressed in the R light-irradiated seeds. In contrast, an ABA deactivating gene (CYP707A2) is upregulated by R light irradiation. These regulations are also observed in the ga1 mutant background, suggesting that the changes in gene expression are not secondary effects accompanied by germination. In contrast to wild-type and ga1 mutant seeds, ABA biosynthetic genes are constitutively repressed in the pil5 ga1 double mutant irrespective of light condition, while an ABA deactivating gene is expressed higher. Consistent with gene expression patterns, endogenous ABA levels are constitutively low in the pil5 mutant background irrespective of light conditions. Taken together, the results indicate that PIL5 reciprocally regulates GA and ABA levels by regulating the expression of GA and ABA metabolic genes (Oh et al. 2007; Fig. 7).
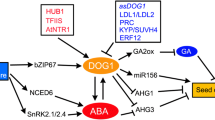
Regulation of GA and ABA signaling by PIL5 and SOM. PIL5 is a negative regulator of seed germination. PIL5 protein is degraded upon interaction with active phytochrome. PIL5 acts as a transcriptional activator that binds directly to the promoter of SOM, GAI and RGA. SOM negatively regulates seed germination in part through regulation of GA and ABA levels. GAI and RGA are Arabidopsis DELLA proteins that act as negative regulators in the GA response pathway. GAI and RGA proteins are further degraded by GA activity (see Fig. 3). Arrows indicate positive regulation and T-bars indicate negative regulation. Active elements and regulation in response to inductive light treatment are shown in red
Light also enhances GA responsiveness in imbibed Arabidopsis seeds via phytochromes (Hilhorst and Karssen 1988; Derkx and Karssen 1993; Yang et al. 1995; Oh et al. 2007). Germination frequency of a severe GA-deficient ga1 mutant is increased by inductive light treatment in response to exogenous GA. However, the pil5 ga1 double mutant constitutively shows high GA responsiveness and R light does not further increase the GA responsiveness, suggesting that PIL5 regulates the GA responsiveness as well as GA metabolism (Oh et al. 2007). Arabidopsis has five genes encoding DELLA proteins (RGA, GAI, RGL1, RGL2 and RGL3), negative regulators in the GA response pathway (Fig. 3). The increase in GA responsiveness by light is likely due in part to the transcriptional repression of two DELLA genes (RGA and GA1). Transcript levels of RGA and GAI decrease when PhyA or PhyB is activated even in the ga1 mutant background. In the pil5 mutant seeds, their expression levels are constitutively very low. The double-loss-of-function mutant of GAI and RGA (gai-t6 rga-28) is hypersensitive to R light for seed germination, supporting the functional significance of the two DELLA genes in repressing seed germination. In contrast to Arabidopsis, however, GA signaling is not affected by light in lettuce seeds, and expression of genes encoding DELLA proteins is not significantly affected by light (Sawada et al. 2008b).
The double loss-of-function gai-t6 rga-28 mutant, though hypersensitive to light, still requires light for seed germination, whereas the triple loss-of-function mutant of GAI, RGA, and RGL2 germinates irrespective of light condition, indicating that light must inactivate RGL2 to promote seed germination (Tyler et al. 2004; Cao et al. 2005). Since GA activates the degradation of DELLA proteins, the increased bioactive GA level by light is likely to decrease the amount of RGL2 protein in light-treated seeds. Therefore, the results indicate that phytochrome and PIL5-mediated light signaling activates GA signaling both by repressing transcription of two DELLA genes and by increasing bioactive GA levels in imbibed seeds (Fig. 7).
ChIP analysis has shown that PIL5 binds to promoters of RGA and GAI genes, but does not bind to promoters of any GA and ABA metabolic genes, indicating that PIL5 directly regulates GA signaling genes but indirectly regulates metabolic genes. However, GAI and RGA are not the major PIL5 direct target genes mediating the regulation of GA and ABA metabolic genes. Expression of all GA and ABA metabolic genes tested, except for AtGA3ox1, is similarly regulated between wild type and the gai-t6 rga-28 double loss-of-function mutant seeds in response to light (Oh et al. 2007). The expression of AtGA3ox1 is still induced by light in the double mutant, but the expression level is relatively higher in the double mutant compared to wild type. Taken together, the results suggest that the regulation of GA and ABA metabolic genes by PIL5 is largely independent of GAI and RGA (Oh et al. 2007).
SOM
The indirect regulation of GA and ABA metabolic genes by PIL5 independently of GAI and RGA suggests that other PIL5 direct target genes regulate the expression of the metabolic genes. Recent analysis of somnus (som) mutant suggests that SOM regulates GA and ABA metabolic genes downstream of PIL5. The som mutant was isolated based on its ability to germinate under complete darkness (Kim et al. 2008). The SOM gene encodes a CCCH-type zinc finger protein that probably functions as an RNA binding protein. The som mutant contains higher levels of GA4 and lower levels of ABA, with consistent changes in expression levels of GA and ABA metabolic genes. ChIP analysis indicates that PIL5 directly binds to the promoter of SOM and activates the expression of it. Taken together, these data suggest that PIL5 regulates GA and ABA metabolic genes partly through SOM (Kim et al. 2008; Fig. 7). How SOM regulates these metabolic genes needs to be further investigated. Unlike PIL5, SOM does not regulate the expression of DELLA genes, and GA responsiveness appears to be normal in som mutant seeds (Kim et al. 2008; Fig. 7).
IMB1
In addition to PIL5 and SOM, IMBIBITION-INDUCIBLE1 (IMB1) is implicated to play a role in linking PhyA signaling to ABA signaling. IMB1 is a member of the bromodomain protein family, which probably function as transcription factors. IMB1 expression levels are very low in dry seeds and elevated upon imbibition (Duque and Chua 2003). A loss-of-function imb1 mutant is defective in PhyA-dependent seed germination and hypersensitive to exogenous ABA in terms of inhibition of cotyledon greening during seed germination. The mutant phenotypes can be partially explained by the higher accumulation of ABI5 transcripts in response to exogenous ABA during germination (Duque and Chua 2003). The exact molecular mechanism by which IMB1 mediates the PhyA signaling during seed germination needs to be further investigated.
Conclusions and future perspectives
It has long been suggested that the balance of GA and ABA levels determines the germination capacity. This theory is supported by the fact that ABA-deficient mutants have been found as suppressors of non-germinating GA-deficient mutants (Koornneef et al. 1982; Bentsink and Koornneef 2002). This means that the amount of GA required for seed germination is decreased when endogenous ABA levels are reduced. Thus, the germination capacity is not determined by the levels of one hormone alone, but by the balance of the two antagonistic hormones. This hormone balance theory predicts that it should be an efficient way for seeds to regulate both GA and ABA levels in response to a signal in making a decision to germinate or not. Recent studies have proven that this is actually the case for light-regulation of seed germination in Arabidopsis, as discussed here. Light regulates concentrations of two antagonistic hormones, GA and ABA, and also GA responsiveness. Light signals are transmitted to GA and ABA-related genes via a phytochrome-interacting protein, PIL5, which functions as a transcription factor that directly or indirectly regulates the hormone-related genes. Recent papers have reported that the phytochrome-interacting proteins PIF3 and PIF4 physically interact with DELLA proteins in regulating gene expression during seedling growth (de Lucas et al. 2008; Feng et al. 2008). It has yet to be determined whether PIL5 also regulates seed germination through a similar mechanism, besides regulating transcription of DELLA genes. SOM is a direct target of PIL5 and regulates GA and ABA metabolic genes downstream of PIL5 via an as yet unknown mechanism. Elucidation of the molecular function of SOM is required to draw a whole picture of how light signals modulate hormone levels in seeds. There is evidence that the levels of GA and ABA in seeds are negatively regulated by each other. This regulatory loop would facilitate an efficient change in the ratio of the two antagonistic hormones in response to a signal. How one hormone regulates the levels of the other in this regulatory circuit requires further studies.
In the field condition, there are multiple environmental factors that change time to time and affect seed germination. Thus, plants must integrate different external cues to make a decision to germinate or not. Temperature is another critical factor that affects seed germination. High temperature inhibits germination (thermoinhibition), while exposure of imbibed seeds to cold temperature (stratification) stimulates germination in many plant species. Recent studies have shown that temperature modulates GA (and ABA) levels during thermoinhibition and stratification of Arabidopsis seeds through regulation of GA and ABA metabolic genes (Yamauchi et al. 2004; Toh et al. 2008). A PIL5-related bHLH protein, SPATULA (SPT), regulates GA biosynthetic genes in response to stratification (Penfield et al. 2005). Our improved understanding of light-induced seed germination and the identification of regulators of GA and ABA metabolic/signaling genes have started to allow us to study in further detail the mechanism by which multiple external cues are integrated to hormone metabolic/signaling pathways in seeds.
References
Bae G, Choi G (2008) Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol 59:281–311. doi:10.1146/annurev.arplant.59.032607.092859
Bentsink L, Koornneef M (2002) Seed dormancy and germination: April 4, 2002. The Arabidopsis book. American Society of Plant Biologists, Rockville, MD. doi: 10.1199/tab.0050, http://www.aspb.org/publications/arabidopsis/
Borthwick HA, Hendricks SB, Parker MW, Toole EH, Toole VK (1952) A reversible photoreaction controlling seed germination. Proc Natl Acad Sci USA 38:662–666. doi:10.1073/pnas.38.8.662
Cao D, Hussain A, Cheng H, Peng J (2005) Loss of function of four DELLA genes leads to light- and gibberellin-independent seed germination in Arabidopsis. Planta 21:1–9
de Lucas M, Daviere JM, Rodriguez-Falcon M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blazquez MA, Titarenko E, Prat S (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451:480–484. doi:10.1038/nature06520
Derkx MPM, Karssen CM (1993) Effects of light and temperature on seed dormancy and gibberellin-stimulated germinatin in Arabidopsis thaliana: studies with gibberellin-deficient and -insensitive mutants. Physiol Plant 89:360–368. doi:10.1111/j.1399-3054.1993.tb00167.x
Duque P, Chua NH (2003) IMB1, a bromodomain protein induced during seed imbibition, regulates ABA- and phyA-mediated responses of germination in Arabidopsis. Plant J 35:787–799. doi:10.1046/j.1365-313X.2003.01848.x
Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, Schafer E, Fu X, Fan LM, Deng XW (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451:475–479. doi:10.1038/nature06448
Finkelstein RR, Gampala SSL, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell Suppl:S15–S45
Finkelstein R, Reeves W, Ariizumi T, Steber C (2008) Molecular aspects of seed dormancy. Annu Rev Plant Biol 59:387–415. doi:10.1146/annurev.arplant.59.032607.092740
Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, Gong F, Phillips AL, Hedden P, Sun TP, Thomas SG (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18:3399–3414. doi:10.1105/tpc.106.047415
Hilhorst HWM, Karssen CM (1988) Dual effect of light on the gibberellin-stimulated and nitrate-stimulated seed-germination of Sisymbrium officinale and Arabidopsis thaliana. Plant Physiol 86:591–597
Hirano K, Ueguchi-Tanaka M, Matsuoka M (2008) GID1-mediated gibberellin signaling in plants. Trends Plant Sci 13:192–199. doi:10.1016/j.tplants.2008.02.005
Hirayama T, Shinozaki K (2007) Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci 12:343–351. doi:10.1016/j.tplants.2007.06.013
Iuchi S, Suzuki H, Kim YC, Iuchi A, Kuromori T, Ueguchi-Tanaka M, Asami T, Yamaguchi I, Matsuoka M, Kobayashi M, Nakajima M (2007) Multiple loss-of-function of Arabidopsis gibberellin receptor AtGID1s completely shuts down a gibberellin signal. Plant J 50:958–966. doi:10.1111/j.1365-313X.2007.03098.x
Kahn A, Goss JA, Smith DE (1957) Effect of gibberellin on germination of lettuce seed. Science 125:645–646. doi:10.1126/science.125.3249.645
Khan AA (1968) Inhibition of gibberelic acid-induced germination by abscisic acid and reversal by cytokinins. Plant Physiol 43:1463–1465
Kim DH, Yamaguchi S, Lim S, Oh E, Park J, Hanada A, Kamiya Y, Choi G (2008) SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell 20:1260–1277. doi:10.1105/tpc.108.058859
Ko JH, Yang SH, Han KH (2006) Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J 47:343–355. doi:10.1111/j.1365-313X.2006.02782.x
Koornneef M, Jorna ML, Brinkhorst-van der Swan DLC, Karssen CM (1982) The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) Heynh. Theor Appl Genet 61:385–393
Lee S, Cheng H, King KE, Wang W, He Y, Hussain A, Lo J, Harberd NP, Peng J (2002) Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev 16:646–658. doi:10.1101/gad.969002
Lee KH, Piao HL, Kim HY, Choi SM, Jiang F, Hartung W, Hwang I, Kwak JM, Lee IJ, Hwang I (2006) Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126:1109–1120. doi:10.1016/j.cell.2006.07.034
Mathews S (2006) Phytochrome-mediated development in land plants: red light sensing evolves to meet the challenges of changing light environments. Mol Ecol 15:3483–3503. doi:10.1111/j.1365-294X.2006.03051.x
McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun TP, Steber CM (2003) The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15:1120–1130. doi:10.1105/tpc.010827
Nakaminami K, Sawada Y, Suzuki M, Kenmoku H, Kawaide H, Mitsuhashi W, Sassa T, Inoue Y, Kamiya Y, Toyomasu T (2003) Deactivation of gibberellin by 2-oxidation during germination of photoblastic lettuce seeds. Biosci Biotechnol Biochem 67:1551–1558. doi:10.1271/bbb.67.1551
Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56:165–185. doi:10.1146/annurev.arplant.56.032604.144046
North HM, De Almeida A, Boutin JP, Frey A, To A, Botran L, Sotta B, Marion-Poll A (2007) The Arabidopsis ABA-deficient mutant aba4 demonstrates that the major route for stress-induced ABA accumulation is via neoxanthin isomers. Plant J 50:810–824. doi:10.1111/j.1365-313X.2007.03094.x
Oh E, Kim J, Park E, Kim JI, Kang C, Choi G (2004) PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16:3045–3058. doi:10.1105/tpc.104.025163
Oh E, Yamaguchi S, Kamiya Y, Bae G, Chung WI, Choi G (2006) Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J 47:124–139. doi:10.1111/j.1365-313X.2006.02773.x
Oh E, Yamaguchi S, Hu JH, Yusuke J, Jung B, Paik I, Lee HS, Sun TP, Kamiya Y, Choi G (2007) PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19:1192–1208. doi:10.1105/tpc.107.050153
Penfield S, Josse EM, Kannangara R, Gilday AD, Halliday KJ, Graham IA (2005) Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr Biol 15:1998–2006. doi:10.1016/j.cub.2005.11.010
Sankhla N, Sankhla D (1968) Reversal of (±)-abscisin II induced inhibition of lettuce seed germination and seedling growth by kinetin. Physiol Plant 21:190–195. doi:10.1111/j.1399-3054.1968.tb07243.x
Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Jeong DH, An G, Kitano H, Ashikari M, Matsuoka M (2003) Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299:1896–1898. doi:10.1126/science.1081077
Sawada Y, Aoki M, Nakaminami K, Mitsuhashi W, Tatematsu K, Kushiro T, Koshiba T, Kamiya Y, Inoue Y, Nambara E, Toyomasu T (2008a) Phytochrome- and gibberellin-mediated regulation of abscisic acid metabolism during germination of photoblastic lettuce seeds. Plant Physiol 146:1386–1396. doi:10.1104/pp.107.115162
Sawada Y, Katsumata T, Kitamura J, Kawaide H, Nakajima M, Asami T, Nakaminami K, Kurahashi T, Mitsuhashi W, Inoue Y, Toyomasu T (2008b) Germination of photoblastic lettuce seeds is regulated via the control of endogenous physiologically active gibberellin content, rather than of gibberellin responsiveness. J Exp Bot 59:3383–3393. doi:10.1093/jxb/ern192
Schwechheimer C (2008) Understanding gibberellic acid signaling—are we there yet? Curr Opin Plant Biol 11:9–15. doi:10.1016/j.pbi.2007.10.011
Seo M, Hanada A, Kuwahara A, Endo A, Okamoto M, Yamauchi Y, North H, Marion-Poll A, Sun TP, Koshiba T, Kamiya Y, Yamaguchi S, Nambara E (2006) Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J 48:354–366. doi:10.1111/j.1365-313X.2006.02881.x
Shen YY, Wang XF, Wu FQ, Du SY, Cao Z, Shang Y, Wang XL, Peng CC, Yu XC, Zhu SY, Fan RC, Xu YH, Zhang DP (2006) The MG-chelatase H subunit is an abscisic acid receptor. Nature 443:823–826. doi:10.1038/nature05176
Shinomura T (1997) Phytochrome regulation of seed germination. J Plant Res 110:151–161. doi:10.1007/BF02506854
Shinomura T, Nagatani A, Chory J, Furuya M (1994) The induction of seed germination in Arabidopsis thaliana is regulated principally by phytochrome B and secondarily by phytochrome A. Plant Physiol 104:363–371
Steber CM, Cooney SE, McCourt P (1998) Isolation of the GA-response mutant sly1 as a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics 149:509–521
Sun TP, Gubler F (2004) Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol 55:197–223. doi:10.1146/annurev.arplant.55.031903.141753
Toh S, Imamura A, Watanabe A, Nakabayashi K, Okamoto M, Jikumaru Y, Hanada A, Aso Y, Ishiyama K, Tamura N, Iuchi S, Kobayashi M, Yamaguchi S, Kamiya Y, Nambara E, Kawakami N (2008) High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiol 146:1368–1385. doi:10.1104/pp.107.113738
Toledo-Ortiz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15:1749–1770. doi:10.1105/tpc.013839
Toyomasu T, Tsuji H, Yamane H, Nakayama M, Yamaguchi I, Murofushi N, Takahashi N, Inoue Y (1993) Light effects on endogenous levels of gibberellins in photoblastic lettuce seeds. J Plant Growth Regul 12:85–90. doi:10.1007/BF00193238
Toyomasu T, Yamane H, Murofushi N, Inoue Y (1994) Effects of exogenously applied gibberellin and red light on the endogenous levels of abscisic acid in photoblastic lettuce seeds. Plant Cell Physiol 35:127–129
Toyomasu T, Kawaide H, Mitsuhashi W, Inoue Y, Kamiya Y (1998) Phytochrome regulates gibberellin biosynthesis during germination of photoblastic lettuce seeds. Plant Physiol 118:1517–1523. doi:10.1104/pp.118.4.1517
Tyler L, Thomas SG, Hu JH, Dill A, Alonso JM, Ecker JR, Sun TP (2004) DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol 135:1008–1019. doi:10.1104/pp.104.039578
Ueguchi-Tanaka M, Nakajima M, Motoyuki A, Matsuoka M (2007) Gibberellin receptor and its role in gibberellin signaling in plants. Annu Rev Plant Biol 58:183–198. doi:10.1146/annurev.arplant.58.032806.103830
Wang XF, Zhang DP (2008) Abscisic acid receptors: multiple signal-perception sites. Annal Bot 101:311–317. doi:10.1093/aob/mcm284
Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann EM, Maier A, Schwechheimer C (2007) The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19:1209–1220. doi:10.1105/tpc.107.051441
Yamaguchi S (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59:225–251. doi:10.1146/annurev.arplant.59.032607.092804
Yamaguchi S, Smith MW, Brown RG, Kamiya Y, Sun TP (1998) Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 10:2115–2126
Yamashino T, Matsushika A, Fujimori T, Sato S, Kato T, Tabata S, Mizuno T (2003) A link between circadian-controlled bHLH factors and the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol 44:619–629. doi:10.1093/pcp/pcg078
Yamauchi Y, Ogawa M, Kuwahara A, Hanada A, Kamiya Y, Yamaguchi S (2004) Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16:367–378. doi:10.1105/tpc.018143
Yamauchi Y, Takeda-Kamiya N, Hanada A, Ogawa M, Kuwahara A, Seo M, Kamiya Y, Yamaguchi S (2007) Contribution of gibberellin deactivation by AtGA2ox2 to the suppression of germination of dark-imbibed Arabidopsis thaliana seeds. Plant Cell Physiol 48:555–561. doi:10.1093/pcp/pcm023
Yang YY, Nagatani A, Zhao YJ, Kang BJ, Kendrick RE, Kamiya Y (1995) Effects of gibberellins on seed germination of phytochrome-deficient mutants of Arabidopsis thaliana. Plant Cell Physiol 36:1205–1211
Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, Fleet CM, Jikumaru Y, Nambara E, Kamiya Y, Sun TP (2007) Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19:3037–3057. doi:10.1105/tpc.107.054999
Acknowledgements
GC and SY are supported in part by Korea Science and Engineering Foundation (R0A-2007-000-20024-0) and the Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research (20570049), respectively.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Seo, M., Nambara, E., Choi, G. et al. Interaction of light and hormone signals in germinating seeds. Plant Mol Biol 69, 463–472 (2009). https://doi.org/10.1007/s11103-008-9429-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-008-9429-y