Abstract
Microbial resistance to available drugs is a growing health threat imposing the need for the development of new drugs. The scaffold of plant defensins, including their γ-cores, are particularly good candidates for drug design. This work aimed to improve the antifungal activity of a previous design peptide, named A36,42,44γ32–46VuDef (for short DD) against yeasts by altering its biochemical parameters. We explore the correlation of the biological activity and structure of plant defensins and compared their primary structures by superimposition with VuDef1 and DD which indicated us the favorable position and the amino acid to be changed. Three new peptides with modifications in charge, hydrophobicity (RR and WR) and chirality (D-RR) were designed and tested against pathogenic yeasts. Inhibition was determined by absorbance. Viability of mammalian cells was determined by MTT. The three designed peptides had better inhibitory activity against the yeasts with better potency and spectrum of yeast species inhibition, with low toxicity to mammalian cells. WR, the most hydrophobic and cationic, exhibited better antifungal activity and lower toxicity. Our study provides experimental evidence that targeted changes in the primary structure of peptides based on plant defensins γ-core primary structures prove to be a good tool for the synthesis of new compounds that may be useful as alternative antifungal drugs. The method described did not have the drawback of synthesis of several peptides, because alterations are guided. When compared to other methods, the design process described is efficient and viable to those with scarce resources.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microbial resistance to available drugs is a growing public health threat and fungal infections, such as candidiasis and aspergillosis, caused by resistant fungi are among the most worrying (Jensen 2016; Fisher et al. 2018). In addition, infections caused by multi-resistant Candida to commercial antifungals have been increasing in recent decades (Bongomin et al. 2017; Fisher et al. 2018). Especially those caused by C. auris that is an emerging yeast species, which has a phenotype of multiple resistance to the main classes of antifungals clinically available, and has caused serious invasive infections in hospitalized patients (Forsberg et al. 2019). In addition to the multi-resistance problem that severely limits treatment, C. auris also spreads very easily in hospital environment aggravating the problem (Forsberg et al. 2019). Therefore, the need for the research and development of new substances that act as an alternative to existing antifungals become evident, to overcome the problem of widespread resistance among infectious agents, to be new options for the treatment of fungal diseases in view of the scarcity of antifungals compared to antibacterial agents (Costa-de-Oliveira and Rodrigues 2020), and to combat new emerging pathogens such as C. auris. Among candidate substances are the antimicrobial peptides (AMPs).
All extant organisms, ranging from bacteria to mammals, encode and express AMPs. AMPs are peptides composed of l-amino acids in chains of up to 100 units, in a wide variety of primary structures, organized in amphiphilic positively charged three-dimensional structures (Koehbach and Craik 2019). The amphiphilicity property enables them to be soluble in aqueous environment and to interact with lipid membranes, explaining their antimicrobial activity (Brogden 2005; Ghosh et al. 2018; Ciumac et al. 2019). AMPs are known since the 40s’ (Balls and Hale 1940), but only at the beginning of the 80s’ had become clear their pivotal role in innate immunity of different organisms like cecropin from insects (Steiner et al. 1981), defensins of mammals (Selsted et al. 1985) and magainin in amphibians (Zasloff 1987). The antimicrobial activity of AMPs was demonstrated against different microorganisms such as viruses, bacteria, fungi, and protozoa, and extends beyond antimicrobial activity being also able to inhibit the growth of cancer cells (Deslouches and Di 2017). The recognition of the broad spectrum of antimicrobial activity along with their clear participation in innate immunity in metazoan boosted the interest on AMPs, which culminated in the discovery of many AMPs in different organisms (Koehbach and Craik 2019). Several of them with specific biological activities (Whittington and Belov 2007; Sunagar et al. 2013). Moreover, AMPs have some characteristics that classify them as possible therapeutic agents: (1) their broad antimicrobial activity which means that one AMP inhibits different microorganisms (Greco et al. 2019); (2) their small size and high stability to harsh environments (e.g. pH extremes, high temperature, and protease resistance) (Beer and Vivier 2008; Colgrave and Craik 2004); (3) amenability to peptide engineering by molecular biology or chemical synthesis (De Samblanx et al. 1997). This last technique has the advantage to allow the incorporation into primary structures of AMP of unusual amino acids, like non-proteinogenic or d-enantiomers (Ding et al. 2020; Greco et al. 2019), improving their activity, specificity and stability, decreasing their toxicity and protecting them from proteolytic degradation in the target organism (Li et al. 2016). Based on these characteristics, some authors claimed that AMPs could be interesting scaffolds for the development of antimicrobial substances for therapeutic use in humans to refrain the upsurge of microbial resistance (Zasloff 1987; Zhang et al. 2019). Thus, many AMPs from different sources are in clinical trials (Ghosh et al. 2018).
Plants produce several AMPs that have the potential to be used in the development of new antimicrobial therapeutics. Plant AMPs participate, as in other organisms, in their innate immunity, protecting them from viruses, bacteria and fungi attacks (Campos et al. 2018). One of the best characterized plant AMPs are the defensins which are known since the 90′s (Colilla et al. 1990; Méndez et al. 1990). Since then, the characterization of many peptides belonging to this family had given great knowledge about their biology. These molecules are synthesized as a protein precursor processed in the endoplasmic reticulum yielding mature peptides of 45–54 l-amino acid residues in their primary structure that are arranged in a tertiary structure composed of three anti-parallel β-strands and one α-helix (Carvalho and Gomes 2011). This tertiary structure is highly conserved among the family (Carvalho and Gomes 2011) with some known exceptions like VrD1 (from Vigna radiata, mung bean) that has a 310 type helix between the β1 strand and the α-helix (Liu et al. 2006), and Sd5 (from sugarcane) that has an unstructured C-terminal extension (De Paula et al. 2011). Amidst the amino acids that constitute the primary structure are eight strictly positional conserved cysteine residues that bind in specific pairs to form four disulfide bridges with C1–C8, C2–C5, C3–C6, and C4–C7 arrangement (Thomma et al. 2002; Shafee et al. 2017) with the exception of PhD1 and PhD2 (from Petunia hybrida, ornamental petunia), which have five disulfide bridges (Lay et al. 2003). These bridges turn the molecule globular and compact by holding tight together the secondary structural elements and are responsible for the high physicochemical stability of plant defensins, especially the C1–C8 bridge turn the molecule pseudocyclic (Carvalho and Gomes 2011). Two of the disulfide bridges, the C3–C6 and C4–C7 that bind the α-helix to the β3 strand, are part of the structural arrangement denominated cysteine stabilized αβ (CSαβ) motif (Thomma et al. 2002; Carvalho and Gomes 2011). Plant defensins were included in the superfamily of cis-defensins, because the disposition of those two disulfide bridges, i.e. between the α-helix and the β3 strand, that bind the same secondary structural elements and also because they are organized at the same side of their three-dimensional structures (Shafee et al. 2017). Another amino acid residue that is well conserved in the primary structure of plant defensins is a glycine at approximately position 32 [all numbers cited in the text refer to the position of the amino acid in the primary structure of the molecule, and considering the alignment made in the Supplementary Table 1 the corresponding position is 38 including the gaps (−)]. This glycine is part of another well conserved motif, the γ-core, which occurs in other AMPs as well Yount and Yeaman (2004). For plant defensins, the γ-core is in the dextromeric formula NH2–[X1-3]–[GXC]–[X3-9]–[C]–COOH, X being any amino acid (Yount and Yeaman 2004).
With the exception of those conserved residues (e.g. C and G) in the primary structure of plant defensins, other amino acids that compose it show high variation. The greatest variations between amino acid residues occur in the loops of the plant defensins, especially in the loop region between the β2 and β3 strands, which comprises the γ-core itself (Supplementary Table 1; Carvalho and Gomes 2011). This amino acid diversity in the loop regions drives variations in the three-dimensional position and length of the loops and additionally many amino acids that compose them are exposed to the molecule surface (Liu et al. 2006; Sagaram et al. 2013; Machado et al. 2018). For these reasons, they are free to interact with other proteins or targets and this may be the explanation for the broad biological activities already described for plant defensins. In fact, for plant defensins, their inhibitory activities on protein translation (Méndez et al. 1990, 1996) and α-amylases (Bloch and Richardson 1991; Lin et al. 2007; Pelegrini et al. 2008; Dos Santos et al. 2010), tolerance to heavy metal (Mirouze et al. 2006), blockage of ion channels (Spelbrink et al. 2004), antimicrobial inhibitory spectra (Carvalho and Gomes 2011), their capacity to bind to membrane lipids (phospholipids and sphingolipids) (Baxter et al. 2015; Gonçalves et al. 2012; De Paula et al. 2011; Poon et al. 2014), dimerization (Lay et al. 2012; Song et al. 2011) and antifungal inhibitory mechanisms (Coninck et al. 2013; Parisi et al. 2019) are well described. Also, it is startling that many of these abilities are linked to the amino acid stretch that compose the γ-core which is a desirable characteristic for drug design as explained in the follow paragraph (Supplementary Table 1).
Since the γ-core has a fundamental role in the antimicrobial activity of AMPs (Yount and Yeaman 2004) including plant defensins, it has become an attractive region for targeted modifications. Because the restriction of the biological activity to a minimal stretch of amino acids, such as the γ-core in plant defensins, is important for miniaturization of the biologically active sequence that is a desirable characteristic for drug development, which might be further manipulated to improve stability, decrease toxicity toward host and, moreover, lower the production costs (Ramesh et al. 2016). Foremost, plant defensins present another desirable characteristic for drug development as these peptides are, in general, not active against mammalian cells, at least at the concentrations that causes inhibition of microorganisms (Carvalho and Gomes 2011; Vriens et al. 2015). This safety to mammalian cells is reinforced by in vivo tests in murine models where Rs-AFP2 (defensin from Raphanus sativus, radish) was as active as fluconazole in reducing candidiasis caused by C. albicans (Tavares et al. 2008) and NoD173 (defensin from Nicotiana occidentalis, tobacco) that inhibited the growth of solid B16-F1 melanoma tumor (Lay et al. 2019). Both defensins presented no toxicity to the animal hosts. Taking the aforementioned data together, the scaffold of plant defensins, including their γ-cores, are particularly good candidates for drug design.
Our research group is focused on the VuDef1 (this peptide is now renamed from Vu-Def to VuDef1 following the nomenclature rule proposed for plant defensins by Sathoff et al. 2019), a plant defensin isolated from Vigna unguiculata (cowpea) seeds, that presented in combination with another AMP, a lipid transfer protein, inhibitory activities against filamentous fungi (Carvalho et al. 2001), α-amylases from insects (Dos Santos et al. 2010), and against Leishmania amazonesis (Souza et al. 2018). We had also synthesized a peptide containing VuDef1 γ-core, named A36,42,44γ32–46VuDef (in which A36,42,44 denotes the positions where the three C were replaced by A; γ32–46 denotes the position of amino acids in the primary sequence of VuDef1 where the γ-core is found, and for short, this peptide will be called in the following text DD, because it has two adjacent aspartic acids in its primary structure) which also presented activity against L. amazonesis as VuDef1 (Souza et al. 2019). The strong activity of DD against L. amazonesis impelled us to test the peptide against pathogenic yeasts. Nonetheless, DD showed no significant activity against them. This result further prompted us to improve its activity against yeasts by altering its biochemical parameters through amino acid substitutions. There are some approaches to design new peptides (Fjell et al. 2012). However, these methods present the drawback of the necessity of production of several peptides that need to be tested, and in many cases, they present lower activity than the original peptide (Schaaper et al. 2001; Misawa et al. 2017) turning the process inefficient and imposes a limiting factor to those with scarce resources. In this work, we explore the correlation of the biological activity and primary structures of plant defensins for design new peptides with improved antifungal activity. Accordingly, we search for articles correlating the structure and activity of plant defensins, compiled them in a table, analyzed their primary structures and their biological activities. Based on this analysis and to test our hypothesis that this correlation study could indicate both the favorable position and the amino acid to be changed, we designed three new peptides with targeted variations in charge, hydrophobicity and chirality. The new synthetic peptides were tested against the same yeasts as the original DD. Our results indicated that the three new designed peptides based on this approach had better inhibitory activity against the pathogenic yeasts and one of them was the best in potency and in the spectrum of yeast species inhibition. Additionally, all three new peptides had low toxicity to mammalian cells.
Materials and methods
Database analysis, peptide and biophysical properties analysis
We searched for articles on data banks at National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/pubmed), Science Direct (https://www.sciencedirect.com/), and Google Scholar (https://scholar.google.com.br/) using a combination of the following keywords as search parameters: “defensin”, “plant”, “improved”, “variant”, “motif”, “mutational”, “domain”, and “mechanism of action”. The retrieved articles had the primary structures of plant defensins or their derived peptides analyzed. Their primary structures were first aligned by Clustal Omega multiple sequence alignment with default settings (https://www.ebi.ac.uk/Tools/msa/clustalo/; Sievers et al. 2011) and the amino acid alterations were marked in the aligned sequences, as well as the effect resulting from the performed changes on the biological activity. These data are presented in Supplementary Table 1.
We also analyzed the hydrophobicity and net charge of the modified defensins or their derived peptides (Supplementary Table 1). Hydrophobicity was calculated by PepDraw which uses the hydrophobicity Wimley-White scale (http://pepdraw.com/), and net charge was calculated by PepCalc (https://pepcalc.com/).
To compare the correlation between the potency of the inhibitory activity on fungi with charge and hydrophobicity of plant defensins and their derived peptides, we selected articles and extract the pieces of information: charge and hydrophobicity (from Supplementary Table 1) and the concentration in μM that inhibits 50% of the tested fungus (IC50). To give a unifying parameter in these correlation analyzes we chose the same fungus. The data were organized in graphics with the parameters of charge and hydrophobicity classified in increasing order using Excell.
Based on the analysis of the Supplementary Table 1, we generated three new peptides derived from the previous peptide DD (Souza et al. 2019) altering its net charge, hydrophobicity and chirality.
Peptide chemical synthesis
DD and the three new designed peptides were acquired commercially by Aminotech. All peptides were dissolved in pure sterilized water at 2 µg/mL (approximately 1000 µM, depending on the peptide molecular weight) and stored in aliquots at – 70 ºC. Peptides purity was assured as ≥ 95% as determined by reversed-phase high-pressure liquid chromatography and mass spectrometry analyzes (Supplementary Figs. 1, 2, 3, and 4).
Yeasts and antimicrobial assay
The yeasts Candida albicans (CE022), Candida buinensis (4674), Candida parapsilosis (CE002), Candida pelliculosa (3974), Candida tropicalis (CE017) and Saccharomyces cerevisiae (1038) were grown in Sabouraud agar (10 g/L peptone, 20 g/L D( +)glucose, 17 g/L agar, Merck) at 30 ºC for 24 h and then stocks of each yeast were maintained at 4 °C and transferred to a new medium every 3 months.
The yeasts were grown on new Sabouraud agar at 30 ºC for 24 h. After the growth period, a colony was resuspended in Sabouraud broth (5 g/L peptone from meat, 5 g/L peptone from casein, 20 g/L D( +)glucose, 17 g/L agar, Merck) and the cells were counted in a Neubauer chamber (Laboroptik) in an optical microscope (Axio Imager.A2, Zeiss). The assay was performed on a polystyrene 96-well microplate (Nunc, Thermo Scientific) and was composed of 2000 cells/mL, 18.5 μM of each peptide filter-sterilized (0.22 μm, Millex-GV, Millipore) and 100 μL (final volume) of Sabouraud broth. Yeast growth was determined after incubation for 24 h at 30 °C by absorbance at 620 nm (EZ Read 400, Biochrom) as described by Broekaert et al. (1990). Samples in which no peptides were added were considered as controls (100% of growth). Wells containing only Sabouraud broth were considered blanks. Gray scale images of the bottom of the wells were obtained by Galaxy Note 9 camera (Samsung) at 24 h.
Determination of minimal inhibitory concentration
After determining the inhibitory activity against the yeasts for each peptide at the fixed concentration of 18.5 μM, we chose the combination of the most sensitive yeast and the strongest peptide and determined their minimal inhibitory concentration (MIC100). The assay was performed as described in item Yeasts and antimicrobial assay, with the modification that different concentrations of the peptides were used: 14, 18.5, 23, 27.5, 32, and 36.5 μM, depending on the original inhibition of each peptide determined at the first antimicrobial assay. MIC100 was defined as visually the lowest tested concentration of peptides in μM that caused the complete yeast growth inhibition under the conditions the assay was done (30 ºC for 24 h in Sabouraud broth) (Broekaert et al. 1990; Wiegand et al. 2008).
Determination of yeast cell viability and lethal dose
After the MIC100 determination assay, the content of the wells were washed once in Sabouraud broth and evenly spread with a Drigalski spatula on a Petri dish containing Sabouraud agar and incubated at 30 ºC for 24 h to allow the development of colonies. After the formation of the colonies, they were analyzed in relation to the control sample, which was considered 100% viable. Gray scale images were acquired as described in item Yeasts and antimicrobial assay. Viability was defined as the ability of the yeasts cells to divide and thereof to forming colonies in appropriated conditions (30 ºC for 24 h in Sabouraud broth). This peptide concentration in μM that causes the death of all cell population in the original assay was defined as the lethal dose (LD100).
After LD100 determination for each peptide, a control with fluconazole (Sigma-Aldrich) with the same LD100 concentration determined for the peptides was tested. Fluconazole was resuspended in dimethyl sulfoxide (DMSO, Sigma-Aldrich) at a concentration of 2 μg/μL (stock). The control, without adding fluconazole, was done with 0.4% DMSO, the same final concentration of DMSO in the fluconazole treated samples. After 24 h, images of the well bottom were acquired as described in item Yeasts and antimicrobial assay.
Mammalian cell viability assay of design peptides
RAW 264.7 murine macrophages (American Type Culture Collection, ATCC TIB-71) and THP-1 human monocytes (ATCC TIB-202) were cultured in DMEM-F12 supplemented with 10% fetal bovine serum and gentamicin (50 µg/mL) in 5% CO2 at 37 ºC in the LBR, from CBB, UENF, Campos dos Goytacazes, Rio de Janeiro, Brazil. Cells (5 × 105 cells/mL) were separately seeded in 96-well tissue culture plates and incubated for 24 h at 37 °C in 5% CO2. RAW 264.7 reached 60–80% confluence and THP-1 reached 7 × 105 cells/mL. Then, the designed peptides were added at 14, 18.5, 23, 27.5, 36.5 and 50 µM and incubated for further 24 h at same conditions. After 24 h of incubation with peptides, 10 μL of MTT reagent (5 mg/mL, Sigma-Aldrich) was added and incubated for an additional 2 h at 37 °C in 5% CO2. The MTT solution was removed and 100 μL of acidified isopropanol was added to solubilize the formazan crystals formed. The absorbance was measured at 570 nm and absorbance for background correction was determined at 620 nm. Non-treated cells were used as a positive control (optical density (O.D.) 1.94 ± 0.05, cell viability—98.8 ± 2.1%) and 1% Triton X-100 detergent-treated cells as a negative control (O.D. 0.14 ± 0.03, cell viability—0%). The percentage of cell survival was calculated as follows: % Cell viability = 100 × (experimental well ABS – negative viability control) / (positive viability control). The 50% cytotoxic concentration (CC50) was defined as the concentration (µM) required for the reduction of cell viability by 50%, which were calculated by regression analysis (Moodley et al. 2014).
Statistical analysis
The antimicrobial assays were done in triplicate and repeated three times and the mammalian viability assay was done in duplicate and repeated three times. Graphics were depicted as means with standard deviation of one independent assay for antimicrobial assays or by mean values obtained over three experiments for mammalian viability assay. The data obtained in the assays were statistically tested by the one-way ANOVA test, where P < 0.05 was considered significant, using the GraphPad Prism 8 software.
Results and discussion
Previously, we had synthesized a peptide based on the VuDef1 γ-core, namely A36,42,44γ32–46VuDef (DD), which has the primary structure of VuDef1 γ-core except by the exchange of three C residues by A ones (Souza et al. 2019). The A substitutions turn this peptide slightly less hydrophobic, from + 21.98 to + 20.42 kcal/mol (according to the hydrophobicity scale used, that the more positive the value, the more hydrophilic it is) but it retained the same + 2 net charge comparing the original primary structure of VuDef1 γ-core. Additionally, the A substitutions made disulfide bridges formation impossible and avoided a free C to remain (Table 1). To solve this problem, some authors substituted the C by aminoisobutyric acid (Schaaper et al. 2001). DD had inhibitory activity against L. amazonensis as the entire VuDef1 indicating that the inhibitory activity on L. amazonensis is located within VuDef1 γ-core (Souza et al. 2019). Because of the satisfactory inhibitory activity of DD against L. amazonensis, in this work, we tested it against five yeast species of medical importance and to which therapeutic substances are required, and also against the model yeast S. cerevisiae. We used the same incubation time (24 h) and concentration (18.5 μM) determined for L. amazonensis (Souza et al. 2019) in the beginning of our tests. But DD did not present significant activity (Fig. 1).
Yeast growth in the absence (control) and in the presence of the four synthetic peptides. The percentage of inhibition of fungal growth is shown above the test bars and the image of the bottom wells with the growth pattern of each yeast species is shown below their correspondent bars. The image is representative of one assay out of three. Asterisks indicate statistical significance (****P < 0.0001) and ns non significant from control
Because of DD low activity against the tested yeasts, we planned to improve its activity against yeasts by altering its biochemical properties. Many synthetic peptides had been generated by different methods (Fjell et al. 2012), and for example, in the linguistic model described by Loose et al. (2006), the design had been done based on AMPs of natural occurrence that was interpreted as a grammar, the authors had to analyzed a set of 3 million possible sequences from this grammar from which they choose 42 to be synthesized. From the 42 designed peptides, 2 were insoluble. In addition to these 42 peptides, a further 42 randomized sequences were made as controls so that they bear no resemblance to any grammar, and 4 were insoluble. Two groups of peptides were also synthesized, one comprising eight peptides from the Antimicrobial Peptide Database as positive controls and the other composed of six peptides from non-antimicrobial proteins as negative controls. To optimize resources and time, we do the Supplementary Table 1 (which is supplied as Word.doc to facilitate comparison) that compiled literature analysis of articles that correlate plant defensin primary structures and biological activities. Based on this Supplementary Table 1, we designed three new peptides to test our hypothesis that the correlation of primary structures and biological activities of plant defensins could be used to design new peptides with the clear indication the position and also what type of amino acid should be better choice for substitution. We found 27 articles that correlate the primary structure and biological activity of 19 plant defensins and derived peptides (Supplementary Table 1). Changed amino acids were indicated in the primary structure and also the correlated aftermaths of the change had on the biological activity, i.e. if it increased, decreased or was neutral. It is interesting to note that main changes that influenced the biological activity were in the region that is responsible for plant defensins biological activity, the γ-core (see 4th paragraph of introduction; Supplementary Table 1). Before we start the discussion about the design, we drew important pieces of information about the Supplementary Table 1 that will be discussed below.
First about the correlation of length in amino acid residues and inhibitory activity of plant defensin-derived peptides against fungi. We note, like our DD, the ineffectiveness of very short peptides based on the plant defensin γ-cores against fungi. Peptides encompassing only the γ-core amino acid residues, or smaller, did not have the correspondent inhibitory potency against microorganisms like the original defensin did or have no activity. For example, GMA1, GMA1-L, and GMA4, GMA4-L derived from MsDef1 (defensin from Medicago sativa, alfalfa) and MtDef4 (defensin from Medicago truncatula, lucerne), respectively (Muñoz et al. 2014; Sagaram et al. 2011); C36–C45, derived from Rs-AFP2 (De Samblanx et al. 1996); γ33-41PvD1 derived from PvD1 (defensin from Phaseolus vulgaris, common bean) (Mello et al. 2019) (Supplementary Table 1). Also, smaller peptides had lower inhibitory activity when compared to their bigger counterparts that covers the entire γ-core region alongside some residues of the β2 and β3 strands. For example, GMA1 have no antifungal activity and GMA1-C have antifungal activity; GMA4 and γ33-41PvD1++ has lower antifungal activity than GMA4-C and γ33-45PvD1++ (derived from the γ-core of the defensin PvD1) (Supplementary Table 1). Rekdal et al. (1999) provided similar results for lactoferricin analogs (peptides with antimicrobial activity derived from lactoferrin obtained by gastric digestion), in which the derived peptides should not be shorter than 15 amino acid residues to maintain their antimicrobial activity. According to our analysis, the minimum size for these peptides derived from plant defensins to be active is about nine amino acid residues. In fact, this size limit is observed in nature, the smallest known peptides with broad antimicrobial activity against different microorganisms are the temporins, which have about 10–13 amino acid residues long, obtained from the skin secretion of anuran amphibian Rana temporaria (European common frog) (Mishra et al. 2018) and bactenecin with 13 amino acid residues from bovine neutrophils (Cherkasov et al. 2009).
The reason for the weaker or ineffectiveness of antifungal activity of short peptides is not well understood. One possible explanation for the activity of γ-core derived peptides that does not match the activity of defensin as a whole could be the observations that for some plant defensins there are amino acids important for biological activity tracked outside this motif. For example, the K4 (position 7 in the Supplementary Table 1) in NaD1 (defensin from Nicotiana alata, ornamental tobacco) and the K6 (position 7 in the Supplementary Table 1) in TPP3 (defensin from Lycopersicon esculentum, tomato) are outside those defensin γ-cores. They are essential for binding to the fungal target and dimerization as demonstrated by their replacement by A, which turns both defensins ineffective in their ability to form dimers, which is important for lipid biding, antifungal and antitumor activities (Lay et al. 2012; Baxter et al. 2015). It is also observed that the microorganism species is relevant to the inhibition process, because despite being inactive against yeasts, DD was active against L. amanzonensis (Souza et al. 2019). Corroborating with this suggestion are studies with fungi demonstrating that lipids, especially the negatively charged ones, that are present in the fungal membrane, such as mannosyldiinositol phosphorylceramide (M(IP)2C), phosphatidic acid (PA) and phosphatidylinositol 4,5 bisphosphate (PI(4,5)P2), may be the targets of plant defensins and the cationic and hydrophobic amino acids of the γ-core region are important for this interaction (Parisi et al. 2019). MtDef4, for example, had different mechanism of action on the fungi Neurospora crassa and Fusarium graminearum, because the differential concentration of sphingolipids in those fungi, as suggested by the authors (El-Mounadi et al. 2016). Ramamoorthy et al. (2007) observed that the inhibition of the fungus F. graminearum by the defensin MsDef1 depended on the sphingolipid glucosylceramide (GlcCer), but not the inhibition caused by MtDef4. Likewise, S. cerevisiae that does not have GlcCer in its membrane is resistant to Rs-AFP2, while Pichia pastoris and C. albicans yeasts that have GlcCer are sensitive to Rs-AFP2. Additionally, P. pastoris and C. albicans mutants, without GlcCer in their membranes, exhibited resistance to Rs-AFP2 (Thevissen et al. 2004). The higher affinity and capacity of protegrin-1 (AMP of the cathelicidin class obtained from pig neutrophils) to lyse erythrocyte membranes from mice, rabbits and humans whereas is ineffective to bovine, sheep and porcine erythrocytes (Bellm et al. 2000) are supposedly explained by the differential lipid composition of erythrocyte membranes of those organisms (Nouri-Sorkhabi et al. 1996a, b). Therefore, perhaps the yeasts tested in this work might not have the specific target of DD, which would explain its lack of activity. Besides that, the two negative charged amino acids (D37 and D38; positions 47 and 48 in the Supplementary Table 1, respectively) of DD may result in repulsion from negative charged structures in the fungal cell wall, e.g. manophosphoproteins, or membrane constituents, as explained above, and demonstrated to other peptides where some interaction positions with microbial membrane constituents are not favorable by charge repulsion (Haney et al. 2007), which may resulted in a weak interaction and consequently antifungal activity. This characteristic seems to be more important for the peptide than to the VuDef1 itself, probably because in the entire defensin there are other amino acid interactions that compensate this repulsion.
The second observation is about charge and hydrophobicity. Supplementary Table 1 shows that the alterations that add positively charged and hydrophobic amino acid residues were the most positively influential on the biological activity of plant defensins (Supplementary Figs. 5, 6 and 7). Studies that correlate the structure and mechanism of action of plant defensins show that the attraction of opposite charges between positively charged defensins with the negatively charged membranes of microorganisms ensures the initial interaction. Then, the hydrophobic region of the defensin interacts with the hydrophobic part of the membrane, which can cause its destabilization (Giuliani et al. 2007) as explained above. This oppose charge interaction is supported by the abrogation of plant defensins antifungal effect caused by the addition of divalent cations, especially Mg+2 and Ca+2, in the culture medium which screen electrostatic charges disrupting the initial attraction (Terras et al. 1993; Van der Weerden et al. 2008). Our observation that the more cationic peptides have stronger inhibitory activity is supported by studies with plant defensins isoforms that the more cationic ones had stronger inhibitory activity. For example, Rs-AFP1 and Rs-AFP2 are two natural defensin isoforms from R. sativus seeds, which are nearly identical in their primary structure except by the substitutions of E5 to Q and N27 to R in Rs-AFP2 (position 7 and 29 in Supplementary Table 1). The first substitution removes one negative charge from Rs-AFP2 and the second adds one positive charge to it. Therefore, Rs-AFP2 is more cationic (net charge from + 3.6 to + 5.6) and hydrophobic (from + 36.68 to + 34.78 kcal/mol) and these increments in cationicity and hydrophobicity were correlated with the Rs-AFP2 stronger inhibitory active against fungi than Rs-AFP1, even in media with a high ionic strength that inactivate most of AMPs (Terras et al. 1993; Tam et al. 2002). The isoforms Ph1 and Ph2 (Lay et al. 2003) and VrD1 and VrD2 (Lin et al. 2007) are other examples. Beside these observations, we can mention a chimera of MsDef1, called MsDef1-γ4 in which the DDFQ amino acids of MsDef1 have been replaced by the amino acids that determine MsDef4 antifungal activity, GFRRR. This substitution increased the net charge (from + 2.6 in MsDef1 to + 6.6 in MsDef1-γ4) and hydrophobicity (from + 51.68 kcal/mol MsDef1 to + 49.17 kcal/mol in MsDef1-γ4), resulting in a significant increase in antifungal activity of the chimera when compared to MsDef1 and it was as potent as MsDef4 (Supplementary Table 1). The OefDef1.1_V5 variant also showed an improved antifungal activity when compared to the original OefDef1.1 (defensin from Olea europaea, olive). In this case, replacing the KHYG residues to AAAA decreased the positive charge (from + 8.1 in OefDef1.1 to + 7.0 in OefDef1.1_V5), but increased the hydrophobicity (from + 50.31 kcal/mol in OefDef1.1 to + 46.74 kcal/mol in OefDef1.1_V5). Also, the chimera VrD2c produced by the exchange of RDDFR from VrD2 to GMTRT from VrD1 that increased its net charge (from + 2.7 to + 3.7) and its hydrophobicity (from + 54.42 to + 48.02 kcal/mol) resulted in an increased inhibitory potency against α-amylase. Not only alterations in stretches of amino acids resulted in this effect, but also point changes. For example, to Rs-AFP2 the N36R change (position 41 in Supplementary Table 1) increased the modified peptide net charge from + 5.6 to + 6.6 (although the hydrophobicity decreased from + 34.78 to + 35.74 kcal/mol), this net charge increasing augmented its antimicrobial activity. A similar alteration, Q40R (position 50 in Supplementary Table 1) (increased net charge from -1.1 to -0.1, although a slight decreased in hydrophobicity from + 46.84 to + 47.88 kcal/mol) in MtDef2 had the same effect. Likewise, amino acid exchanges that decreased the positive charge worsened the biological activity of defensins. For example, Rs-AFP2 has K at position 43 (position 50 in Supplementary Table 1), and when it was replaced by Q (decrease net charge from + 5.6 to + 4.6 and hydrophobicity from + 34.78 to + 32.75 kcal/mol) the inhibitory activity of the defensin decreased; the same is observed for R38Q (position 50 in Supplementary Table 1) (which decrease net charge from + 2.6 to + 1.6 and increased hydrophobicity from + 51.68 to + 50.64 kcal/mol) in MsDef1, and RR to AA (position 49 and 50 in Supplementary Table 1) in MtDef4RGFRRR/RGFAA (with decrease net charge from + 5.8 to + 3.8 and increased hydrophobicity from + 49.16 to + 46.54 kcal/mol) (Supplementary Table 1). Still in regard to charge, it is important to mention that the position of charge insertion is important for activity. For example, the inclusion of positively charged amino acids by substitution of V38R and A41R (positions 43 and 48 in Supplementary Table 1, respectively) in Rs-AFP2 decreased its antimicrobial activity (Supplementary Table 1). Other substitution outside the γ-core, such as S12R, I26R, L28R, I46R (positions 15, 29, 31, 53 in Supplementary Table 1, respectively) also decreased its antimicrobial activity. In addition, the removal of those hydrophobic residues, concomitantly reduced hydrophobicity (+ 36.13, + 37.71, + 37.84 and + 37.71 kcal/mol, respectively) compared to the original Rs-AFP2 (+ 34.78 kcal/mol), which may also have contributed to decreased antifungal activity. These observations reinforce that not only positive charge increase is important, but also the position where the positive charge is inserted, probably to avoid charge repulsion as explained in the 4th paragraph of the Results and discussion section, along with the hydrophobicity for biological activity.
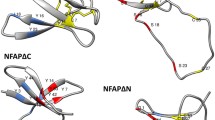
Based in all pieces of information aforementioned, we specifically compared the previous sequence of VuDef1 and DD with other defensins and their derived peptides and acknowledged that in the γ-core region, highlighted in a red box in Supplementary Fig. 1, there are some amino acids exchanged in some plant defensins or derived peptides that resulted in variation in the biological activity that were considered interesting to change in the original sequence of DD. The positive charges at positions 41 and 50 (reference position indicated in the Supplementary Table 1) that are conserved and important for the biological activity of plant defensins are of special interest (Supplementary Fig. 8). Please refer to the 5th paragraph of the Results and discussion section for the discussion of these charges in Rs-AFP2, MsDef1, MtDef2, MtDef4RGFRRR/RGFRAA. Also OsAFP1 (defensin from Oryza sativa, rice) has a K35 and K42 (positions 41 and 50 in the Supplementary Table 1) that were replaced by A (which decreased the antifungal activity in both variants, and also increased hydrophobicity from + 51.24 to + 48.94 and + 48.94 kcal/mol, respectively). At those correspondent positions DD has positively charged amino acids, R, therefore, we maintained them in our design approach. In relation to the substitution of negative charges, MsDef1 has the sequence in its γ-core RDD36FR (position 48 in the Supplementary Table 1) which was replaced by the sequence of the MtDef4 γ-core RGFR39RR (position 48 in the Supplementary Table 1) generating the peptide MsDef1-γ4, with increased antifungal activity (and hydrophobicity increased from + 51.68 to + 49.17 kcal/mol), and DD has a very similar sequence to MsDef1, RDD38VR (position 48 in the Supplementary Table 1). Also in peptides derived from the γ-core of defensin PvD1 where the replacement of two negative residues in γ31-45PvD1 (RSGRARD37D38FRAWATK) (position 47 and 48 in the Supplementary Table 1) by two positive residues in γ31-45PvD1++ (RSGRARR37R38FRAWATK, which also increased its hydrophobicity from + 24.78 to + 21.12 kcal/mol), improved its antifungal activity. The same was observed with the defensin MtDef4, which lost its antifungal activity when the F38R39 residues, corresponding to our D37 and D38 (positions 48 and 49 in the Supplementary Table 1) were replaced by A. In this case, a decrease in both net charge (from + 5.8 to + 4.8) and hydrophobicity (from + 49.16 to + 50.06 kcal/mol) were observed (Supplementary Table 1). Therefore, we selected D37 and D38 (positions 48 and 49 in Supplementary Table 1) to be substituted by R. Based on this charge analysis, we designed two new peptides. In the first, the R36 and R40 (positions 46 and 50 in Supplementary Table 1), flanking the VuDef1 γ-core sequence, were kept and D37 and D38 were replaced by R, and it was called A36,42,44R37,38γ32–46VuDef (R37,38 indicate the position of the change of DD for RR, positions 48 and 49 in Table 1. For short, we will call this peptide RR because of the two R that replaced the two D). The second peptide had the exact RR sequence except all amino acids are d-enantiomers, thus it was named D-A36,42,44R37,38γ32–46VuDef (D-RR to indicate the change in chirality), and both peptides had its net charge increased from + 2 to + 6 and also its hydrophobicity was increased from + 21.98 to + 18.32 kcal/mol in regard to DD (Table 1). We choose to synthesize a peptide composed of D-amino acids, because studies had shown that they are more resistant to degradation by proteases while retaining antimicrobial activity and are in general less toxic to mammalian cells (Hamamoto et al. 2002; Braunstein et al. 2004).
The third new designed peptide we maintained the charge (+ 6) but has increased its hydrophobicity since we realized that in the region of the γ-core in Rs-AFP2 the change of Y38G (position 42 in Supplementary Table 1) decreased its hydrophobicity from + 34.78 to + 36.64 kcal/mol (without charge variation) as well as its antimicrobial activity. The same is observed for SPE10 (defensin from Pachyrhizus erosus), the change of D38F39 to ND (position 48 and 49 in Supplementary Table 1) the hydrophobicity drop from + 59.01 to + 61.57 kcal/mol (both retained + 0.7 charge) and the peptide C36-C45(Y38A) (the change of Y38A, position 42 in Supplementary Table 1) derived from Rs-AFP2 the hydrophobicity drop from + 11.60 to + 12.81 kcal/mol. It is interesting to note the presence of an aromatic amino acid residue in this region (Y38 Rs-AFP2 and F39 in SPE10) (position 42 and 49 in Supplementary Table 1, respectively) that seemingly contributes to both hydrophobicity and antimicrobial activity. DD γ-core region has V39 in correspondent position (Supplementary Table 1), thus it was selected to be substituted. Another position was the A36 (position 40 in Supplementary Table 1) which replaced a C in that position in the original VuDef1 γ-core sequence. As it is a hydrophobic amino acid it was also chosen to be substituted. For the substitutions, we chose an aromatic residue, and W was chosen. We decided to keep the positive charges conserved at positions 41 and 50 (Supplementary Table 1), as described in the 5th paragraph of the Results and discussion section, and the peptide was called A42,44R37,38W36,39γ32–46VuDef (where W36,39 indicate the position of the change of A36 and V39 for W, for short WR to summarize the two amino acids exchanged) (Table 1). The substitutions A36W and V39W ensured greater hydrophobicity to WR (+ 14.10 kcal/mol) when compared to RR and D-RR (+ 18.32 kcal/mol) and DD (+ 21.98 kcal/mol). And the same + 6 net charge of RR and D-RR. Higher hydrophobic character of all three new synthesized peptides was confirmed by the increased retention time in reversed-phase C-18 column in HPLC, especially for WR (Supplementary Figs. 1, 2, 3 and 4). W was chosen, because it is a neutral amino acid found in high proportion in short biologically active AMPs such as indolicidin (AMP from bovine neutrophil) and tritrpticin (AMP from pig bone marrow) (Mishra et al. 2018). Its uncharged aromatic side chain has the ability to form hydrogen bonds and insert itself uniquely in microbial membranes (Strøm et al. 2000; Bi et al. 2013). In addition, tryptophan-rich peptides with biological activity also contain arginine residues. The positive charge of this amino acid assists in the initial electrostatic attraction and formation of hydrogen bonds between the peptide and the target membrane. Then, W is anchored to the membrane with high affinity (Chan et al. 2006). Studies carried out with indolicidin, tritrpticin, bovine lactoferricin and temporins provided evidences that the unique properties of the side chains of the W and R residues render short peptides highly active, with antibacterial, antifungal, antiviral and antitumor, albeit with low hemolytic activity (Lawyer et al. 1996; Strøm et al. 2000; Bi et al. 2013; Shagaghi et al. 2016).
Next, the antifungal activity of the three new synthetic peptides was tested using the same conditions as DD. We found that RR inhibited 47.5, 100, and 72.1% of the growth of C. albicans, C. buinensis, and C. tropicalis, respectively, however, it was unable to inhibit the growth of S. cerevisiae, C. parapsilosis and C. pelliculosa as DD (Fig. 1). This inhibition, obtained by measuring cell cultures optical density, can also be observed visually, since there was a clear decrease in the mass of yeast cells at the microplate well bottom and there was a change in the fungal growth pattern for C. albicans and also for C. parapsilosis, albeit they were not inhibited, when compared to their respective controls (Fig. 1). As expected, only the increase in the positive net charge from + 2 to + 6 and in specific positions in RR was sufficient for an improvement in its antifungal activity when compared to the original DD. D-RR, which has the same biochemical characteristics of RR but in D configuration, inhibited the same yeasts as RR, 84.9, 99.7, and 100% of the growth of C. albicans, C. buinensis, and C. tropicalis, respectively, but with more potency, and was unable to inhibit the growth of S. cerevisiae, C. parapsilosis and C. pelliculosa (Fig. 1) as RR. Corroborating the high inhibition activity, there was no visible mass of cells at the well bottom of the inhibited yeasts captured by the camera (Fig. 1). Since both RR and D-RR had inhibitory activity against the same yeasts suggests that their mechanism of action is independent of chirality. The higher potency of D-RR against the inhibited yeast may be related to protease resistance (Hamamoto et al. 2002; Braunstein et al. 2004) that will be further investigated. WR inhibited 26.1, 96.2, 98.5, and 58.2% of the growth of S. cerevisiae, C. albicans, C. buinensis, and C. tropicalis, respectively, and was unable to inhibit the growth of C. parapsilosis and C. pelliculosa (Fig. 1). However, to C. parapsilosis and C. pelliculosa, there was a change in the fungal growth pattern, despite they were not inhibited, when compared to their respective controls (Fig. 1). The increase in positive net charge (from + 2 to + 6) and hydrophobicity (from + 21.98 to + 14.1 kcal/mol) made WR with the best antifungal activity when compared to the other three peptides tested. Our results clearly indicated that augmented charge and hydrophobicity by targeted modification of amino acids in the designed peptides improved their antimicrobial activity. All designed peptides inhibited more yeast species and with greater potency than DD, although C. parapsilosis and C. pelliculosa were not inhibited by all tested peptides in our conditions (Fig. 1 and Table 2).
Based on the first antimicrobial assays, the combinations of the most sensitive and medically relevant yeast and the most active synthetic peptide were chosen for MIC100 and cell viability determinations (LD100). Depending on the percentage of inhibition observed for the 18.5 µM concentration of the first assays, we decreased or increased it in 4.5 μM increments to find the MIC100 and LD100 for each chosen peptide-yeast combination. For RR and C. tropicalis the initial 18.5 μM concentration was increased until 36.5 μM. At the 23 μM, there was a small mass of yeast cells visible at the bottom of the microplate well that was not captured by the camera. At the concentration from 27.5 to 36.5 μM, there was no visible growth at the bottom of the well, and as we had defined our MIC100 as the concentration that completely inhibits the visible growth, 27.5 μM was the MIC100 (Fig. 2a). In cell viability assay, no colony developed indicating that this concentration was also the lethal dose (LD100) (Fig. 2b). For D-RR and C. albicans, we rose the initial 18.5 μM concentration up to 36.5 μM and already at 23 μM, there was no visible growth at the well bottom, and it was determined as the MIC100 (Fig. 2c). The viability test indicated that no colony developed at 36.5 μM and it was determined as the LD100 (Fig. 2d). For the other lower concentrations, few colonies developed (Supplementary Fig. 9). For D-RR and C. tropicalis we decreased the initial concentration to 14 μM, and there was no visible growth at the well bottom, and this concentration was determined as the MIC100 (Fig. 2e). However, for the viability assay, few colonies developed at this concentration (Supplemented Fig. 9). Therefore, to determine the LD100, we seeded the yeast cells treated with the higher concentrations into a new fresh medium, at 18 μM few colonies developed (Supplementary Fig. 9), and from 23 to 36.5 μM no colony developed, therefore, the LD100 was 23 μM (Fig. 2f). For WR and C. albicans, we decreased the concentration for 14 μM since 18.5, 23, 27.5 and 36.5 μM no growth could be observed at the bottom of the wells, although they were not captured by the camera (Fig. 2g). However, at 14 μM, a small mass of yeast cells was observed which was not captured by the camera, and therefore, 18.5 μM was determined as the MIC100. In the viability assay, for the 14, 18.5 and 23 μM concentrations, few colonies developed, and for the 27.5 μM, no colony developed, indicating this concentration as the LD100 (Fig. 2h). The effect of the synthetic peptides on the viability of the yeasts species was fungicide and their effect were confronted to a widely used fungice, fluconazole (Sapampinato and Leonardi, 2013), used at corresponding concentration of peptides DL100. For all tested yeasts, the inhibitory effect of fluconazole was weaker than that the synthetic peptides because was observed a cell mass at the bottom of the wells (Fig. 2b, d, f and h). As consequence of this partial inhibition, to all tested yeasts the antifungal effect of fluconazole was fungistatic (Fig. 2b, d, f and h). Thevissen et al. (2004) demonstrated in vitro that the defensins Hs-AMP1 (defensin from Heuchera sanguinea, coralbell), Rs-AFP2 and Dm-AMP1 (defensin from Dahlia merki, dahlia) were fungicidal for Candida species tested and more efficient in killing yeasts when compared to commercial antifungals, among them fluconazole. Our peptides showed fungicidal action, which reduces the chances of selection of resistant strains, an important characteristic for new drugs (Levy and Marshall 2004; Thevissen et al. 2004).
Determination of the minimal inhibitory concentration (MIC100) (a, c, e and g) and cell viability (LD100) (b, d, f and h) assays for the combination of the chosen peptides and yeasts. a, c, e and g Images of the bottom of the wells at the end of the assay (24 h) and the arrow points to MIC100. Peptide concentration above this one pointed by the arrow presents a small mass of cells growth which was not captured by the camera and below this indicated concentration no growth is observed. b, d, f and h In the viability assay is indicated the concentration of the MIC100 assay that kills all initial cell population and therefore, indicating a fungicide action at this concentration and also the LD100. In the well treated with fluconazole, at the same LD100 concentration determined to the peptides, it was possible to see a small fungal growth and, after plating, there was the growth of colonies, demonstrating fungistatic action. The images are representative of one assay out of three
Additionally, we tested the toxicity of the three new peptides toward two lineages of mammalian cells. All three peptides had low toxicity (Fig. 3a, b) as indicated by the low reduction of metabolic activity. For RR, at the LD100 determined for C. tropicalis (27.5 μM), murine macrophages and human monocytes were metabolically inhibited by 14.7 and 14.9%, respectively. For D-RR, at the LD100 determined for C. albicans (36.5 μM) and C. tropicalis (23 μM), murine macrophages and human monocytes were metabolically inhibited by 17.2, 14.9, 20.5 and 14.6%, respectively. For WR, at the LD100 determined for C. albicans (27.5 μM), murine macrophages and human monocytes were metabolically inhibited by 3.6 and 10.2%, respectively. Even at the highest tested concentration of 50 μM, which is much higher than the LD100 for the tested yeasts, the most toxic peptide, D-RR, only lowered the metabolic activity of murine macrophages and human monocytes by 23.9 and 25.1%, respectively (Fig. 3a, b). As seen in Fig. 3, the non-adherent cell line (human monocytes) was slightly more sensitive than the adherent cell line (murine macrophages). The reason for that observation is because the surface area exposed to peptides is greater in the non-adherent cell line than in the adherent one. By testing toxicity with adherent and non-adherent cell lines we have a more real view of toxicity to tissues and blood cells. Additionally, both cell lines are important immune cells that act as the first defense line in the immune response to pathogens, and for this reason, they should not be seriously affected by the peptides to fulfill their primordial defense function. The 50% cytotoxic concentration (CC50) for all three peptides for the two cell lineages were higher than 50 μM (Fig. 3c) indicating a discriminating capability between yeasts and mammalian cells by the peptides which is a good feature for a drug candidate. The low toxicity of our peptides is in agreement of studies that showed low toxicity of plant defensins to mammalian cells (Tavares et al. 2008; Carvalho and Gomes 2011; Vriens et al. 2015; Lay et al. 2019). Altogether, our results are summarized in Table 2.
Viability of RAW 264.7 murine macrophage (a) and THP-1 human monocite (b) cells in response to the synthetic peptides after 24 h treatment. Cell viability was assayed by the colorimetric MTT based assay. Cell viability percentage was calculated in relation to the positive control (O.D. 1.94 ± 0.05, cell viability—98.8 ± 2.1%), untreated macrophages, and to the negative control, macrophages culture treated with 1% (v/v) Triton X-100 (0.14 ± 0.03, cell viability—0%). The bars for each sample refer to concentrations tested in ascending order. The results presented are mean values obtained over three experiments, each done in duplicate. c 50% cytotoxic concentration (CC50) required for the reduction of cell viability by 50%, which were calculated by regression analysis. ***P < 0.001 compared to untreated group (0 µM) determined by Tukey test
As predicted, the increase in net positive charge and hydrophobicity in the WR resulted in the best antifungal activity when compared to the other three peptides tested. However, only the increase in the positive charge in RR was enough for an improvement in its antifungal activity when compared to DD (Fig. 1). A similar result was obtained by Mello et al. (2019) with peptides derived from the γ-core of the PvD1 as explained in the 6th paragraph of the Results and discussion section about the replacement of two negative residues (DD) in γ31-45PvD1 (RSGRARDDFRAWATK) by two positive residues (RR) in γ31-45PvD1++ (RSGRARRRFRAWATK). The γ31-45PvD1++ also has a higher hydrophobicity (+ 17.39 kcal/mol) than γ31-45PvD1 (+ 21.12 kcal/mol), which may have contributed to its antifungal activity improvement (Supplementary Table 1). In regard to the antimicrobial spectra of plant defensins, some fungal species are inhibited while others are not (Fig. 1 and Table 2). This can be partially explained by the peptide sequence and structure. Furthermore, the opposite charge attraction can also partially explain the plant defensins and their derived peptides capability of interact and inhibit microorganisms, because the difference in the composition of the fungal membrane, as explained in the 4th paragraph of the Results and discussion section. Additionally, the RGFRRR-positive amino acid sequence present in the γ-core of MtDef4 is essential for the binding of the defensin to PA in the target membrane, cell entry and induction the fungal death. Mutants in which the positive sequence was exchanged to AAARR and RGFRAA lost their ability to bind to the PA, were unable to enter in the fungal cell and kill the fungus F. graminearum (Sagaram et al. 2013). Such results indicate that the fungal inhibition caused by defensins is species dependent, that is, they depend on a specific target presented by the fungal species. It is likely that the directional amino acid changes made to the WR peptide reinforced its interaction with the target membrane and consequently increased its antifungal activity. This was also observed by Saravanan et al. (2014) where the authors synthesized seven peptides from ten residues derived from HBD-28 C-terminal (human β-defensin), with modifications that increased net charge, by adding R, and hydrophobicity, by adding W. Their results showed that peptides with a higher positive net charge and higher hydrophobicity, such as RWKRWWRRKK-NH2 (charge + 7.1 and hydrophobicity + 20.45 kcal/mol) and RKKRWWRRKK-NH2 (charge + 8.1 and hydrophobicity + 25.34 kcal/mol) had the best antimicrobial activity, without increasing cytotoxicity for mammalian cells. In addition, it was shown that these peptides interacted strongly with the microbial membrane, destabilized it and caused its permeabilization. Such results show the importance of positively charged and hydrophobic amino acids in the AMP interaction with microorganisms, not only for plant defensins, but also for other AMPs and/or peptides derived from them as well.
Conclusion
Despite plant defensins huge primary structure variation which difficult a straightforward analysis (Lacerda et al. 2014), our data clearly demonstrated that the correlation of biological activity and primary structure of plant defensins that some position of preference for certain amino acid residues, especially R in positions 41 and 50, and aromatic residue approximately in position 42. Thus, we explored the correlation of the biological activity and structure of plant defensins and design new peptides with improved antifungal activity. Importantly, as this approach in mainly focused on the γ-core that plant defensins share with many other AMPs (Yount and Yeman 2004), it has the potential to be applied to other γ-core containing AMPs. This approach has the advantages of: (1) being fast, once the table presented in the Supplementary Fig. 1 is ready and primary structures contained in it can be easily compare with other AMPs, (2) there is no need to synthesize peptide libraries, because the modifications are guided by studies that demonstrate activity or loss of activity due to specific amino acid exchange. Our results indicate that the targeted modifications in the RR, D-RR and WR peptides resulted in improved antifungal activity by inhibiting more yeast species with high potency, when compared to the original DD, with WR, the most hydrophobic and cationic peptide, exhibited better antifungal activity among the four and also was the less toxic to mammalian cells in our conditions. So far, we showed that the three new peptides have antifungal activity, depending on charge, hydrophobicity and yeast species. However, more research is still needed to understand the mechanisms of action of these molecules and to identify their possible cellular targets, which is of fundamental importance for the design and development of new peptides with further improved biological activity. Therefore, our study provides experimental evidence that targeted changes in the primary structure of peptides based on plant defensins γ-core primary structures prove to be a good tool for the synthesis of new compounds that may be useful as alternative antifungal drugs.
Abbreviations
- DD:
-
A36,42,44γ32–46VuDef
- RR:
-
A36,42,44R37,38γ32–46VuDef
- D-RR:
-
D-A36,42,44R37,38γ32–46VuDef
- WR:
-
A42,44R37,38W36,39γ32–46VuDef
- AMP:
-
Antimicrobial peptides
- CC50 :
-
50% Cytotoxic concentration
- DL100 :
-
Lethal dose
- GlcCer:
-
Glucosylceramide
- PA:
-
Phosphatidic acid
- PI(4,5)P2 :
-
Phosphatidylinositol 4,5 bisphosphate
- M(IP)2C:
-
Mannosyldiinositol phosphorylceramide
- MIC100 :
-
Minimal inhibitory concentration
References
Balls AK, Hale WS (1940) A sulphur-bearing constituent of the petroleum ether extract of wheat flour. Cereal Chem 17:243–245
Baxter AA, Richter V, Lay FT, Poon IK, Adda CG, Veneer PK, Phan TK, Bleackley MR, Anderson MA, Kvansakul M, Hulett MD (2015) The tomato defensin TPP3 binds phosphatidylinositol (4,5) bisphosphate via a conserved dimeric cationic grip conformation to mediate cell lysis. Mol Cell Biol 35(11):1964–1978. https://doi.org/10.1128/MCB.00282-15
Beer A, Vivier MA (2008) Vv-AMP1, a ripening induced peptide from Vitis vinifera shows strong antifungal activity. BMC Plant Biol. https://doi.org/10.1186/1471-2229-8-75
Bellm L, Lehrer RI, Ganz T (2000) Protegrins: new antibiotics of mammals origin. Expert Opin Investig Drugs 9(8):1731–1742. https://doi.org/10.1517/13543784.9.8.1731
Bi X, Wang C, Ma L, Sun Y, Shang D (2013) Investigation of the role of tryptophan residues in cationic antimicrobial peptides to determine the mechanism of antimicrobial action. J Appl Microbiol 115(3):663–672. https://doi.org/10.1111/jam.12262
Bloch C Jr, Richardson M (1991) A new family of small (5 kDa) protein inhibitors of insect α-amylases from seeds of sorghum (Sorghum bicolor (L) Moench) have sequence homologies with wheat γ-purothionins. FEBS Lett 279(1):101–104. https://doi.org/10.1016/0014-5793(91)80261-z
Bongomin F, Gago S, Oladele RO, Denning DW (2017) Global and multi-national prevalence of fungal diseases-estimate precision. J Fungi 3(4):57. https://doi.org/10.3390/jof3040057
Braunstein A, Papo N, Shai Y (2004) In vitro activity and potency of an intravenously injected antimicrobial peptide and its DL amino acid analog in mice infected with bacteria. Antimicrob Agents Chemother 48(8):3127–3129. https://doi.org/10.1128/AAC.48.8.3127-3129.2004
Broekaert WF, Terras FRG, Cammue BPA, Vanderleyden J (1990) An automated quantitative assay for fungal growth inhibition. FEMS Microbiol Lett 69(1–2):55–59. https://doi.org/10.1111/j.1574-6968.1990.tb04174.x
Brogden KA (2005) Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol 3(3):238–250. https://doi.org/10.1038/nrmicro1098
Campos ML, de Souza CM, de Oliveira KBS, Dias SC, Franco OL (2018) The role of antimicrobial peptides in plant immunity. J Exp Bot 69(21):4997–5011. https://doi.org/10.1093/jxb/ery294
Carvalho AO, Gomes VM (2011) Plant defensins and defensin-like peptides—biological activities and biotechnological applications. Curr Pharm Des 17(38):4270–4293. https://doi.org/10.2174/138161211798999447
Carvalho AO, Machado OLT, Da Cunha M, Santos IS, Gomes VM (2001) Antimicrobial peptides and immunolocalization of a LTP in Vigna unguiculata seeds. Plant Physiol Biochem 39(2):137–146. https://doi.org/10.1016/S0981-9428(00)01230-4
Chan DI, Prenner EJ, Vogel HJ (2006) Tryptophan- and arginine-rich antimicrobial peptides: Structures and mechanisms of action. Biochim Biophys Acta 1758(9):1184–1202. https://doi.org/10.1016/j.bbamem.2006.04.006
Cherkasov A, Hilpert K, Jenssen H, Fjell CD, Waldbrook M, Mullaly SC, Volkmer R, Hancock RE (2009) Use of artificial intelligence in the design of small peptide antibiotics effective against a broad spectrum of highly antibiotic-resistant superbugs. ACS Chem Biol 4(1):65–74. https://doi.org/10.1021/cb800240j
Ciumac D, Gong H, Hu X, Lu JR (2019) Membrane targeting cationic antimicrobial peptides. J Colloid Interf Sci 537:163–185. https://doi.org/10.1016/j.jcis.2018.10.103
Colgrave ML, Craik DJ (2004) Thermal, chemical, and enzymatic stability of the cyclotide kalata B1: The importance of the cyclic cystine knot. Biochemistry 43(20):5965–5975. https://doi.org/10.1021/bi049711q
Colilla FJ, Rocher A, Mendez E (1990) γ-Purothionins: amino acid sequence of two polypeptides of a new family of thionins from wheat endosperm. FEBS Lett 270(1–2):191–194. https://doi.org/10.1016/0014-5793(90)81265-p
Coninck B, Cammue BPA, Thevissen K (2013) Modes of antifungal action and in planta functions of plant defensins and defensin-like peptides. Fungal Biol Rev 26(4):109–120. https://doi.org/10.1016/j.fbr.2012.10.002
Costa-de-Oliveira S, Rodrigues AG (2020) Candida albicans antifungal resistance and tolerance in bloodstream infections: the triad yeast-host-antifungal. Microorganisms 8(2):154. https://doi.org/10.3390/microorganisms8020154
De Paula VS, Razzera G, Barreto-Bergter E, Almeida FC, Valente AP (2011) Portrayal of complex dynamic properties of sugarcane defensin 5 by NMR: Multiple motions associated with membrane interaction. Structure 19(1):26–36. https://doi.org/10.1016/j.str.2010.11.011
De Samblanx GW, Fernandez del Carmen A, Sijtsma L, Plasman HH, Schaaper WM, Posthuma GA, Fant F, Meloen RH, Broekaert WF, van Amerongen A (1996) Antifungal activity of synthetic 15-mer peptides based on the Rs-AFP2 (Raphanus sativus antifungal protein 2) sequence. Pept Res 9(6):262–268
De Samblanx GW, Goderis IJ, Thevissen K, Raemaekers R, Fant F, Borremans F, Acland DP, Osborn RW, Patel S, Broekaert WF (1997) Mutational analysis of a plant defensin from radish (Raphanus sativus L.) reveals two adjacent sites important for antifungal activity. J Biol Chem 272(2):1171–1179. https://doi.org/10.1074/jbc.272.2.1171
Deslouches B, Di YP (2017) Antimicrobial peptides with selective antitumor mechanisms: prospect for anticancer applications. Oncotarget 8(28):46635–46651. https://doi.org/10.18632/oncotarget.16743
Ding Y, Ting JP, Liu J, Al-Azzam S, Pandya P, Afshar S (2020) Impact of non-proteinogenic amino acids in the discovery and development of peptide therapeutics. Amino Acids 52:1207–1226. https://doi.org/10.1007/s00726-020-02890-9
Dos Santos IS, Carvalho AO, de Souza-Filho GA, do Nascimento VV, Machado OL, Gomes VM (2010) Purification of a defensin isolated from Vigna unguiculata seeds, its functional expression in Escherichia coli, and assessment of its insect α-amylase inhibitory activity. Protein Expr Purif 71(1):8–15. https://doi.org/10.1016/j.pep.2009.11.008
El-Mounadi K, Islam KT, Hernández-Ortiz P, Read ND, Shah DM (2016) Antifungal mechanisms of a plant defensin MtDef4 are not conserved between the ascomycete fungi Neurospora crassa and Fusarium graminearum. Mol Microbiol 100(3):542–559. https://doi.org/10.1111/mmi.13333
Fisher MC, Hawkins NJ, Sanglard D, Gurr SJ (2018) Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science 360(6390):739–742. https://doi.org/10.1126/science.aap7999
Fjell CD, Hiss JA, Hancock RE, Schneider G (2012) Designing antimicrobial peptides: form follows function. Nat Rev Drug Discov 11(2):168. https://doi.org/10.1038/nrd3591
Forsberg K, Woodworth K, Walters M, Berkow EL, Jackson B, Chiller T, Vallabhaneni S (2019) Candida auris: The recent emergence of a multidrug-resistant fungal pathogen. Med Mycol 57(1):1–12. https://doi.org/10.1093/mmy/myy054
Ghosh C, Sarkar P, Issa R, Haldar J (2018) Alternatives to conventional antibiotics in the era of antimicrobial resistance. Trends Microbiol 27(4):323–338. https://doi.org/10.1016/j.tim.2018.12.010
Giuliani A, Pirri G, Nicoletto SF (2007) Antimicrobial peptides: An overview of a promising class of therapeutics. Cent Eur J Biol 2:1–33. https://doi.org/10.2478/s11535-007-0010-5
Gonçalves S, Teixeira A, Abade J, de Medeiros LN, Kurtenbach E, Santos NC (2012) Evaluation of the membrane lipid selectivity of the pea defensin Psd1. Biochim Biophys Acta 1818 5:1420–1426. https://doi.org/10.1016/j.bbamem.2012.02.012
Greco I, Hansen JE, Jana B, Molchanova N, Oddo A, Thulstrup PW, Damborg P, Guardabassi L, Hansen PR (2019) Structure–activity study, characterization, and mechanism of action of an antimicrobial peptide D2 and its D- And L-peptide analogues. Molecules 24(6):1121. https://doi.org/10.3390/molecules24061121
Hamamoto K, Kida Y, Zhang Y, Shimizu T, Kuwano K (2002) Antimicrobial activity and stability to proteolysis of small linear cationic peptides with D-amino acid substitutions. Microbiol Immunol 46(11):741–749. https://doi.org/10.1111/j.1348-0421.2002.tb02759.x
Haney EF, Lau F, Vogel HJ (2007) Solution structures and model membrane interactions of lactoferrampin, an antimicrobial peptide derived from bovine lactoferrin. Biochim Biophys Acta 1768(10):2355–2364. https://doi.org/10.1016/j.bbamem.2007.04.018
Jensen RH (2016) Resistance in human pathogenic yeasts and filamentous fungi: Prevalence, underlying molecular mechanisms and link to the use of antifungals in humans and the environment. Dan Med J 63(10):B5288
Koehbach J, Craik DJ (2019) The vast structural diversity of antimicrobial peptides. Trends Pharmacol Sci 40(7):517–528. https://doi.org/10.1016/j.tips.2019.04.012
Lacerda AF, Vasconcelos EA, Pelegrini PB, Grossi de Sa MF (2014) Antifungal defensins and their role in plant defense. Front Microbiol 5:116. https://doi.org/10.3389/fmicb.2014.00116
Lawyer C, Pai S, Watabe M, Borgia P, Mashimo T, Eagleton L, Watabe K (1996) Antimicrobial activity of a 13 amino acid tryptophan-rich peptide derived from a putative porcine precursor protein of a novel family of antibacterial peptides. FEBS Lett 390(1):95–98. https://doi.org/10.1016/0014-5793(96)00637-0
Lay FT, Brugliera F, Anderson MA (2003) Isolation and properties of floral defensins from ornamental tobacco and petunia. Plant Physiol 131(3):1283–1293. https://doi.org/10.1104/pp.102.016626
Lay FT, Mills GD, Poon IK, Cowieson NP, Kirby N, Baxter AA, Van der Weerden NL, Dogovski C, Perugini MA, Anderson MA, Kvansakul M, Hulett MD (2012) Dimerization of plant defensin NaD1 enhances its antifungal activity. J Biol Chem 287(24):19961–19972. https://doi.org/10.1074/jbc.M111.331009
Lay FT, Ryan GF, Caria S, Phan TK, Veneer PK, White JA, Kvansakul M, Hulett MD (2019) Structural and functional characterization of the membrane-permeabilizing activity of Nicotiana occidentalis defensin NoD173 and protein engineering to enhance oncolysis. FASEB J 33(5):6470–6482. https://doi.org/10.1096/fj.201802540R
Levy SB, Marshall B (2004) Antibacterial resistance worldwide: causes, challenges and responses. Nature Med 10(S12):S122-S129
Li H, Anuwongcharoen N, Malik AA, Prachayasittikul V, Wikberg JE, Nantasenamat C (2016) Roles of D-amino acids on the bioactivity of host defense peptides. Int J Mol Sci 17(7):1023. https://doi.org/10.3390/ijms17071023
Lin KF, Lee TR, Tsai PH, Hsu MP, Chen CS, Lyu PC (2007) Structure-based protein engineering for α-amylase inhibitory activity of plant defensin. Proteins 68(2):530–540. https://doi.org/10.1002/prot.21378
Liu YJ, Cheng CS, Lai SM, Hsu MP, Chen CS, Lyu PC (2006) Solution structure of the plant defensin VrD1 from mung bean and its possible role in insecticidal activity against bruchids. Proteins 63(4):777–786. https://doi.org/10.1002/prot.20962
Loose C, Jensen K, Rigoutsos I, Stephanopoulos G (2006) A linguistic model for the rational design of antimicrobial peptides. Nature 443(7113):867–869. https://doi.org/10.1038/nature05233
Machado LESF, De Paula VS, Pustovalova Y, Bezsonova I, Valente AP, Korzhnev DM, Almeida FCL (2018) Conformational dynamics of a cysteine-stabilized plant defensin reveals an evolutionary mechanism to expose hydrophobic residues. Biochemistry 57(40):5797–5806. https://doi.org/10.1021/acs.biochem.8b00753
Mello EO, Taveira GB, Carvalho AO, Gomes VM (2019) Improved smallest peptides based on positive charge increase of the γ-core motif from PνD1 and their mechanism of action against Candida species. Int J Nanomed 14:407–420. https://doi.org/10.2147/IJN.S187957
Méndez E, Moreno A, Colilla F, Pelaez F, Limas GG, Mendez R, Soriano F, Salinas M, de Haro C (1990) Primary structure and inhibition of protein synthesis in eukaryotic cell-free system of a novel thionin, γ-hordothionin, from barley endosperm. Eur J Biochem 194(2):533–539. https://doi.org/10.1111/j.1432-1033.1990.tb15649.x
Méndez E, Rocher A, Calero M, Girbés T, Citores L, Soriano F (1996) Primary structure of ω-hordothionin, a member of a novel family of thionins from barley endosperm, and its inhibition of protein synthesis in eukaryotic and prokaryotic cell-free systems. Eur J Biochem 239(1):67–73. https://doi.org/10.1111/j.1432-1033.1996.0067u.x
Mirouze M, Sels J, Richard O, Czernic P, Loubet S, Jacquier A, François IE, Cammue BP, Lebrun M, Berthomieu P, Marquès L (2006) A putative novel role for plant defensins: A defensin from the zinc hyper-accumulating plant, Arabidopsis halleri, confers zinc tolerance. Plant J 47(3):329–342. https://doi.org/10.1111/j.1365-313X.2006.02788.x
Misawa T, Imamura M, Ozawa Y, Haishima K, Kurihara M, Kikuchi Y, Demizu Y (2017) Development of helix-stabilized antimicrobial peptides composed of lysine and hydrophobic α, α-disubstituted α-amino acid residues. Bioorg Med Chem Lett 27(17):3950–3953. https://doi.org/10.1016/j.bmcl.2017.07.074
Mishra B, Wang X, Lushnikova T, Zhang Y, Golla RM, Narayana JL, Wang C, McGuire TR, Wang G (2018) Antibacterial, antifungal, anticancer activities and structural bioinformatics analysis of six naturally occurring temporins. Peptides 106:9–20. https://doi.org/10.1016/j.peptides.2018.05.011
Moodley S, Koorbanally NA, Moodley T, Ramjugernath D, Pillay M (2014) The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay is a rapid, cheap, screening test for the in vitro anti-tuberculous activity of chalcones. J Microbiol Meth 104:72–78. https://doi.org/10.1016/j.mimet.2014.06.014
Muñoz A, Chu M, Marris PI, Sagaram US, Kaur J, Shah DM, Read ND (2014) Specific domains of plant defensins differentially disrupt colony initiation, cell fusion and calcium homeostasis in Neurospora crassa. Mol Microbiol 92(6):1357–1374. https://doi.org/10.1111/mmi.12634
Nouri-Sorkhabi MH, Agar NS, Sullivan DR, Gallagher C, Kuchel PW (1996a) Phospholipid composition of erythrocyte membranes and plasma of mammalian blood including australian marsupials; quantitative 31P NMR analysis using detergent. Comp Biochem Physiol B Biochem Mol Biol 113(2):221–227. https://doi.org/10.1016/0305-0491(95)02011-x
Nouri-Sorkhabi MH, Wright LC, Sullivan DR, Kuchel PW (1996b) Quantitative 31P Nuclear magnetic resonance analysis of the phospholipid of erythrocyte membranes using detergent. Lipids 31(7):765–770. https://doi.org/10.1007/BF02522893
Ochiai A, Ogawa K, Fukuda M, Ohori M, Kanaoka T, Tanaka T, Taniguchi M, Sagehashi Y (2018) Rice defensin OsAFP1 is a new drug candidate against human pathogenic fungi. Sci Rep 8:11434. https://doi.org/10.1038/s41598-018-29715-w
Parisi K, Shafee TMA, Quimbar P, Van der Weerden NL, Bleackley MR, Anderson MA (2019) The evolution, function and mechanisms of action for plant defensins. Semin Cell Dev Biol 88:107–118. https://doi.org/10.1016/j.semcdb.2018.02.004
Pelegrini PB, Lay FT, Murad AM, Anderson MA, Franco OL (2008) Novel insights on the mechanism of action of α-amylase inhibitors from the plant defensin family. Proteins 73(3):719–729. https://doi.org/10.1002/prot.22086
Poon IK, Baxter AA, Lay FT, Mills GD, Adda CG, Payne JA, Phan TK, Ryan GF, White JA, Veneer PK, Van der Weerden NL, Anderson MA, Kvansakul M, Hulett MD (2014) Phosphoinositide-mediated oligomerization of a defensin induces cell lysis. Elife 3:e01808. https://doi.org/10.7554/eLife.01808
Ramamoorthy V, Cahoon EB, Li J, Thokala M, Minto RE, Shah DM (2007) Glucosylceramide synthase is essential for alfalfa defensin-mediated growth inhibition but not for pathogenicity of Fusarium graminearum. Mol Microbiol 66(3):771–786. https://doi.org/10.1111/j.1365-2958.2007.05955.x
Ramesh S, Govender T, Kruger HG, de la Torre BG, Albericio F (2016) Short antimicrobial peptides (SAMPs) as a class of extraordinary promising therapeutic agents. J Pept Sci 22(7):438–451. https://doi.org/10.1002/psc.2894
Rekdal O, Andersen J, Vorland LH, Svendsen JS (1999) Construction and synthesis of lactoferricin derivatives with enhanced antibacterial activity. J Pept Sci 5(1):32–45. https://doi.org/10.1002/(SICI)1099-1387(199901)5:1%3c32::AID-PSC172%3e3.0.CO;2-9
Sagaram US, Pandurangi R, Kaur J, Smith TJ, Shah DM (2011) Structure-activity determinants in antifungal plant defensins MsDef1 and MtDef4 with different modes of action against Fusarium graminearum. PLoS ONE 6(4):e18550. https://doi.org/10.1371/journal.pone.0018550
Sagaram US, El-Mounadi K, Buchko GW, Berg HR, Kaur J, Pandurangi RS, Smith TJ, Shah DM (2013) Structural and functional studies of a phosphatidic acid-binding antifungal plant defensin MtDef4: Identification of an RGFRRR motif governing fungal cell entry. PLoS ONE 8(12):e82485. https://doi.org/10.1371/journal.pone.0082485
Saravanan R, Li X, Lim K, Mohanram H, Peng L, Mishra B, Basu A, Lee JM, Bhattacharjya S, Leong SS (2014) Design of short membrane selective antimicrobial peptides containing tryptophan and arginine residues for improved activity for improved activity, salt-resistance, and biocompatibility. Biotechnol Bioeng 111(1):37–49. https://doi.org/10.1002/bit.25003
Sathoff AE, Velivelli S, Shah DM, Samac DA (2019) Plant defensin peptides have antifungal and antibacterial activity against human and plant pathogens. Phytopathology 109(3):402–408. https://doi.org/10.1094/PHYTO-09-18-0331-R
Schaaper WMM, Posthuma GA, Plasman HH, Sijtsma L, Fant F, Borremans FAM, Thevissen K, Broekaert WF, Meloen RH, van Amerongen A (2001) Synthetic peptides derived from the β2-β3 loop of Raphanus sativus antifungal protein 2 that mimic the active site. J Peptide Res 57(5):409–418
Selsted ME, Brown DM, DeLange RJ, Harwig SS, Lehrer RI (1985) Primary structures of six antimicrobial peptides of rabbit peritoneal neutrophils. J Biol Chem 260(8):4579–4584
Shafee TM, Lay FT, Phan TK, Anderson MA, Hulett MD (2017) Convergent evolution of defensin sequence, structure and function. Cell Mol Life Sci 74(4):663–682. https://doi.org/10.1007/s00018-016-2344-5
Shagaghi N, Palombo EA, Clayton AH, Bhave M (2016) Archetypal tryptophan-rich antimicrobial peptides: properties and applications. World J Microbiol Biotechnol 32(2):31. https://doi.org/10.1007/s11274-015-1986-z
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. https://doi.org/10.1038/msb.2011.75
Song X, Zhang M, Zhou Z, Gong W (2011) Ultra-high resolution crystal structure of a dimeric defensin SPE10. FEBS Lett 585(2):300–306. https://doi.org/10.1016/j.febslet.2010.12.039
Souza GS, de Carvalho LP, de Melo EJT, Gomes VM, Carvalho AO (2018) The toxic effect of Vu-Defr, a defensin from Vigna unguiculata seeds, on Leishmania amazonensis is associated with reactive oxygen species production, mitochondrial dysfunction, and plasma membrane perturbation. Can J Microbiol 64(7):455–464. https://doi.org/10.1139/cjm-2018-0095
Souza GS, de Carvalho LP, de Melo EJT, da Silva FCV, Machado OLT, Gomes VM, Carvalho AO (2019) A synthetic peptide derived of the β2-β3 loop of the plant defensin from Vigna unguiculata seeds induces Leishmania amazonensis apoptosis-like cell death. Amino Acids 51(10–12):1633–1648. https://doi.org/10.1007/s00726-019-02800-8
Spelbrink RG, Dilmac N, Allen A, Smith TJ, Shah DM, Hockerman GH (2004) Differential antifungal and calcium channel-blocking activity among structurally related plant defensins. Plant Phisiol 135(4):2055–2067. https://doi.org/10.1104/pp.104.040873
Steiner H, Hultmark D, Engström A, Bennich H, Boman HG (1981) Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 292(5820):246–248. https://doi.org/10.1038/292246a0
Strøm MB, Rekdal O, Svendsen JS (2000) Antibacterial activity of 15-residue lactoferricin derivatives. J Pept Res 56(5):265–274. https://doi.org/10.1034/j.1399-3011.2000.00770.x
Sunagar K, Undheim EA, Chan AH, Koludarov I, Muñoz-Gómez SA, Antunes A, Fry BG (2013) Evolution stings: the origin and diversification of scorpion toxin peptide scaffolds. Toxins (Basel) 5(12):2456–2487. https://doi.org/10.3390/toxins5122456
Tam JP, Lu YA, Yang JL (2002) Correlations of cationic charges with salt sensitivity and microbial specificity of cystine-stabilized β-strand antimicrobial peptides. J Biol Chem 277(52):50450–50456. https://doi.org/10.1074/jbc.M208429200
Tavares PM, Thevissen K, Cammue BP, François IE, Barreto-Bergter E, Taborda CP, Marques AF, Rodrigues ML, Nimrichter L (2008) In vitro activity of the antifungal plant defensin Rs-AFP2 against Candida isolates and its in vivo efficacy in prophylactic murine models of candidiasis. Antimicrob Agents Chemother 52(12):4522–4525. https://doi.org/10.1128/AAC.00448-08
Terras FR, Torrekens S, Van Leuven F, Osborn RW, Vanderleyden J, Cammue BP, Broekaert WF (1993) A new family of basic cysteine-rich plant antifungal proteins from Brassicaceae species. FEBS Lett 316(3):233–240. https://doi.org/10.1016/0014-5793(93)81299-f
Thevissen K, Warnecke DC, François IE, Leipelt M, Heinz E, Ott C, Zähringer U, Thomma BP, Ferket KK, Cammue BP (2004) Defensins from insects and plants interact with fungal glucosylceramides. J Biol Chem 279(6):3900–3905. https://doi.org/10.1074/jbc.M311165200
Thomma BP, Cammue BP, Thevissen K (2002) Plant defensins. Planta 216(2):193–202. https://doi.org/10.1007/s00425-002-0902-6
Van der Weerden NL, Lay FT, Anderson MA (2008) The plant defensin, NaD1, enters the cytoplasm of Fusarium oxysporum hyphae. J Biol Chem 283(21):14445–14452. https://doi.org/10.1074/jbc.M709867200
Vriens K, Cools TL, Harvey PJ, Craik DJ, Spincemaille P, Cassiman D, Braem A, Vleugels J, Nibbering PH, Drijfhout JW, De Coninck B, Cammue BP, Thevissen K (2015) Synergistic activity of the plant defensin HsAFP1 and caspofungin against Candida albicans biofilms and planktonic cultures. PLoS ONE 10(8):e0132701. https://doi.org/10.1371/journal.pone.0132701
Whittington CM, Belov K (2007) Platypus venom: a review. Aust Mamm 29(1):57–62. https://doi.org/10.1071/AM07006
Wiegand I, Hilpert K, Hancock REW (2008) Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3(2):163–175. https://doi.org/10.1038/nprot.2007.521
Yount NY, Yeaman MR (2004) Multidimensional signatures in antimicrobial peptides. Proc Natl Acad Sci USA 101(19):7363–7368. https://doi.org/10.1073/pnas.0401567101
Zasloff M (1987) Magainins, a class of antimicrobial peptides from Xenopus skin: Isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc Natl Acad Sci USA 84(15):5449–5453. https://doi.org/10.1073/pnas.84.15.5449
Zhang D, He Y, Ye Y, Ma Y, Zhang P, Zhu H, Xu N, Liang S (2019) Little antimicrobial peptides with big therapeutic roles. Protein Pept Lett 26(8):564–578. https://doi.org/10.2174/1573406415666190222141905
Funding
The work was supported by the Brazilian agencies Conselho Nacional de Desenvolvimento Cientifico e Tecnologico (CNPq), Fundação de Amparo a Pesquisa do Estado do Rio de Janeiro (FAPERJ, process nº. E-26/202.760/2018-Bolsa) and Coordenacao de Aperfeiçoamento de Pessoal de Nível Superior (Finance Code 001) as well as the Universidade Estadual do Norte Fluminense Darcy Ribeiro. We acknowledge Luiz Carlos Dutra de Souza and Valeria Miguelote Kokis for their technical support.
Author information
Authors and Affiliations
Contributions
EBT participated in the peptide design, antifungal assays, and contributed to the writing of the manuscript, DRL participated in the peptide design and antifungal assays, TLBVS and SDC performed the mammalian viability assay and data analysis, EL and MFM participated in mammalian viability assay experimental design and data analysis, FZD performed statistical analyses, VMG performed experimental design, AOC participated in conception, experimental design and data analysis, contributed to the writing of the manuscript, and revised the manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare any conflicts of interest.
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling editor: J. D. Wade.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Toledo, E.B., Lucas, D.R., Simão, T.L.B.V. et al. Design of improved synthetic antifungal peptides with targeted variations in charge, hydrophobicity and chirality based on a correlation study between biological activity and primary structure of plant defensin γ-cores. Amino Acids 53, 219–237 (2021). https://doi.org/10.1007/s00726-020-02929-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-020-02929-x