Abstract
Amyloid beta peptide (Aβ), the main component of senile plaques of Alzheimer’s disease brains, is produced by sequential cleavage of amyloid precursor protein (APP) and of its C-terminal fragments (CTFs). An unanswered question is how amyloidogenic peptides spread throughout the brain during the course of the disease. Here, we show that small lipid vesicles called exosomes, secreted in the extracellular milieu by cortical neurons, carry endogenous APP and are strikingly enriched in CTF-α and the newly characterized CTF-η. Exosomes from N2a cells expressing human APP with the autosomal dominant Swedish mutation contain Aβ peptides as well as CTF-α and CTF-η, while those from cells expressing the non-mutated form of APP only contain CTF-α and CTF-η. APP and CTFs are sorted into a subset of exosomes which lack the tetraspanin CD63 and specifically bind to dendrites of neurons, unlike exosomes carrying CD63 which bind to both neurons and glial cells. Thus, neuroblastoma cells secrete distinct populations of exosomes carrying different cargoes and targeting specific cell types. APP-carrying exosomes can be endocytosed by receiving cells, allowing the processing of APP acquired by exosomes to give rise to the APP intracellular domain (AICD). Thus, our results show for the first time that neuronal exosomes may indeed act as vehicles for the intercellular transport of APP and its catabolites.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Exosomes represent a subset of extracellular vesicles with an approximate diameter of 100 nm secreted by all mammalian cells. They originate from multivesicular bodies (MVBs), which are late endosomes filled with vesicles budding from the limiting membrane into the lumen of endosomes [1]. These vesicles can be degraded inside lysosomes after fusion of MVBs with lysosomes or be released after fusion of MVBs with the plasma membrane into the extracellular milieu, where they are referred to as exosomes [2, 3]. Several populations of MVBs exist [4, 5] and intralumenal vesicles (ILVs) can be formed by different mechanisms [6,7,8,9], thus explaining the existence of heterogeneous exosome populations [10, 11].
Exosomes allow intercellular communication as they can be taken up by other cells in which their cargoes modify protein, lipid, and gene expression [12,13,14]. Whole brain and cerebrospinal fluid can be used as a source of exosomes with different cell origins [15, 16]. Indeed, cultured glial cells, i.e., oligodendrocytes [17], astrocytes [18], and microglia [19], as well as neurons [20], were all found to secrete exosomes. Release of exosomes by neurons is tightly regulated by synaptic activity, suggesting a genuine role in neuronal physiology [21,22,23]. Furthermore, we have demonstrated that neuronal exosomes bind specifically to and are taken up only by neurons, in contrast to neuroblastoma-derived exosomes, which are indifferently internalized by neurons and glial cells [24]. Thus, an elaborate intercellular communication network seems to rely on exosomes in the brain. Based on studies of prion propagation, it has been hypothesized that this network represents the substrate for the spreading within the brain of pathogenic proteins causing neurodegenerative diseases [21, 25, 26]. For example, aggregated α-synuclein, SOD1, and TDP43 are found in exosomes and would act as prions propagating Parkinson’s disease and Amyotrophic Lateral Sclerosis [27, 28]. Local injections in the brain of Aβ peptides or of hyperphosphorylated Tau lead to the formation and intracerebral spreading of amyloid plaques and neurofibrillary tangles, two hallmarks of Alzheimer’s disease but so far the mechanism of spreading remains unknown [25, 29].
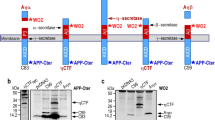
Aβ peptides are produced after sequential cleavage of the transmembrane amyloid precursor protein (APP) [30]. Main cleavages and fragments are summarized in Fig. 1a. One cleavage in the extracellular domain by the β-secretase BACE-1 occurs in endosomes, leading to a 99 amino-acid long C-terminal fragment (C99, called CTF-β herein), which contains the entire Aβ domain and accumulates in endosomes thereby leading to their dysfunction [31, 32]. CTF-β can in turn be cleaved within its transmembrane domain by γ-secretase releasing Aβ peptides and the APP intracellular domain (AICD). The latter is released in the cytosol and enters the nucleus, where it regulates transcription. APP is also cleaved at the cell surface by an α-secretase to produce a 83 amino-acid long CTF (C83, called CTF-α herein), a substrate of γ-secretase giving rise to non-amyloidogenic peptides p3 and to AICD. Recently, the extracellular domain of APP was shown to be also split by a membrane bound metalloproteinase into a long 25–30 kDa fragment referred to as CTF-η which is a privileged substrate for BACE-1 [33, 34].
APP processing is thought to mainly occur in endosomes in which exosomes are formed [4, 35,36,37]. Accordingly, APP, CTF-α, CTF-β, and Aβ have been reported in varying amounts in exosomes from various cell lines overexpressing APP [38,39,40] and on extracellular vesicles of unknown origin isolated from mouse brain [15]. However, there is today no clear consensus about which of the different APP fragments are associated with exosomes secreted by neurons and there is still no evidence that exosomes actually allow the transport of these proteins to other neurons.
Here, for the first time, we have identified the endogenous APP protein and fragments associated with exosomes released by mature neurons. Exosomes secreted by rat cortical neurons during 20 min carry APP and are strikingly enriched in endogenous CTF-α and another CTF of 26 kDa, which likely corresponds to CTF-η. Glutamatergic activation of the neurons increases the release of exosomes and thereby of APP and its fragments without changing their proportions. We then used N2a cells expressing human APP to study if a familial Alzheimer’s disease mutation can influence APP fragments exiting cells through exosomes. As in the case of cortical neurons, CTF-α and CTF-η were strikingly enriched in exosomes from N2a cells expressing non-mutated APP (APPwt). In contrast, CTF-α, CTF-β but not CTF-η, were present in exosomes secreted by N2a cells expressing APP with the Swedish mutation (APPswe) [41]. Aβ40 and Aβ42 were also highly enriched in exosomes, particularly in the case of APPswe-expressing cells. In addition, we showed that exosomes containing APP specifically target neurons in contrast to another subpopulation of exosomes containing CD63, which interact with both neurons and glial cells. Finally, using a luciferase reporter assay, we reveal that APP/CTF carried by exosomes is processed by the γ-secretase of recipient cells. Thus, our study demonstrates that a subpopulation of exosomes which bind to and are selectively endocytosed by neurons concentrate CTFs and Aβ peptides and could thus contribute to the increase of APP-derived pathological products in endosomes of receiving neurons.
Materials and methods
Chemicals
Products for cell culture (DMEM #41966052, HBSS #14175053, trypsin–EDTA #25200056, Neurobasal #21103049, B27 #17504044, PBS #14190169, l-glutamine #25030024, Minimal Essential Medium #21430) were from Life Technologies except for poly-d-lysine (P7280) and cytosine-β-d-arabinofuranoside hydrochloride (AraC, C6645), soybean trypsin inhibitor (T6522) which were from Sigma-Aldrich. Bicuculline methiodide (14343), l-glutamate (G2128), MK801 (M107), γ-secretase inhibitor N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester (DAPT, D5942) were from Sigma-Aldrich. BACE inhibitor IV was from EMD Millipore (BACE-IV, 565788). JetPEI transfection reagent was from Polyplus transfection.
Plasmids
cDNA coding for human APP695 isoform tagged with myc at its N-terminus (myc-APPwt) was originally in pRK5 vector (a generous gift from P. Paganetti, Tavern, Switzerland) and was subcloned in pCDNA3.1 between BamHI and EcoRI restriction sites. One HA tag was introduced at the C-terminus of APP in myc-APPwt construct (myc-APP-HA). myc-APPwt-GFP construct was a generous gift from P. Paganetti. APP-Gal4 vector contains a cDNA encoding for human APP695 fused in-frame at its C-terminus to the yeast transcription factor Gal4 (APP-Gal4). It was kindly provided by R. Williams and M. Perkinton (University of Bath, United Kingdom) as well as the pFR-luciferase reporter vector containing the Firefly luciferase gene (from beetle Photinus pyralis) under the control of a synthetic promoter consisting of five tandem repeats of the yeast GAL4 activation sequence upstream of a minimal TATA box. To obtain the construct coding the Nuclear Localization Sequence (“NLS”, PKKKRKV) fused in frame to Gal4 sequence at its N-terminus (NLS-Gal4), APP sequence has been removed from APP-Gal4 and replaced by Kozak plus NLS sequences by inverse PCR using the following oligonucleotides:
forward: 5′ GCCTAAAAGAAGCGTAAAGTCAAGCTACTGTCTTCTATCGAACAAG 3′reverse: 5′ CGCTTCTTTTTAGGCATGGTGGCAAGCTTGGGTCTCCCTATAGTG 3′
phRL thymidine kinase vector containing the sea pansy (Renilla reniformis) luciferase gene was used to normalize Firefly luciferase activity and was a gift from R. Williams and M. Perkinton. Swedish point mutations (KM670-671NL) have been inserted in myc-APPwt constructs with In-Fusion technology (Clontech) using the following oligos:
forward: 5′-TCTGAAGTGAATCTAGATGCAGAATTCCGACATGAC-3′;
reverse: 5′-TAGATTCACTTCAGAGATCTCCTCCGTCTTGATATT-3′.
pSuper-mCherry has been described previously [42]. Rab5Q79L-mCherry construct was a generous gift from J. Gruenberg’s laboratory (Geneva, Switzerland).
Antibodies
Rabbit polyclonal anti-myc was from Santa Cruz (sc789; 1/500 for immunofluorescence “IF”). Rabbit polyclonal anti-APP C-terminus (CT-20, 171610; 1/30000 for Western-blot “WB”) raised against the last 676–695 amino acids of APP, mouse monoclonal 22C11 recognizing amino acids 66–81 in the N-terminus of APP (MAB348; 1/1000 for WB), mouse monoclonal anti-MAP2 (MAB3418; 1/200 for IF), were from Millipore. Rat monoclonal anti-mouse CD63 from MBL (D263-3; 1/1000 for WB) was used in WB with non-reducing sample buffer. Mouse monoclonal anti-HA tag (6E2, 2367S; 1/200 for IF) was from Cell signaling. Mouse monoclonal anti-Flotillin-1 antibody was from BD Bioscience (610820; for WB). Mouse monoclonal anti-GFP (ab3277; 1/1000 for WB) was from Abcam. Homemade rabbit polyclonal anti-Alix has been described previously [43] and is sold by Covalab (pab0204). Secondary Horse-Radish-Peroxidase (HRP)-tagged antibodies were from Jackson laboratories. Fluorescently tagged secondary antibodies were from Life technologies.
Neuroblastoma cells
Neuroblastoma Neuro2a cells (N2a) and primary neurons were cultured in a humidified atmosphere in an incubator at 37 °C under 5% CO2. N2a were maintained in complete culture medium composed of DMEM containing 10% fetal calf serum, 2 mM l-glutamine. Cells were stripped with trypsine–EDTA and divided twice a week. N2aGFP–CD63 stable cell line was described previously and maintained identically [24].
Primary culture of neurons
In accordance with the policy of the Institut des Neurosciences de Grenoble (GIN) and the French legislation, experiments were done in compliance with the European Community Council Directive of 24 November, 1986 (86/609/EEC). Animals were handled and killed in conformity with European law and internal regulations of INSERM. Every effort was made to minimize the number of animals used. Embryos (E18) were removed from pregnant OFA rats from Charles River, at 18 days of gestation, under isoflurane anesthesia. The cortices and hippocampi were dissected. Cells were dissociated by mechanical pipetting and trypsin digestion before counting. Neurons were seeded on poly-d-lysine-coated dishes at 33,000 cortical neurons/cm2 or 15,600 hippocampal neurons/cm2, in DMEM with 10% horse serum. The medium was changed after 16 h for complete culture medium (Neurobasal medium with 2% B27, 2 mM l-glutamine, 1 mM sodium pyruvate). After 4 Days In Vitro (DIV), 25% of new complete medium was added to feed neurons. At the same time, 5 μM AraC was added to cortical neurons to inhibit glial cell proliferation.
Cell transfections
N2a cells were transfected 24 h after plating. For one 58 cm2 dish, DNA mix (3 μg plasmids in 300 μl 150 mM NaCl) was added to JetPEI (15 μl JetPEI in 300 μl 150 mM NaCl) and left for 20 min at room temperature before addition to cells. Hippocampal neurons were grown on poly-d-lysine-coated, 14 mm glass coverslips in 3.5 cm diameter dishes and transfected at 10 DIV by calcium phosphate precipitation. Briefly, neuron conditioned medium was collected, kept at 37 °C and replaced by transfection medium (Minimal Essential Medium, Eagle’s salts with 0.22% (w/v) sodium bicarbonate, 20 mM d-glucose, 0.5 mM l-glutamine, and 2% B27). Calcium chloride (5 μl, 2 M) was added drop by drop to 45 μl distilled water containing 2 μg DNA. This mixture was then dropped onto 50 μl 2 × BES buffer saline (14280, Sigma) before vortexing for 3 s. After 15 min at room temperature, the solution was added to neurons in transfection medium. After 1 h 30 min, neurons were washed once in transfection medium and returned to the conditioned medium.
Exosome secretion and purification
-
From cortical neurons
Cortical neurons at 15 DIV were gently washed in warm Neurobasal and were incubated in secretion medium for 20 min at 37 °C. Secretion medium was either Neurobasal alone or containing 40 μM bicuculline or 50 μM l-glutamate or 50 μM l-glutamate plus 5 μM MK801 as indicated in figures. Secretion medium was collected and exosomes were purified by differential centrifugation as described in Laulagnier et al. [44]. In brief, media were centrifuged at 2000×g, 10 min at 4 °C followed by 20,000×g 30 min at 4 °C. Supernatants were filtered through 0.2 μm Millex GV PVDF filters (Millipore) and finally ultracentrifuged at 110,000×g, 1 h 30 min at 4 °C with SW41Ti or SW32 Ti rotors.
-
From N2a cells
Exosome-free medium was made by ultracentrifuging complete culture medium at 150,000×g for 18 h followed by sterilization through 0.2 μm filters. It replaced the complete medium used for plating N2a cells after 48 h. When required, 5 μM BACE inhibitor was added to exosome-free medium. After a 20 h secretion period, supernatants were collected and exosomes were purified by differential centrifugation as above described. Depending on their use, 110,000×g pellets were resuspended either:
-
in reducing sample buffer (2% SDS, 2.5% β-mercaptoethanol, 125 mM Tris–HCl pH 6.8, 10% glycerol, 2.5% bromophenol blue) for Western-blot analysis,
-
or in lysis buffer (20 mM Tris, 150 mM NaCl, 1 mM EDTA, 1% NP40 and protease inhibitor cocktail from Roche #05 056 489 001) for anti-Aβ40, Aβ42 ELISA, BCA protein dosage,
-
or in PBS to perform nanoparticle tracking analysis,
-
or in 8% sucrose, 3 mM imidazole, pH 7.4 for sucrose gradients.
-
BCA dosage was performed as recommended by the manufacturer (Thermo Fisher). When cell lysates were required, neurons or N2a cells in one 58 cm2 dish were lysed in 1 ml lysis buffer for 30 min on ice and centrifuged at 20,000×g, 10 min at 4 °C before use.
Sucrose gradient
Exosomes were loaded on the top of a continuous gradient from 8 to 60% (0.3–1.4 M) sucrose and centrifuged for 18 h at 200,000×g [44]. Ten 1 ml fractions were collected, homogenized, and their sucrose densities determined by refractometry. Each fraction was diluted in 10 ml 3 mM imidazole, pH 7.4, and ultracentrifuged for 2 h at 200,000×g with a SW41Ti rotor. Pellets were resuspended in sample buffer for Western-blot analysis or in lysis buffer for ELISA.
Nanoparticle tracking analysis (NTA)
NTA was performed on Nanosight NS300 (Malvern) equipped with a syringe pump. N2a exosomes from 2.5 × 106 cells resuspended in PBS were diluted in 1 ml PBS and loaded in the syringe. Pump speed was set to 20, camera level was at 11 and detection threshold at 6. Five 60 s videos were captured, processed, and averaged for each type of exosomes using the NTA 3.0 software.
Western-blot analysis
Unless otherwise stated each lane contained the total amount of exosomes secreted during 20 h by 2 × 107 N2a cells. Note that the amount of protein loaded was about 25 μg, but we estimated that at least 50% of the proteins originated from the medium containing the 10% exosome-free FCS used for the culture. 20 μg of cell lysate proteins corresponding to 105 secreting cells were loaded in parallel lanes. For experiments with neurons, each lane contained exosomes secreted during 20 min by 7.8 × 106 cortical cells (at the time of plating) representing about 5 μg of exosome proteins. The amount of the cell lysates proteins was 7 μg, approximately corresponding to 1.3 × 104 cells.
Electrophoresis in glycine buffer on 8 or 10% acrylamide glycine gels was performed to detect full-length APP (APP), Alix, Flotillin-1 and GFP-CD63. Proteins were transferred in semi-dry conditions on 0.45 μm PVDF membranes (Millipore) during at least 1 h. 16% acrylamide tricine gels were used as described [45] to separate APP-CTFs and/or APP. After electrophoresis, semi-dry transfer was done on 0.2 μm nitrocellulose membranes (GE Healthcare Amersham) during 5 h. Membranes were blocked in 3% milk in Tris-buffered saline with 0.1% Tween (TBS-Tween) and incubated with primary antibodies for 1 h at room temperature or overnight at 4 °C. HRP-bound secondary antibodies were diluted 1/15,000 in TBS-Tween and incubated 1 h on membranes at room temperature. Detection reagent was 50% of 250 mM luminol and 50% of 90 mM coumaric acid with 0.015% H2O2 added extemporarily. Signal quantifications were performed using the Plot profile plugin of ImageJ software.
ELISA
Anti-human Aβ42 or human Aβ40 ELISA kits were from Anaspec (#55551 and 55552). Exosomes from 1 × 107 N2a cells expressing APPwt or APPswe and 10% of the corresponding cell lysates were analyzed according to the manufacturer’ instructions.
Electron microscopy
1.5 × 107 GFP-CD63 N2a cells were transfected with myc-APP-HA construct and exosomes were purified from their supernatants. The exosomal pellet was resuspended in 32 μl 2% paraformaldehyde in 0.1 M phosphate buffer and 4 µl were deposited on carbon-coated formvar nickel grids. After 20 min, the grids were transferred to PBS containing 50 nM glycine overnight at 4 °C and then permeabilized by 0.05% saponin in PBS-BSA for 10 min. Double immunolabelling was performed sequentially. HA was detected using mouse anti-HA followed by an anti-mouse Fab 1.4 nm (Nanoprobe, 1/100 for 1 h), finally revealed by HQ silver enhancement (also Nanoprobe) for 6 min in the dark. For CD63 labelling, grids were incubated on rat anti-CD63 for 1 h followed by biotinylated anti-rat IgG (Dako, 1/100, 1 h) revealed by streptavidin gold 6 nm (Electron Microscopy Sciences, 1/20, 30 min). At the end of the procedure, the exosomes were fixed with 1% glutaraldehyde and negatively stained with uranyl acetate. Control exosomes were treated either for only HA or CD63, or with the whole procedure omitting the primary antibodies. Observations were made using a transmission electron microscope (JEOL JEM 1200 EX) at 80 kV, equipped with a digital camera (Veleta, SIS).
Immunofluorescence labelling of exosome preparations
7 × 106 GFP-CD63 N2a cells were transfected with myc-APP-HA construct and exosomes purified from their supernatants. The exosome pellet was resuspended in PBS and allowed to adhere to coverslips previously ionized using a Plasmacleaner FEMTO (Diener electronic) for 2 min at 0.7 mbar. Coverslips were washed twice in PBS to remove unbound exosomes and then incubated with 4% PFA for 20 min. Exosomes were permeabilized using 0.05% saponin in PBS containing 0.5% BSA and 5% goat pre-immune serum. Immunostaining was performed using anti-HA mAb and Alexa596 secondary antibodies. The coverslips were washed and mounted in Mowiol (Calbiochem) for observation using a 63 × oil objective of an AxioVert 200 M inverted microscope (Zeiss). Colocalization was quantified using JACoP of ImageJ [46].
Binding of exosomes to hippocampal neurons and immunofluorescence
Purified exosomes of 1 × 107 N2a secreting cells were resuspended in Neurobasal medium and incubated on DIV17 hippocampal cells grown on a 13 mm glass coverslip. After incubation (2 or 20 h), neurons were washed with prewarmed Neurobasal medium. Neurons were fixed in 4% paraformaldehyde, 4% sucrose in PBS 20 min at room temperature and permeabilized with 0.025% w/v saponin (Sigma) in PBS containing 3% w/v of bovine serum albumin (PBS-BSA) as blocking agent. Coverslips were then incubated with indicated primary antibodies diluted in PBS-BSA during 1 h, washed in PBS, and incubated with secondary antibodies linked to Alexa488, Cyanin3, or Cyanin5 fluorochromes diluted at 1/500 in PBS-BSA. Cells were washed and incubated in Hoechst 33258 (Sigma) to label nuclei and finally mounted in Mowiol for analysis.
Microscopy and image analysis
Images were acquired with an LSM710 confocal microscope under 63 × or 40 × oil objectives. Pinholes were left at 1 airy unit for all lasers. To insure that acquisition of exosome labelling after immunofluorescence was not due to non-specific fixation of antibodies, parameters were first set up using cells not treated with exosomes but submitted to immunofluorescence with primary and secondary antibodies.
To quantify the percentage of exosomes bound to MAP2-labelled dendrites and cell bodies (Fig. 3c), the total number of fluorescent dots corresponding to labelled exosomes was first counted using Image J plugins “Analyze particles”. Thereafter, the number of each class of particles, either APPwt or APPswe or GFP-CD63 exosomes, bound to MAP2-positive structures was counted manually. In Figs. 3d and 5, neuron cultures transfected with mCherry were incubated with exosomes during 20 h before immunofluorescence against myc and MAP2. To quantify the relative binding of exosomes on dendrites and axons, Z-stacks were acquired with intervals between slices of 0.42 μm to image the entire axons (three slices) or dendrites (four slices). Pixel size was kept at 80 nm for all acquisitions. Myc-positive exosomes on axons or dendrites were counted manually on the projection of maximum intensities and length of mCherry-positive dendrite or axon was measured with Analyze/Measure tool of Image J. Pictures of APP–exosome labelling in Fig. 5 have been obtained after deconvolution using Diffraction PSF 3D plugin followed by Iterative Deconvolve 3D plugin of Image J.
Exosome APP transfer assay: luciferase reporter gene activity assay for quantification of γ-secretase-mediated cleavage of APP695-Gal4
The assay was adapted from [47]. N2a cells were plated in four-well plates (1.8 × 104 cells per well) and transfected the next day with Firefly reporter vector and Renilla normalization vector. 37 ng pFr-Firefly reporter vector, 3 ng phRL-Renilla, and 260 ng of empty pCDNA3.1 vector in 50 μl NaCl were mixed with 2 μl JetPEI in 50 μl NaCl, left 20 min at room temperature, and added to cells in one well. Purified exosomes from 4 × 107 N2a expressing APPwt-Gal4 were resuspended in 30 μl of PBS and added on one well of receiving cells 24 h after their transfection. As a positive control of the assay, one well of N2a cells was transfected with pFr-Firefly Luciferase, phRL-Renilla plus 260 ng APPwt-GAL4 without exosome addition. When required, γ-secretase inhibitor DAPT was added at 10 μM, 24 h before luciferase measurement. APP-Gal4 exosomes were left 20 h on receiving cells and luciferase activities measured using the Dual-Glo luciferase activity assay (Promega). Briefly, receiving cells were washed once in PBS and lysed in 100 μl of furnished lysis buffer 15 min at room temperature under agitation. 20 μl lysate was transferred to 96 well plates and 70 μl of Firefly activity reagent was added per well. Lumminescence was measured with Pherastar FS plate reader (BMG Labtech) during 5 s. 70 μl Stop&Glo reagent was then added to switch off Firefly activity and induce Renilla luciferase activity which was measured during 3 s. Ratios of Firefly over Renilla activity were calculated and results were shown as a fold increase of this ratio compared to mock treated cells transfected with pFr-Luci and phRL vectors without added exosomes.
Results
CTF-α is greatly enriched in exosomes secreted by cortical neurons
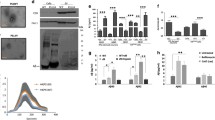
We first tested whether exosomes secreted by mature neurons contained endogenous APP and its cleavage products. For this, we used dissociated rat cortical neurons after 15 DIV, which have mature synapses and make active networks. Culture medium was removed and exosomes isolated from fresh medium incubated on neurons for 20 min. To activate both synaptic and extrasynaptic glutamate receptors, neurons were treated during this time with 40 μM bicuculline to indirectly activate glutamatergic synapses or with 50 μM glutamate. In accordance with our previous observations [23], both treatments activated the release of exosomes as revealed by the general Flotillin-1 marker, which was increased in the exosomes but not in the cell lysate fractions (Fig. 1b upper panel, d).
CTFs are enriched in neuronal exosomes. a Schematic drawing of APP and its proteolytic fragments described in the text. α, β, γ, and η refer to cleavage sites by the relevant secretases. b Exosomes released by DIV 15 cortical neurons contain APP and are enriched in CTF-α and a 26 kDa CTF proposed to be CTF-η. Western blot (WB) analysis of Flotillin-1 (Flot-1), full-length APP (APP) and C-terminal fragments (CTF-η and CTF-α) in cells and exosomes. Exosomes were harvested from supernatants of cortical neurons incubated during 20 min in control medium (−), or medium containing 40 μM bicuculline (Bic) or 50 μM glutamate (Glu). c Glutamate receptor activation increases exosomal release of APP-CTF-α and CTF-η. Densitometry of WB shows the glutamate-induced increase of APP, CTF-α, and Flotillin-1 in exosome pellets but not in cells. Means of three experiments ± Standard Error Mean “SEM” (*p < 0.05; unpaired Student t test). In the case of CTF-η, quantification is the mean of two experiments; the values of the relative protein levels are: 2.3 and 1.9 for bicuculline, and 2.9 and 2.5 for glutamate. d Inhibition of NMDA receptors by MK801 blocks glutamate-induced increase in exosomal release of APP and CTF-α. WB of exosomes released by cortical neurons incubated in control medium (−), added of 50 μM glutamate (Glu), or 50 μM glutamate together with 5 μM MK801 (Glu + MK). Note that the film showing CTF-α has been cut to remove irrelevant lanes of the same gel
CT-20 antibody directed against the last nine C-terminal amino acids of APP revealed the presence of endogenous full-length APP (APP) as well as two major CTFs running at 13 and 26 kDa (Fig. 1b). The 13 kDa fragment corresponds to CTF-α produced by the α-secretase cleavage of APP which according to Hoey et al. is by far the major form of CTFs found in cell lysates of cortical cultures [47]. The other fragment most likely corresponds to the recently characterized CTF-η (see below) [33, 34]. APP fragments described in this study are represented in Fig. 1a.
Bicuculline and glutamate both approximately doubled the amount of APP, CTF-η, and CTF-α in exosomes secreted within 20 min, without changing their apparent relative ratios. This increase was similar to that seen with the exosome marker Flotillin-1 (Fig. 1b, c). The increase in the amount of Flotillin-1, APP, and CTF-α in exosomes was abolished by MK801, an NMDA receptor antagonist (Fig. 1d). Thus, the amount of CTFs and APP in neuron supernatant is directly linked to exosome short-term release and thereby increased by NMDA receptor activation.
One striking feature of the two neuronal CTFs was their very low amount in cells and massive enrichment in exosomes (Fig. 1b). This was not the case for APP showing that the vast majority of endogenous CTFs produced by APP processing in cortical neurons is selectively sorted into exosomes. Noteworthy is that neither the cell lysates nor exosomes produced any band that could possibly correspond to CTF-β.
The APP cleavage products present in exosomes are changed by the Swedish mutation
We next assessed whether a mutation favouring amyloidogenesis could modify export of APP products by exosomes and/or influence exosome secretion. For this, we turned to mouse neuroblastoma N2a cells overexpressing the human APP695 isoform, either wild type (APPwt) or carrying the autosomal dominant Swedish mutation KM670NL (APPswe). This mutation is known to increase the processing of APP by β secretase and thereby the production of CTF-β and Aβ peptides [41, 48, 49]. As shown in Fig. 2a, cells expressed mainly APP and low amounts of CTFs. CTF-α was detected in equal amounts in APPwt and APPswe N2a cells. An extra CTF running just above CTF-α was identified as CTF-β, since it was only detectable in APPswe cells. Exosomes secreted by both cell types contained similar amounts of APP and CTF-α, while CTF-β was detected only in exosomes of APPswe cells. Instead of CTF-β, exosomes from APPwt cells contained a CTF just above 26 kDa, corresponding to the molecular weight of CTF-η (Fig. 2a). This fragment was almost undetectable in APPwt N2a cells and in exosomes from APPswe cells. Based on the immunoreactivity of the different CTFs, the amount of the 26 kDa fragment in APPwt exosomes was equivalent to that of CTF-β in APPswe exosomes. This reinforces the possibility that this fragment is indeed CTF-η, since it is a known substrate of β-secretase and the swe mutation enhances the processing of APP substrates by β-secretase [33, 34]. Accordingly, incubation of APPswe-expressing N2a cells with a BACE-1 inhibitor almost abolished the presence of CTF-β while increasing CTF-η in exosomes (Fig. 2b). As expected, the amount of CTF-α was not affected by this treatment. Similar to our observation with cortical neurons, all CTFs but not APP holoproteins were strikingly enriched in exosomes secreted by N2a cells overexpressing APPwt and APPswe (Fig. 2a, b). This enrichment was comparable to that seen with Alix and CD63. Alix is a regulator of the Endosomal Sorting Complexes Required for Transport (ESCRT) system, while CD63 is a tetraspanin protein. Both proteins participate in the formation of and concentrate inside ILVs and are, therefore, often used as exosome markers.
Exosomes released by N2a cells expressing human APPwt and APPswe are enriched in CTFs and Aβ peptides. a Exosomes contain APP and are enriched in CTF-α and two distinct CTFs depending on the APP form expressed. The upper panel is a WB of cells and exosomes from N2a cells (−), or N2a expressing APPwt (APPwt) or APPswe (APPswe) labelled with an antibody against the N-terminal part of APP to reveal full-length APP. CTFs were revealed using antibodies against the C-terminal part of APP. The lower panels are WB of cells and exosomes from N2a cells or APPwt- or APPswe-expressing N2a cells showing that expression of these proteins does not apparently change the amount of two exosome markers, Alix or CD63. Note that for both WB of Alix and CD63, the film have been cut to remove irrelevant lanes of the same gel. b BACE-1 inhibition (BI) blocks the appearance of CTF-β and increases the amount of CTF-η. Alix was used as an exosome marker. c Aβ peptides are enriched in exosomes compared to cells. Aβ42 and Aβ40 were quantified by ELISA. Means of 4–5 experiments ± SEM. *p < 0.05 **p < 0.01 ***p < 0.001. d–f Aβ40, CTF-α, and CTF-β are present in the exosomal fractions containing CD63. Exosomes released by GFP-CD63 N2a cells transfected with APPwt (d) or APPswe (e, f) were separated on sucrose density gradients. Two fractions were collected and analyzed by WB (d, e). ELISA was also used to detect Aβ40 in the fractions (f). Sucrose concentration of exosome-containing fractions is indicated in g/mL
Using ELISA, we found that, like CTFs, both amyloid peptides Aβ42 and Aβ40 were present and enriched in exosomes compared to cell extracts of APP-overexpressing N2a cells (Fig. 2c). In the case of APPswe-overexpressing cells, Aβ40 was seven times more concentrated in exosomes than in cells (Fig. 2c). Noteworthy, the amount of Aβ peptides tended to be higher in cell lysates and exosomes of APPswe than of APPwt, with Aβ40 being 4.9 times more concentrated in exosomes of APPswe, than of APPwt N2a.
To confirm the presence of Aβ in exosomes, we floated the vesicles on linear sucrose gradients. In this case, we used exosomes secreted by N2a cells stably expressing GFP-CD63 [24]. Exosomes secreted by GFP-CD63 N2a cells expressing APPwt (Fig. 2d) or APPswe (Fig. 2e) were separated according to their density on sucrose gradients and fractions analyzed by Western blots. CTF-α was detected in GFP-CD63-exosome-containing fractions in the case of APPwt cells. CTF-α and CTF-β were present in similar exosome-containing fractions in the case of vesicles secreted by APPswe cells. In Fig. 2f, ELISA analysis of sucrose gradient fractions revealed the presence of Aβ40 in the exosome fractions, suggesting that the hydrophobic part of the peptide is still inserted in the lipid bilayer of exosomes or that the peptide is tightly bound to a receptor of the exosomal membrane.
Noteworthy is that overexpression of APPswe had no detectable effects on the production or release of exosomes. Indeed, neither APPwt nor APPswe expressions noticeably changed the amount of exosomes released, as monitored by immunodetection of two exosomal markers, Alix, and CD63 (Fig. 2a). In addition, nanoparticle tracking analysis used to quantify nanovesicles revealed that the number of particles secreted by N2a cells was not significantly different between non transfected, APPwt-, or APPswe-transfected cells (Sup Fig. 1a). Furthermore, exosomes secreted by control, APPwt-, and APPswe-N2a cells had similar sizes of 98.5, 100, and 96.3 nm, respectively (Sup Fig. 1b). Thus, APP overexpression or amyloidogenesis induced by APPswe had no obvious effect on the number and size of exosomes.
APP–exosomes bind specifically to neurons
The next step of our study was to follow the possible binding of APP-containing exosomes to neural cells. Here, we used exosomes secreted by N2a cells expressing APP with both myc- and HA-tags on the N-terminal and C-terminal parts respectively (myc-APP-HA). Incubation of APP–exosomes on hippocampal primary cell cultures followed by anti-myc and anti-HA antibody staining revealed double immunostained exosomes only around cells looking like neurons (Fig. 3a). Using MAP2 immunostaining to selectively label neurons, we confirmed that APP–exosomes bound to neuronal soma and dendrites. Importantly, we did not detect any obvious difference in the binding specificity of exosomes carrying APPwt and those carrying APPswe (Fig. 3b). GFP-CD63-expressing N2a cells can serve as a source of fluorescent exosomes. In contrast to APP exosomes, but as expected from our previous work [24], GFP-CD63 exosomes bound indifferently to neurons and glial cells present in the cultures (Fig. 3b). A few GFP-CD63 exosomes may also have bound directly to the poly-d-lysine-coated substrate. 80–90% APP–exosomes bound to MAP2-positive neuronal soma or dendrites. This specificity of binding was not significantly different between exosomes containing wt or swe APP. In sharp contrast, only 30% of GFP-CD63 exosomes bound to MAP2-positive structures (Fig. 3c). We also used hippocampal cultures transfected with mCherry, in which the fluorescent protein is sometimes expressed in, and delineates, flat glial cells. This allowed discrimination between MAP2-positive dendrites and underlying glial cells and observation of APP–exosomes decorating dendritic processes but not glial cells (Fig. 3d). Thus, our results indicate that exosomes secreted by N2a cells overexpressing APPwt or APPswe preferentially interact with neurons, in sharp contrast to those secreted by GFP-CD63 N2a, which bind to both neurons and glial cells.
APP exosomes bind specifically to neurons. a Exosomes purified from myc-APP-HA-expressing N2a supernatants were incubated during 2 h on DIV 17 hippocampal cells. Double immunofluorescence was used to localize the myc-tagged N-terminal- (green) and the HA-tagged C-terminal (magenta) parts of APP on exosomes. Colocalizations of the tags appear as white dots and demonstrate full-length APP-containing exosomes labelling of neurites. b APP-containing exosomes bind specifically to neurites in contrast to CD63 containing exosomes which bind to all cells. Exosomes purified from supernatants of GFP-CD63 N2a (GFP-CD63 exosomes) or N2a cells expressing myc-APPwt (APPwt exos) or mycAPPswe (APPswe exos) were incubated on hippocampal cultures and APP exosomes localized by immunofluorescence with anti-myc antibody (green). Co-staining with anti-MAP2 (magenta) shows that APP exosomes label MAP2-positive dendrites in contrast to GFP-CD63 exosomes which bind to all cells of the culture. c Percentage of each class of exosomes bound to MAP2-positive neuron cell bodies and dendrites. d Hippocampal cells transfected with mCherry were incubated with APP exosomes which were revealed by myc immunostaining (APPwt exos). Neurons were labelled using anti-MAP2 (Magenta). The picture shows APP exosomes bound to MAP2-positive dendrites but not to the underlying mCherry-expressing, MAP2-negative glial cell. Nuclei were stained with Hoechst (blue in a–c). Bars 10 μm
We searched for the reason for this different binding specificity using exosomes purified from N2a cells expressing both GFP-CD63 and APP. Sucrose gradient separation of such exosomes showed that APP, CTF-α and GFP-CD63 were present in the same exosome-containing fractions (Figs. 2d, e and 4a). However, electron microscopic observation of exosome secreted by N2a cells expressing myc-APP-HA and GFP-CD63 using double immunogold labelling for CD63 and HA indicated that CD63 and APP were present almost exclusively on different exosome-like vesicles (Fig. 4b). This lack of colocalization of APP and CD63 on exosomes was confirmed by immunofluorescence of the same exosome preparation using anti-HA (Fig. 4c).
APP and CD63 define exosome subpopulations with different target cells. Exosomes were purified from culture supernatants of N2a cells expressing both GFP-CD63 and myc-APPwt (a, d) or myc-APPwt-HA (b, c). a Exosomes were separated on a linear sucrose gradient, as shown in Fig. 2. Myc-APPwt, Flotillin-1, and GFP-CD63 appear in exosomal fractions containing 1.11–1.15 g/mL of sucrose. b Co-immunogold staining with anti-HA (6 nm gold particles) and anti-CD63 (irregular silver deposits > 10 nm) demonstrates the presence of myc-APP-HA and GFP-CD63, respectively, in different vesicles. Bar 100 nm. c Immunofluorescence staining of purified exosomes with anti-HA confirms that APP and GFP-CD63 are localized on different objects (Pearson’s correlation coefficient between APP staining and GFP calculated on 20,000 objects with JACoP of ImageJ: r = 0.01) Bar 5 μm d APP exosomes [APP exos (anti-myc), red] and CD63 exosomes have different binding specificities towards neural cells. Exosomes were incubated on hippocampal cells during 2 h. On the transmitted light picture, circles delineate nuclei of neurons (black) and glial cells (white). Bar 10 μm
APP/GFP-CD63 exosomes were incubated on hippocampal cells and exosomes were revealed by immunofluorescence. GFP-containing-dots similar to those seen with exosomes from GFP-CD63 N2a were observed on both neurons and glial cells (Fig. 4d). In contrast, immunostaining of APP revealed fluorescent dots only on neurites, similar to those seen using exosomes from APPwt N2a. Noteworthy, GFP-CD63 and APP were almost never co-localized (Fig. 4d) showing that in most cases, APP and CD63 were present in two distinct exosome populations having different binding specificities towards neural cells (Fig. 6).
In hippocampal neurons expressing mCherry (Fig. 5a), the fluorescent protein delineates all processes. Among these, the thin and smooth MAP2-negative axons (arrowheads) can be distinguished from the spiny MAP2-positive dendrites. This revealed that APP–exosomes mainly bound to dendrites (Fig. 5b–d) but had no apparent preference for any dendrite sub-compartment, localizing indifferently on dendritic shafts, spine necks or spine heads (Fig. 5e).
APP exosomes mainly bind to dendrites and can be endocytosed by hippocampal neurons. Hippocampal neurons expressing mCherry were incubated for 20 h with APP exosomes. a APP exosomes were revealed with anti-myc [APPexos (anti-myc), green] and dendrites labelled with anti-MAP2 (magenta). Arrowheads point to thin mCherry-filled processes identified as axons because of their MAP2-negativity. b, c Higher magnifications illustrate that APP exosomes bind less to axons (b) than to dendrites (c). Bars 20 μm d quantification of the number of APP exosomes counted per μm of dendrites or axons. Mean of four experiments ± SEM (p** < 0.01, unpaired Student t test). e APP exosomes revealed by anti-myc [APP exos (anti-myc), green] bound to mCherry-expressing dendrites. Photographs illustrate APP exosomes localized on dendritic spine necks (empty arrowhead), dendritic spine heads (white arrowheads), or dendritic shaft (white arrow). Bar 10 μm. f Endocytosis of APP exosomes by hippocampal neurons. Myc-APP-GFP exosomes were incubated for 2 h on Rab5-Q79L-mCherry-expressing neurons (red) and immunostained with anti-myc (magenta). Rab5-Q79L-mCherry delineates enlarged endosomes in which GFP can be detected (green). g–j Higher magnifications illustrating individual dilated Rab5-Q79L-mCherry endosomes containing GFP–exosomes. g shows a Z-stack of four slices into an enlarged endosomes. h–j are individual Z slices of enlarged endosomes. Note that colocalized myc and GFP spots appearing as white dots are only detected on the cell surface, outside endosomes (white empty arrowheads in g, i, j). Bars 10 μm
Exosomal CTFs are endocytosed by receiving neurons
In order to demonstrate that APP–exosomes can be endocytosed after their binding to the neuronal surface, we incubated APP–exosomes on hippocampal neurons expressing Rab5-Q79L-mCherry. This dominant positive mutant of Rab5 does not impede early steps of endocytosis from plasma membrane to endosomes, but inhibits the maturation of endosomes from early to late endosomes/lysosomes [50, 51]. Rab5-Q79L expression induces formation of aberrantly enlarged endosomes which accumulate endocytosed material and thus clearly delineates the early endosome limiting membrane and their lumen (Fig. 5f) [51]. Exosomes were from N2a cells expressing APP with myc- and GFP on the N-terminal and C-terminal parts, respectively (myc-APP-GFP). This alternative C-terminal tag did not change the binding specificity of exosomes as GFP spots decorated the surface of neurons similar to the situation observed using myc-APP-HA-containing exosomes. The staining was also different from that observed with GFP-CD63 exosomes, indicating that the lack of specificity seen with the latter is not a particularity of GFP. However, the number of spots per unit area of neurite was less abundant, in accordance with the lower incorporation of myc-APP-GFP in exosomes compared to that of myc-APP-HA (Williamson et al. in preparation). Strikingly, GFP dots could be detected within Rab5-Q79L-positive endosomes most likely representing endocytosed exosomes blocked during their traffic inside early endosomes (Fig. 5g–j). Exosomes bound to the surface were often labelled with both GFP and myc, demonstrating the presence of intact APP in exosomes (open arrowheads in Fig. 5g, i, j). Such colocalization was very rarely seen on GFP spots detected inside endosomes, suggesting cleavage of the extracellular domain of exosomal APP inside endosomes. Alternatively, the fact that some GFP-containing exosomes from the cell surface were not labelled by myc might suggest that exosomes containing only CTF are more efficiently endocytosed than those containing the holoprotein (Fig. 5f, j). In summary, these observations show that exosomal APP and CTFs bound to the neuronal surface can be endocytosed and traffic through neuronal endosomes (Fig. 6).
Schematic illustration of the results obtained. APP is endocytosed (step 1) and processed into CTFs inside endosomes which also contain CD63 (step 2). APP-CTFs are sorted into different populations of intraluminal vesicles accumulating inside late endosomes called multivesicular bodies (MVBs). MVBs can fuse with the plasma membrane thereby releasing the intraluminal vesicles which once outside are referred to as exosomes (step 3). CD63–exosomes indifferently bind to neurons and glial cells whereas APP–exosomes bind specifically to, and can be endocytosed by neurons (steps 4 and 5). Note that for simplification, we have represented a differential sorting of APP and CD63 into ILV of the same MVB, but the possibility remains that APP and CD63 are sorted into different MVB populations. Note that our work cannot exclude that APP fragments are sorted into vesicles budding from the cell surface (not represented here)
After endocytosis, exosomal CTFs can be processed by γ-secretase in the receiving cell
We next examined if some of the APP and CTFs acquired through exosomes might be processed in the same way as endogenously expressed APP. CTF cleavage by γ-secretase gives rise to a cytosolic fragment (AICD) released into the cytosol and might enter the nucleus to activate transcription. We used a Gal4-reporting system which allows demonstration of the appearance of AICD. APP is fused at its C-terminal part with the yeast Gal4 transcription factor (APP-Gal4) and the Gal4 reporter is a luciferase expression vector driven by the UAS (Upstream Activation Sequence) [47, 52]. As expected, in N2a cells co-transfected with both APP-Gal4 and UAS-luciferase, luciferase expression was detected by luminescence, reflecting the presence of AICD-Gal4. Luminescence was almost completely abolished by the γ-secretase inhibitor DAPT, demonstrating that luciferase expression faithfully reflects CTF-GAL4 processing by the γ-secretase (Fig. 7a). DAPT had no effect on luciferase activity induced by overexpression of Gal4 transcription factor directly targeted to the nucleus (NLS-Gal4, Sup Fig. 2). We next demonstrated that N2a cells expressing only APP-Gal4 secrete exosomes containing full-length APP-Gal4 and CTF-Gal4 (Fig. 7b). These exosomes were then incubated on target N2a cells expressing only the reporter plasmid UAS-luciferase. After 20 h of incubation, luminescence was significantly higher compared to control cells not incubated with exosomes (Fig. 7c). In addition, this increased luciferase activity due to APP-Gal4 exosomes was abolished upon co-incubation with the γ-secretase inhibitor DAPT. These results demonstrate that at least a fraction of APP or APP-CTF carried by exosomes has been inserted in endosomal membranes of the receiving cells and cleaved down to the final AICD product (Fig. 7d).
Exosomal CTFs can be processed in receiving cells. a Luciferase activation in N2a cells co-transfected with APP fused with Gal4 (APP-Gal4) and the UAS-Firefly luciferase reporter demonstrates APP cleavage by γ-secretase into AICD-Gal4. UAS-Firefly luciferase was transfected alone (Mock) or together with APP-Gal4. Renilla luciferase was co-expressed to normalize Firefly activity and luciferase activities measured 48 h after the transfections. γ-secretase inhibitor (DAPT) was added 20 h before measurement. Average of five experiments for APP-Gal4 and three experiments for DAPT conditions are shown ± SEM. (**p < 0.01, unpaired Student t test). b WB analysis of exosomes secreted by N2a cells expressing APP-Gal4 demonstrates the presence of APP-Gal4 and CTF-Gal4 in exosomes. c APP-Gal4 exosomes incubated for 20 h on N2a transfected with the UAS-Firefly luciferase reporter only, activate luciferase. The γ-secretase inhibitor DAPT blocks this activation indicating cleavage of CTF-Gal4 into AICD in the recipient cells. Firefly luciferase activity was measured as in a. Average of four experiments ± SEM. *p < 0.05, unpaired Student t test. d Schematic representation of the way exosomes containing APP-Gal4 would induce luciferase activity in cells transfected with UAS-Firefly luciferase only. Exosomes containing CTF-Gal4 are endocytosed by UAS-luciferase transfected cells. Once inside endosomes, exosomes fuse with the limiting endosomal membrane. CTF-Gal4 is thereby inserted into the membrane and accessible to cleavage by γ-secretase. AICD-Gal4 is released and enters the nucleus to activate transcription of the luciferase gene
Discussion
Cellular mechanisms underlying the slow progression of Alzheimer’s disease through the brain are still poorly understood. One current hypothesis is that pathogenic proteins including Aβ peptides spread via exosomes throughout the parenchyma. APP and its proteolytic products have already been reported in exosomes secreted by several cell lines overexpressing APP [38,39,40, 53] and in vesicles from unknown origin purified from total mouse brains [15]. Neurons differ from cell lines by their highly compartmentalized organisation which requires sophisticated sorting systems. It was, therefore, crucial to describe how APP processing into exosomes occurs in mature neurons as well as to understand whether exosomes can bind and transfer APP and its fragments to other neural cells. To study APP processing into exosomes, we chose 2-week-old cultures of dissociated cortex which contain both glutamatergic and GABAergic neurons making functional networks. We have previously used such cultures to demonstrate that a short increase in glutamatergic activity triggered MVB fusion with the plasma membrane and thereby exosome release [23]. Here, we found that these exosomes contain full-length APP and two CTFs, one running around 13 kDa, the other fainter one just above 26 kDa (Fig. 1b). APP products with similar molecular weights were detected in exosomes of N2a cells expressing human APP. In the case of APPswe-expressing N2a, however, exosomes contained only two low molecular weight CTFs, the lower one running around 13 kDa (Fig. 2). The upper fragment most probably corresponds to CTF-β, the other one being CTF-β, as the Swedish mutation is known to enhance cleavage of APP by BACE-1, thereby increasing the concentration of CTF-β [48]. The upper band of the doublet disappeared from lysates and exosomes after cells was incubated with a BACE-1 inhibitor confirming this interpretation. Therefore, exosomes secreted by N2a cells overexpressing APP or by primary cortical cultures both contain CTF-α but no CTF-β. Instead, they contain a CTF with a molecular weight around 26 kDa, similar to the recently described CTF-η which is a product of the metallo-protease MT5-MMP and a substrate of BACE-1 [33, 34]. The level of this fragment was increased in exosomes of APPswe N2a cells by the treatment with the BACE-1 inhibitor, further suggesting that it indeed corresponds to CTF-η. Interestingly, all three CTFs were highly enriched in exosomes compared to the APP holoprotein which was equally detected in cells and exosomes. This is consistent with the previous observations using immuno-electron microscopy that MVBs of neurons from APP-PS1 transgenic mice are filled with CTFs and Aβ but lack detectable N-terminal APP fragments [54]. Thus, our results show for the first time that in mature cortical neurons, APP is mainly processed into CTF-α and CTF-η, most of which is sorted into intraluminal vesicles of MVBs.
Short-term activation of glutamatergic synaptic receptors increased secretion of this ready releasable pool of CTFs. Neurons which lack lysosomes in distal dendrites might heavily rely on the fusion of MVBs to the plasma membrane to eliminate intracellular Aβ and avoid the deleterious accumulation of CTFs in endosomes. This could be particularly crucial at synapses, where APP processing is enhanced by glutamatergic activity [55, 56]. However, one possible pitfall of such an elimination process is that cleavage of CTF-η by α-secretase also leads to a soluble synaptotoxic Aη peptide [33]. The α-secretase ADAM-10 is known to be associated with exosomes [40, 57] and further studies should test if CTF-η inserted in exosomes released by dendrites can give rise to toxic Aη in the synaptic vicinity. Aβ peptides were enriched in exosomes compared to cells, and as expected from previous work [58,59,60], the level of amyloid peptides tended to be higher in APPswe than in APPwt-expressing N2a cells. Accordingly, there was nearly five times more Aβ40 in exosomes from APPswe- than in those of APPwt-expressing N2a (Fig. 2c). Importantly, density separation showed for the first time that Aβ peptides float together with exosomes, demonstrating their tight association with membrane vesicles. One explanation for this is that the hydrophobic part of Aβ produced after γ-secretase cleavage of CTFs remains embedded in the exosome membrane. Another possibility is that soluble Aβ oligomers are bound to the surface of exosomes by the prion protein PrPc, as recently demonstrated with N2a exosomes using synthetic peptides [61]. In that study, exosomal PrPc was found to accelerate Aβ fibrilization and suggested to protect against the neurotoxic effects of the oligomeric peptide. A similar mechanism might occur in vivo [62] and we have previously shown that PrPc is highly enriched in exosomes purified from cortical neurons [20]. In this case, one function of the exosomes released into parenchymal interstitial space would be to sequester toxic Aβ oligomers released upon fusion of late endosomes to the cell surface in the vicinity of synapses.
The relevance of extracellular versus intracellular Aβ for neuronal toxicity is a source of considerable debate fed by multiple observations of early endosomal abnormalities and progressive intraneuronal Aβ accumulation in the human brain with AD [36, 63]. Endosomal abnormalities could result from accumulation of Aβ or CTF-β [32, 64], normally eliminated by the fusion of late endosomes with the cell membrane. Strikingly SorLA, Bin1, PICALM, and CD2AP, which are linked to increased risk for late onset AD [65, 66], all have functions related to endocytosis and endosome trafficking. Ubelmann et al. have already shown that depletion of CD2AP inhibits APP and/or CTFs sorting towards ILVs and concomitantly increases Aβ production in post-synaptic endosomes [67]. Exosomal proteins released by neurons within few minutes represent an easily accessible sampling of ILVs. Thus, exosomes give unique opportunities to understand how proteins linked to late onset forms of the disease affect endosomal trafficking and sorting especially of APP catalytic products.
Heterogeneity of extracellular vesicles secreted by several cell lines has already been reported [10, 11] and we found that APP and CD63 are sorted into distinct exosome populations. This might be explained by the fact that APP sorting inside ILVs depends on the Endosomal Sorting Complexes Required for Transport (ESCRT) [4], while CD63 can be involved in ESCRT-independent sorting into ILVs [68]. Monitoring the binding of CD63- versus APP-containing exosomes allowed us to demonstrate that two distinct populations of N2a exosomes carrying different cargoes have different binding specificities (Fig. 6). To our knowledge, the possibility that different vesicles secreted by one cell type might target different kinds of cells has never been envisaged before. We have previously used GFP fused to the C-terminal fragment of the heavy chain of Tetanus toxin (TTC) to label exosomes released by cortical neurons. TTC is endocytosed after binding to its receptor of the neuronal surface sorted into intraluminal vesicles of MVBs and released bound to exosomes [23]. Purified GFP-TTC-labelled-neuronal exosomes specifically bound to neurons of mixed hippocampal cultures [24], similar to N2a exosomes containing APP. Thus, APP and CTFs are sorted into a subset of ILVs/exosomes, which specifically bind to neurons and have the capacity to act as vehicles between neurons. However, even if the presence on secreted vesicles of CTF fragments produced inside endosomes is compatible with a possible endosomal origin of the vesicles, we cannot exclude, yet that these latter may be ectosomes directly budding from the plasma membrane. Separation of CD63- from APP-containing vesicles followed by their proteomic characterization may help to resolve this issue.
Further studies will also be required to identify which ligand allows this remarkable binding specificity for neurons of APP-carrying exosomes secreted by cortical neurons or by N2a cells. One candidate is APP itself which has many known neuronal ligands [69] including APP, which is more highly expressed on neurons than on glial cells [70, 71].
Endocytosis is the best accepted way of entry of exosomes in receiving cells. We have blocked trafficking through early endosomes by Rab5-Q79L to show that this is also the case in neurons. After incubation with APP-GFP exosomes, GFP fused with the C-terminal part of APP was detected in patches inside Rab5-Q79L swollen endosomes. Noteworthy, myc staining was almost never colocalized with GFP inside endosomes, suggesting that the extracellular part of APP carried by exosomes had been cleaved. Similar to the reported backfusion of intraluminal vesicles with the endosomal membrane [72], exosome fusion could occur and allow insertion of CTFs in the limiting endosomal membrane. Accordingly, we showed that part of CTFs can be further processed inside receiving cells by γ-secretase to give rise to AICD (Fig. 7d). Further work will be required to test if this mechanism could contribute to the formation of intracellular Aβ. In conclusion, our data demonstrate that CTFs are enriched in exosomes secreted by cortical neurons. This process might allow localized elimination of these potentially toxic fragments from endosomes of synapses far away from lysosomes, particularly in the most distal parts of dendrites. Using neuroblastoma cells, we also show that APP-CTFs and Aβ are selectively sorted into a subpopulation of exosomes that specifically bind to, and are endocytosed by, neurons, thus suggesting that some of these vesicles might contribute to the spreading of pathological fragments throughout the brain. Therefore, our study leads to a better understanding of the molecular mechanisms allowing exosome transfer between neurons, a prerequisite to testing the beneficial or deleterious role of exosomes in AD.
References
Gruenberg J, Stenmark H (2004) The biogenesis of multivesicular endosomes. Nat Rev Mol Cell Biol 5:317–323. doi:10.1038/nrm1360
Denzer K, Kleijmeer MJ, Heijnen HF et al (2000) Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. J Cell Sci 113:3365–3374
Laulagnier K, Schieber NL, Maritzen T et al (2011) Role of AP1 and Gadkin in the traffic of secretory endo-lysosomes. Mol Biol Cell 22:2068–2082. doi:10.1091/mbc.E11-03-0193
Edgar JR, Willén K, Gouras GK, Futter CE (2015) ESCRTs regulate amyloid precursor protein sorting in multivesicular bodies and intracellular amyloid-β accumulation. J Cell Sci 128:2520–2528. doi:10.1242/jcs.170233
White IJ, Bailey LM, Aghakhani MR et al (2006) EGF stimulates annexin 1-dependent inward vesiculation in a multivesicular endosome subpopulation. EMBO J 25:1–12. doi:10.1038/sj.emboj.7600759
Matsuo H, Chevallier J, Mayran N et al (2004) Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science 303:531–534. doi:10.1126/science.1092425
Subra C, Laulagnier K, Perret B, Record M (2007) Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie 89:205–212. doi:10.1016/j.biochi.2006.10.014
Trajkovic K, Hsu C, Chiantia S et al (2008) Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319:1244–1247. doi:10.1126/science.1153124
Wollert T, Hurley JH (2010) Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 464:864–869. doi:10.1038/nature08849
Colombo M, Raposo G, Théry C (2014) Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 30:255–289. doi:10.1146/annurev-cellbio-101512-122326
Kowal J, Arras G, Colombo M et al (2016) Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci USA 113:E968–E977. doi:10.1073/pnas.1521230113
Nanbo A, Kawanishi E, Yoshida R, Yoshiyama H (2013) Exosomes derived from Epstein-Barr virus-infected cells are internalized via caveola-dependent endocytosis and promote phenotypic modulation in target cells. J Virol 87:10334–10347. doi:10.1128/JVI.01310-13
Tian T, Zhu Y-L, Zhou Y-Y et al (2014) Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J Biol Chem 289:22258–22267. doi:10.1074/jbc.M114.588046
Valadi H, Ekström K, Bossios A et al (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9:654–659. doi:10.1038/ncb1596
Perez-Gonzalez R, Gauthier SA, Kumar A, Levy E (2012) The exosome secretory pathway transports amyloid precursor protein carboxyl-terminal fragments from the cell into the brain extracellular space. J Biol Chem 287:43108–43115. doi:10.1074/jbc.M112.404467
Street JM, Barran PE, Mackay CL et al (2012) Identification and proteomic profiling of exosomes in human cerebrospinal fluid. J Transl Med 10:5. doi:10.1186/1479-5876-10-5
Fitzner D, Schnaars M, van Rossum D et al (2011) Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. J Cell Sci 124:447–458. doi:10.1242/jcs.074088
Wang G, Dinkins M, He Q et al (2012) Astrocytes secrete exosomes enriched with proapoptotic ceramide and prostate apoptosis response 4 (PAR-4): potential mechanism of apoptosis induction in Alzheimer disease (AD). J Biol Chem 287:21384–21395. doi:10.1074/jbc.M112.340513
Potolicchio I, Carven GJ, Xu X et al (2005) Proteomic analysis of microglia-derived exosomes: metabolic role of the aminopeptidase CD13 in neuropeptide catabolism. J Immunol 175:2237–2243
Fauré J, Lachenal G, Court M et al (2006) Exosomes are released by cultured cortical neurones. Mol Cell Neurosci 31:642–648. doi:10.1016/j.mcn.2005.12.003
Chivet M, Hemming F, Pernet-Gallay K et al (2012) Emerging role of neuronal exosomes in the central nervous system. Front Physiol 3:145. doi:10.3389/fphys.2012.00145
Chivet M, Javalet C, Hemming F et al (2013) Exosomes as a novel way of interneuronal communication. Biochem Soc Trans 41:241–244. doi:10.1042/BST20120266
Lachenal G, Pernet-Gallay K, Chivet M et al (2011) Release of exosomes from differentiated neurons and its regulation by synaptic glutamatergic activity. Mol Cell Neurosci 46:409–418. doi:10.1016/j.mcn.2010.11.004
Chivet M, Javalet C, Laulagnier K et al (2014) Exosomes secreted by cortical neurons upon glutamatergic synapse activation specifically interact with neurons. J Extracell Vesicles 3:24722. doi:10.3402/jev.v3.24722
Coleman BM, Hill AF (2015) Extracellular vesicles–Their role in the packaging and spread of misfolded proteins associated with neurodegenerative diseases. Semin Cell Dev Biol 40:89–96. doi:10.1016/j.semcdb.2015.02.007
Février B, Vilette D, Laude H, Raposo G (2005) Exosomes: a bubble ride for prions? Traffic 6:10–17. doi:10.1111/j.1600-0854.2004.00247.x
Emmanouilidou E, Melachroinou K, Roumeliotis T et al (2010) Cell-produced alpha-synuclein is secreted in a calcium-dependent manner by exosomes and impacts neuronal survival. J Neurosci 30:6838–6851. doi:10.1523/JNEUROSCI.5699-09.2010
Feiler MS, Strobel B, Freischmidt A et al (2015) TDP-43 is intercellularly transmitted across axon terminals. J Cell Biol 211:897–911. doi:10.1083/jcb.201504057
Jucker M, Walker LC (2013) Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature 501:45–51. doi:10.1038/nature12481
Haass C, Kaether C, Thinakaran G, Sisodia S (2012) Trafficking and proteolytic processing of APP. Cold Spring Harb Perspect Med 2:a006270. doi:10.1101/cshperspect.a006270
Xu W, Weissmiller AM, White JA et al (2016) Amyloid precursor protein–mediated endocytic pathway disruption induces axonal dysfunction and neurodegeneration. J Clin Invest 126:1815–1833. doi:10.1172/JCI82409
Kim S, Sato Y, Mohan PS et al (2016) Evidence that the rab5 effector APPL1 mediates APP-βCTF-induced dysfunction of endosomes in Down syndrome and Alzheimer’s disease. Mol Psychiatry 21:707–716. doi:10.1038/mp.2015.97
Willem M, Tahirovic S, Busche MA et al (2015) η-Secretase processing of APP inhibits neuronal activity in the hippocampus. Nature 526:443–447. doi:10.1038/nature14864
Baranger K, Marchalant Y, Bonnet AE et al (2016) MT5-MMP is a new pro-amyloidogenic proteinase that promotes amyloid pathology and cognitive decline in a transgenic mouse model of Alzheimer’s disease. Cell Mol Life Sci 73:217–236. doi:10.1007/s00018-015-1992-1
Das U, Scott DA, Ganguly A et al (2013) Activity-induced convergence of APP and BACE-1 in acidic microdomains via an endocytosis-dependent pathway. Neuron 79:447–460. doi:10.1016/j.neuron.2013.05.035
Takahashi RH, Milner TA, Li F et al (2002) Intraneuronal Alzheimer abeta42 accumulates in multivesicular bodies and is associated with synaptic pathology. Am J Pathol 161:1869–1879
Sannerud R, Esselens C, Ejsmont P et al (2016) Restricted location of PSEN2/γ-secretase determines substrate specificity and generates an intracellular Aβ pool. Cell 166:193–208. doi:10.1016/j.cell.2016.05.020
Rajendran L, Honsho M, Zahn TR et al (2006) Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc Natl Acad Sci USA 103:11172–11177. doi:10.1073/pnas.0603838103
Vingtdeux V, Hamdane M, Loyens A et al (2007) Alkalizing drugs induce accumulation of amyloid precursor protein by-products in luminal vesicles of multivesicular bodies. J Biol Chem 282:18197–18205. doi:10.1074/jbc.M609475200
Sharples RA, Vella LJ, Nisbet RM et al (2008) Inhibition of gamma-secretase causes increased secretion of amyloid precursor protein C-terminal fragments in association with exosomes. FASEB J 22:1469–1478. doi:10.1096/fj.07-9357com
Citron M, Oltersdorf T, Haass C et al (1992) Mutation of the beta-amyloid precursor protein in familial Alzheimer’s disease increases beta-protein production. Nature 360:672–674. doi:10.1038/360672a0
Belly A, Bodon G, Blot B et al (2010) CHMP2B mutants linked to frontotemporal dementia impair maturation of dendritic spines. J Cell Sci 123:2943–2954. doi:10.1242/jcs.068817
Chatellard-Causse C, Blot B, Cristina N et al (2002) Alix (ALG-2-interacting protein X), a protein involved in apoptosis, binds to endophilins and induces cytoplasmic vacuolization. J Biol Chem 277:29108–29115. doi:10.1074/jbc.M204019200
Laulagnier K, Javalet C, Hemming FJ, Sadoul R (2017) Purification and analysis of exosomes released by mature cortical neurons following synaptic activation. Methods Mol Biol 1545:129–138. doi:10.1007/978-1-4939-6728-5_9
Schagger H (2006) Tricine-SDS-PAGE. Nat Protoc 1:16–22. doi:10.1038/nprot.2006.4
Bolte S, Cordelières FP (2006) A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224:213–232. doi:10.1111/j.1365-2818.2006.01706.x
Hoey SE, Williams RJ, Perkinton MS (2009) Synaptic NMDA receptor activation stimulates alpha-secretase amyloid precursor protein processing and inhibits amyloid-beta production. J Neurosci 29:4442–4460. doi:10.1523/JNEUROSCI.6017-08.2009
Cai XD, Golde TE, Younkin SG (1993) Release of excess amyloid beta protein from a mutant amyloid beta protein precursor. Science 259:514–516
Forman MS, Cook DG, Leight S et al (1997) Differential effects of the swedish mutant amyloid precursor protein on β-amyloid accumulation and secretion in neurons and nonneuronal cells. J Biol Chem 272:32247–32253. doi:10.1074/jbc.272.51.32247
Huotari J, Helenius A (2011) Endosome maturation. EMBO J 30:3481–3500. doi:10.1038/emboj.2011.286
Stenmark H, Parton RG, Steele-Mortimer O et al (1994) Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J 13:1287–1296
Cao X, Südhof TC (2004) Dissection of amyloid-beta precursor protein-dependent transcriptional transactivation. J Biol Chem 279:24601–24611. doi:10.1074/jbc.M402248200
Sullivan CP, Jay AG, Stack EC et al (2011) Retromer disruption promotes amyloidogenic APP processing. Neurobiol Dis 43:338–345. doi:10.1016/j.nbd.2011.04.002
Langui D, Girardot N, El Hachimi KH et al (2004) Subcellular topography of neuronal Abeta peptide in APPxPS1 transgenic mice. Am J Pathol 165:1465–1477
Hoey SE, Buonocore F, Cox CJ et al (2013) AMPA receptor activation promotes non-amyloidogenic amyloid precursor protein processing and suppresses neuronal amyloid-β production. PLoS One 8:e78155. doi:10.1371/journal.pone.0078155
Lesné S, Ali C, Gabriel C et al (2005) NMDA receptor activation inhibits alpha-secretase and promotes neuronal amyloid-beta production. J Neurosci 25:9367–9377. doi:10.1523/JNEUROSCI.0849-05.2005
Stoeck A, Keller S, Riedle S et al (2006) A role for exosomes in the constitutive and stimulus-induced ectodomain cleavage of L1 and CD44. Biochem J 393:609–618. doi:10.1042/BJ20051013
Jämsä A, Belda O, Edlund M, Lindström E (2011) BACE-1 inhibition prevents the γ-secretase inhibitor evoked Aβ rise in human neuroblastoma SH-SY5Y cells. J Biomed Sci 18:76. doi:10.1186/1423-0127-18-76
Lu X, Deng Y, Yu D et al (2014) Histone acetyltransferase p300 mediates histone acetylation of PS1 and BACE1 in a cellular model of Alzheimer’s disease. PLoS One 9:e103067. doi:10.1371/journal.pone.0103067
Mucke L, Masliah E, Yu GQ et al (2000) High-level neuronal expression of abeta 1-42 in wild-type human amyloid protein precursor transgenic mice: synaptotoxicity without plaque formation. J Neurosci 20:4050–4058
Falker C, Hartmann A, Guett I et al (2016) Exosomal cellular prion protein drives fibrillization of amyloid beta and counteracts amyloid beta-mediated neurotoxicity. J Neurochem 137:88–100. doi:10.1111/jnc.13514
An K, Klyubin I, Kim Y et al (2013) Exosomes neutralize synaptic-plasticity-disrupting activity of Aβ assemblies in vivo. Mol Brain 6:47. doi:10.1186/1756-6606-6-47
Cataldo AM, Peterhoff CM, Troncoso JC et al (2000) Endocytic pathway abnormalities precede amyloid beta deposition in sporadic Alzheimer’s disease and Down syndrome: differential effects of APOE genotype and presenilin mutations. Am J Pathol 157:277–286
Almeida CG, Takahashi RH, Gouras GK (2006) β-amyloid accumulation impairs multivesicular body sorting by inhibiting the ubiquitin-proteasome system. J Neurosci 26:4277–4288. doi:10.1523/JNEUROSCI.5078-05.2006
Hollingworth P, Harold D, Sims R et al (2011) Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet 43:429–435. doi:10.1038/ng.803
Naj AC, Jun G, Beecham GW et al (2011) Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet 43:436–441. doi:10.1038/ng.801
Ubelmann F, Burrinha T, Salavessa L et al (2017) Bin1 and CD2AP polarise the endocytic generation of beta-amyloid. EMBO Rep 18:102–122. doi:10.15252/embr.201642738
van Niel G, Charrin S, Simoes S et al (2011) The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev Cell 21:708–721. doi:10.1016/j.devcel.2011.08.019
Deyts C, Thinakaran G, Parent AT (2016) APP Receptor. To Be or Not To Be. Trends Pharmacol Sci. doi:10.1016/j.tips.2016.01.005
Sisodia SS, Koo EH, Hoffman PN et al (1993) Identification and transport of full-length amyloid precursor proteins in rat peripheral nervous system. J Neurosci 13:3136–3142
Wang Y, Ha Y (2004) The X-ray structure of an antiparallel dimer of the human amyloid precursor protein E2 domain. Mol Cell 15:343–353. doi:10.1016/j.molcel.2004.06.037
Le Blanc I, Luyet P-P, Pons V et al (2005) Endosome-to-cytosol transport of viral nucleocapsids. Nat Cell Biol 7:653–664. doi:10.1038/ncb1269
Acknowledgements
We thank M. Jacquier-Sarlin, S. Fraboulet, M. Laporte, and K. Sadoul for critically reading the manuscript. This work was funded by Grants from the Agence Nationale de la Recherche (ANR MALZ program) and from Association France Alzheimer. K. Laulagnier was supported by the Fondation Plan Alzheimer. C. Javalet was supported by the Ministère de l’Enseignement Supérieur et de la Recherche.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
18_2017_2664_MOESM1_ESM.tif
Supplementary Fig. 1 Nanoparticle Tracking Analysis shows that APPwt or APPswe expression affect neither the number nor the size of secreted exosomes. Exosomes purified from supernatants of control N2a, APPwt N2a, and APPswe N2a were resuspended and analyzed using Nanosight. a Number of particles secreted during 24 h by 9 x 106 cells per dish. Averages of five measurements +/- SEM are shown. b Distribution of sizes for each type of exosomes. Average of five measurements is shown with SEM drawn in gray. The mode size is indicated for each population. No significant differences were observed between each type of exosomes. (TIFF 765 kb)
18_2017_2664_MOESM2_ESM.tif
Supplementary Fig. 2 N2a cells were transfected with UAS-Firefly luciferase reporter construct alone (Mock) or together with NLS-Gal4 construct (NLS-Gal4). Renilla luciferase was co-expressed to normalize Firefly activity and luciferase activities measured 48 h after transfection. When indicated, 10 μM of the γ-secretase inhibitor (DAPT) was added 20 h before measurement. In contrast with experiments using APP-Gal4 in Fig. 7, DAPT does not inhibit Firefly luciferase activity. The result represents the mean of two independent cultures. (TIFF 159 kb)
Rights and permissions
About this article
Cite this article
Laulagnier, K., Javalet, C., Hemming, F.J. et al. Amyloid precursor protein products concentrate in a subset of exosomes specifically endocytosed by neurons. Cell. Mol. Life Sci. 75, 757–773 (2018). https://doi.org/10.1007/s00018-017-2664-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-017-2664-0